Abstract
Bacterial blight, caused by the phytopathogen Pseudomonas cannabina pv. alisalensis, is an emerging disease afflicting important members of the Brassicaceae family. The disease is often misdiagnosed as pepper spot, a much less severe disease caused by the related pathogen Pseudomonas syringae pv. maculicola. We have developed a phage-based diagnostic that can both identify and detect the causative agent of bacterial blight and differentiate the two pathogens. A recombinant “light”-tagged reporter phage was generated by integrating bacterial luxAB genes encoding luciferase into the genome of P. cannabina pv. alisalensis phage PBSPCA1. The PBSPCA1::luxAB reporter phage is viable and stable and retains properties similar to those of the wild-type phage. PBSPCA1::luxAB rapidly and sensitively detects P. cannabina pv. alisalensis by conferring a bioluminescent signal response to cultured cells. Detection is dependent on cell viability. Other bacterial pathogens of Brassica species such as P. syringae pv. maculicola, Pseudomonas marginalis, Pectobacterium carotovorum, Xanthomonas campestris pv. campestris, and X. campestris pv. raphani either do not produce a response or produce significantly attenuated signals with the reporter phage. Importantly, the reporter phage detects P. cannabina pv. alisalensis on diseased plant specimens, indicating its potential for disease diagnosis.
INTRODUCTION
Edible plants of the Brassicaceae family are termed cruciferous vegetables. These include broccoli, cauliflower, cabbage and collards (Brassica oleracea L. var.), rapeseed (Brassica napa), arugula (Eruca sativa), rutabaga (Brassica napus), mustard greens (Brassica juncea), and turnip greens (Brassica rapa). Cruciferous vegetables are thus among the most dominant food crops worldwide and are a valuable U.S. commodity. B. oleracea vegetables, for example, have an estimated annual U.S. value of over $1.3 billion. Crucifers are also important nutritionally, since they are an excellent source of vitamin C, nutrients, and soluble fiber as well as compounds with potent health and anticancer properties (e.g., diindolylmethane, glucoraphanin, and indole-3-carbinol) (26, 35). In addition to their use for human consumption as vegetables, crucifers are grown as cover, oil seed, and energy crops, and the crucifer Arabidopsis thaliana is used widely as a model system for plant-pathogen interactions.
Pseudomonas cannabina pv. alisalensis is the causative agent of bacterial blight, a devastating disease of cruciferous vegetables. P. cannabina pv. alisalensis has been documented to cause severe bacterial blight on members of the Brassicaceae in conventional and organic production fields in California, Nevada, New Jersey, and South Carolina (4, 5, 7, 11, 17, 18, 24, 38). A wide geographic range has been recognized for this pathogen; in addition to those that have taken place in the United States, disease outbreaks have been reported in both Europe and Australia (8, 31). Initial disease symptoms are water-soaked flecks on the lower foliage. Over time, these flecks expand and become surrounded by bright yellow borders, which eventually coalesce to form large necrotic areas, rendering the crop unmarketable. The primary source of inoculum for P. cannabina pv. alisalensis is soil adjacent to infected host plants (12). Moreover, pathogens related to P. cannabina pv. alisalensis have been shown to be seed borne. These include Pseudomonas syringae pv. maculicola, causing pepper spot on Brassica spp. (21, 28), and P. syringae pv. tomato, causing bacterial speck on tomato (25).
P. cannabina pv. alisalensis PBSPCA1 bacteriophage, formerly known as PBS1, was isolated from a commercial broccoli raab field that was symptomatic for bacterial blight (11). Previous studies showed that PBSPCA1 infects all P. cannabina pv. alisalensis isolates in various geographical locations and from various symptomatic crucifers (5, 6, 11, 17, 18). PBSPCA1 does not infect P. syringae pv. maculicola, which is important because this pathovar is difficult to distinguish from P. cannabina pv. alisalensis, which causes pepper spot on crucifers that is often confused with bacterial blight. Consequently, phage lysis assays using PBSPCA1 have been utilized to distinguish P. cannabina pv. alisalensis from other pathovars (11). These assays consist of spotting serially diluted phage onto soft agar harboring the bacteria. After overnight incubation, the presence of clearing zones or individual plaques within the bacterial lawn indicates that the bacteria are susceptible to the phage. Thus, the assays require bacterial isolation from the infected plant and subsequent cultivation. Here we describe the development of a bioluminescent reporter phage for the detection of P. cannabina pv. alisalensis. The PBSPCA1::luxAB reporter phage rapidly and sensitively confers a bioluminescent signal response to P. cannabina pv. alisalensis. Importantly, the reporter phage is able to directly detect P. cannabina pv. alisalensis on infected Brassica species, indicating its potential as a bacterial blight diagnostic tool.
MATERIALS AND METHODS
Bacterial strains and phage.
P. syringae pv. maculicola (ATCC 51320, 51321, and 51322), Pectobacterium carotovorum subsp. carotovorum (ATCC 138 and 495), Pseudomonas marginalis (ATCC 10844 and 51281), Xanthomonas campestris pv. campestris (ATCC 33913), and Xanthomonas campestris pv. raphani (ATCC 49079) were obtained from the American Type Culture Collection. P. cannabina pv. alisalensis strains BS91 and T3C were isolated from B. rapa and B. oleracea, respectively (11, 16). Bacteria were grown on Pseudomonas agar F (PAF) (Difco, Becton Dickenson and Co., Sparks, MD) and nutrient agar (NA) plates and in NBY media (nutrient broth [NB] supplemented with yeast extract [2 g/liter], K2HPO4 [2 g/liter], KH2PO4 [0.5 g/liter], 0.5% glucose, and 1 mM MgSO4). Cultures were grown at 28°C for 18 to 24 h with shaking (225 rpm) to generate saturated cultures and then diluted 1:25 into fresh media until the desired optical density at 600 nm (OD600) was reached.
Phage PBSPCA1 was propagated using standard techniques (9). Clonal stocks of PBSPCA1, which were prepared from single plaques, typically harbor 109 to 1010 PFU/ml.
Construction of a luxAB cassette for targeted homologous recombination into the phage genome.
Vibrio harveyi luxA and luxB were utilized as reporter genes. pBluescript II-SK− containing luxAB was used as the parental vector for subsequent steps (33). A strong constitutive promoter was cloned upstream of the reporter (Table 1). PBSPCA1 DNA sequences were obtained using 454 sequencing technology performed by the EnGenCore sequencing service at the University of South Carolina from two separate genomic preparations. It was not possible to join the resulting data into a single genome sequence. However, the phage appeared to carry a putative phoH gene (encoding a phosphate starvation-inducible protein) that we predicted would be nonessential. This gene was therefore targeted for replacement with the luxAB expression cassette. A sequence consisting of the putative phoH gene and some flanking DNA sequences has been deposited in GenBank (see below). The luxAB expression cassette was flanked by PBSPCA1 sequences for targeted integration by homologous recombination. A 270-bp fragment and a 258-bp fragment of PBSPCA1 DNA were PCR amplified using phage DNA as the template and cloned 5′ and 3′ of the luxAB reporter genes, respectively (Table 1 [5′PhoH and 3′PhoH, respectively]). PCR primers were designed to contain compatible restriction endonuclease sites for cloning into the luxAB reporter plasmid. Clonings were performed using Escherichia coli T7 express Iq (New England BioLabs, Ipswich, MA) according to the method of Sambrook et al. (30). The sequences of the cloned fragments were verified by Sanger sequencing using an Applied BioSystems (ABI, Foster City, CA) BigDye Terminator version 1.1 cycle sequencing kit and an ABI Prism 377 sequencer.
Table 1.
PCR primers and oligonucleotide used in this study
| Primer name/type or oligonucleotide | Forward or reverse PCR primer sequence (5′–3′)a | Feature | PCR product size (bp) |
|---|---|---|---|
| 5′PhoH | AATTGAGCTCGCACTCTCTCCTGATGAAATC | 5′ flanking phage sequence for homologous recombination | 270 |
| AATTTCTAGACGACTGGGAGTTACGGTTC | |||
| 3′PhoH | TTAACTCGAGAGGTTGCTTAATGACAGGTG | 3′ flanking phage sequence for homologous recombination | 258 |
| TTAAGCATGCGCACTGGTTTCGGTATGTG | |||
| luxB | ATCGACCAACGGATTCTCAG | luxB screening primers | 184 |
| ACTTCTTTGCTCGTCGCATT | |||
| 5′-INT | CGCAGAAGACAATGTCCTGA | Confirmation of 5′ junction of integration | 667 |
| GCTTTGCCCAGATTAACCAA | |||
| 3′-INT | GTTGCGATTGAGTGTTGTGG | Confirmation of 3′ junction of integration | 516 |
| GTCTTGTGAGACGCGAATGA | |||
| Del | AAGAAGCTGGCGAGAAACTG | phoHb sequence expected to be deleted in the recombinant phage | 416 |
| TTCTTGAACCAGACCGGAAC | |||
| lamH | AATTGAGCTCGTTATGGGGATGCCTATTCG | Phage DNA PCR positive control | 210 |
| AATTTCTAGAGTAGTATTGTTCCGGTAGCTC | |||
| Promoter | CTAGATTGACAATTGTCTAATAAATTTTATAATTTTAAT | Promoter sequence driving luxAB with −10 and −35 hexamers (underlined) | nac |
Bold sequences: GAGCTC, SacI; TCTAGA, XbaI; CTCGAG, XhoI; GCATGC, SphI. For each primer, the sequence in the upper row is the forward primer sequence and the sequence in the lower row is the reverse primer sequence.
phoH, phosphate starvation-inducible gene.
na, not applicable (for the oligonucleotide promoter, the PCR product size is not applicable).
The luxAB expression cassette with the flanking PBSPCA1 sequence was subcloned into the broad-host-range pBBR122 plasmid (MoBiTec GmbH, Goettingen, Germany) and transformed into P. cannabina pv. alisalensis BS91 by the use of the calcium chloride method (15). Transformants were selected on nutrient agar (NA) supplemented with kanamycin (25 μg/ml). Transformants were positive for bioluminescence, indicating that the luxAB reporter was functional in P. cannabina pv. alisalensis (data not shown). Cultures were grown in selective NBY media at 28°C to an OD600 of ∼0.3, infected with PBSPCA1 (multiplicity of infection of ∼0.001), and incubated overnight to allow multiple rounds of phage amplification. The lysate was clarified by centrifugation (4,000 × g for 15 min) and passed through a 0.22-μm-pore-size acrodisc syringe filter (Pall Life Sciences), and the titer was determined using strain BS91 and the agar overlay technique (9) to give nearly confluent lysis results (approximately 2,000 plaques per plate) on ∼50 plates. Phages on each plate were eluted in SM buffer (50 mM Tris-HCl [pH 7.5], 0.1 M NaCl, 8 mM MgSO4 · 7H2O, 0.01% gelatin), and an aliquot was screened by PCR for the presence of luxB. The titers were again determined for lysates from plates that gave positive results and rescreened using successively higher dilutions. This process was repeated for approximately 12 rounds until individual plaques could be screened. Clonal PBSPCA1::luxAB phage was prepared from single plaques and amplified on strain BS91 in the exponential-growth phase. After multiple rounds of amplification, phage lysates were prepared by centrifuging the cultures at 3,000 × g to remove bacterial debris and passing the phage supernatant through a 0.22-μm-pore-size filter. Clonal stocks of PBSPCA1::luxAB typically contained 109 to 1010 PFU/ml and were stored at 4°C.
Recombinant phage identification.
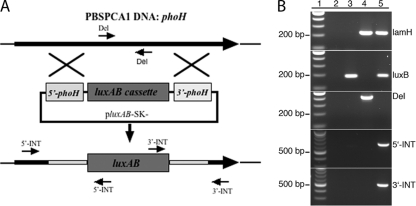
PBSPCA1::luxAB recombinant phage was analyzed by PCR to confirm that luxAB integration had occurred at the correct site in the genome. Primers were designed to detect the presence of luxB (reporter) or to span the 5′ and 3′ integration junction sites (Table 1 and Fig. 1B [5′-INT and 3′-INT]). Each integration primer set was designed to ensure that primer binding occurred both inside and outside the original integration cassette. As a negative control, primers were designed to amplify the segment of DNA predicted to be deleted by the recombination (Table 1 [Del]). PCR analysis was performed as recommended by the manufacturer of Taq DNA polymerase (New England BioLabs).
Fig 1.
Homologous recombination and PCR identification of recombinant PBSPCA1::luxAB. (A) Homologous recombination between plasmid and PBSPCA1 DNA based on a double-crossover event. luxAB was targeted for integration into the phage genome at the phoH gene. As a result of the integration, 798 bp of phoH is predicted to be replaced with 2,159 bp of luxAB. Shaded phoH boxes indicate the 5′ and 3′ homologous DNAs. The positions of the 5′-INT, 3′-INT, and Del forward and reverse primers that were used to verify PBSPCA1::luxAB identity are shown. Positions of the lamH primers are not shown due to their distance from phoH. The diagram is not to scale. (B) PCR primers against PBSPCA1 phage DNA (lamH), the luxB gene (luxB), a PBSPCA1 genomic segment predicted to be deleted in the recombinant phage (Del), and the 5′ and 3′ luxAB integration sites (5′-INT and 3′-INT) were designed. PCR analysis was performed in the absence of template (lane 2) and with the recombination plasmid (lane 3), wild-type PBSPCA1 phage (lane 4), and recombinant PBSPCA1::luxAB (lane 5). The predicted PCR product sizes for lamH, luxB, Del, 5′-INT, and 3′-INT are 210, 184, 416, 667, and 516 bp, respectively. Lane 1, 100-bp ladder.
P. cannabina pv. alisalensis inoculation of Brassica rapa.
The turnip green (B. rapa) cultivar Topper was grown in Metromix 360 soil (The Scott's Co., Maryville, OH) in plastic pots (10 by 10 by 10 cm) in a greenhouse at the USDA Vegetable Laboratory in Charleston, SC. Plants were thinned to one plant per pot at 10 days after seeding and then allowed to grow in the greenhouse. At the third true leaf stage (∼3 weeks after seeding), the plants were inoculated with P. cannabina pv. alisalensis isolate T3C (collected in 2001 from a commercial field in Lexington, SC). This isolate is highly virulent and consistently induces infection over a range of temperatures in greenhouse and growth chamber studies, as well as in the field (37). T3C was grown on PAF medium for 16 h at 27°C. Cells were then harvested and resuspended in sterile distilled water. The cell suspension was adjusted with sterile distilled water to an OD600 of 0.8 (∼1 × 107 CFU/ml) using a model 6136 BioPhotometer (Eppendorf, Hauppauge, NY). Latron B-1956 surfactant (Dow Agrosciences LLC, Indianapolis, IN) was added to the cell suspension at 3.2 μl/ml to enhance leaf coverage. The suspension was applied to the leaves of each plant by the use of a Paashe model H airbrush sprayer at ∼170 kPa until the leaf surface was uniformly covered. Inoculated plants were placed in a humidity chamber at 100% relative humidity for 16 h and then transferred to the greenhouse bench for 10 to 14 days prior to being rated for disease. Greenhouse temperatures ranged from 27 to 32°C.
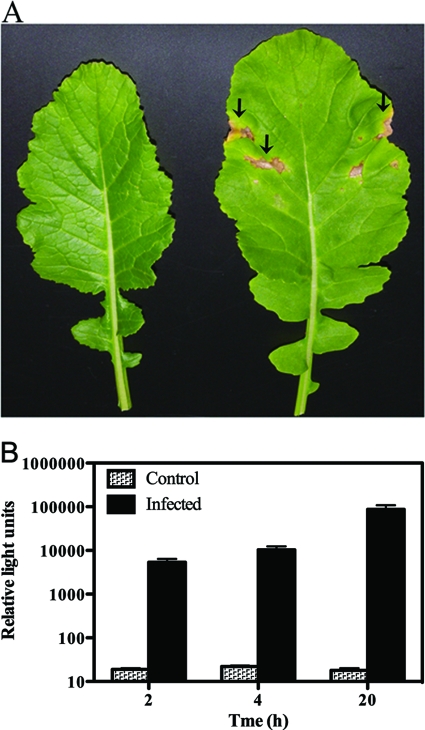
After 10 to 14 days, the presence of bacterial blight was indicated by expanding areas of chlorotic (yellow) and necrotic (brown) areas on the leaves (see Fig. 5A). Individual leaves were harvested from inoculated and noninoculated plants. Three 1-cm2 discs from the chlorotic area of the inoculated and infected leaf tissue were harvested per leaf and pooled. Tissue samples from noninoculated B. rapa were harvested and processed similarly for controls. The tissue was mixed with 1 ml of NBY, subjected to a vigorous vortex procedure, and incubated at 28°C with shaking to allow bacterial elution and outgrowth. After 2, 4, or 20 h, 900 μl of the leaf eluate was placed in a fresh tube, and PBSPCA1::luxAB was added.
Fig 5.
Reporter phage detection of P. cannabina pv. alisalensis from greenhouse-inoculated B. rapa. (A) Control (left leaf) and P. cannabina pv. alisalensis T3C-inoculated (right leaf) B. rapa. Arrows denote areas of brown necrosis indicative of bacterial blight. (B) Phage-mediated detection of T3C from infected leaves. Approximately 1-cm2 discs of control and infected leaf parts were mixed with 1 ml of NBY, subjected to a vigorous vortex procedure, and incubated at 28°C with shaking to allow bacterial elution and outgrowth. At various times, the leaf eluate was placed in a fresh tube and PBSPCA1::luxAB was added. Bioluminescence was measured 120 min after the addition of 2% n-decanal. Numbers represent means ± SE of the results determined with three plants.
Bioluminescence assays.
Unless otherwise stated, PBSPCA1::luxAB phage (∼108 PFU/ml final concentration) and P. cannabina pv. alisalensis cells were mixed and incubated for the times indicated. “Flash” bioluminescence was measured using a Biotek Synergy II multiplate detection reader. Cultures were mixed with n-decanal (0.5%) and read for 10 s. Controls included cells alone and phage alone. Bioluminescence is depicted in relative light units (RLU), and the results are presented as the averages of data from three experiments ± standard deviations (SD) or standard errors (SE). Statistical significance was determined using Student's t test (P < 0.05).
Nucleotide sequence accession number.
A sequence consisting of the putative phoH gene and some flanking DNA sequences has been deposited in GenBank under accession number JQ419755.
RESULTS
Rationale and design of the luxAB reporter cassette for integration into the genome of the P. cannabina pv. alisalensis PBSPCA1 phage.
V. harveyi luxAB was used as the reporter of choice because (i) luxAB has been successfully used as a reporter for phage-mediated detection of Gram-positive and -negative pathogens (3, 19, 22, 32, 33), (ii) bioluminescent signals may be easily visualized by a photon detection device (e.g., luminometer), and (iii) minimal processing of the sample is required (the only requirement is the addition of the substrate n-decanal).
To ensure early and constitutive gene expression, luxAB was placed under the transcriptional control of a strong bacterial promoter (Table 1). The luxAB expression cassette was flanked by phage PBSPCA1 sequence for targeted integration by homologous recombination into the PBSPCA1 genome. In doing so, 798 bp of phage DNA corresponding to a putative phoH-like gene (encoding a phosphate starvation-inducible protein) was predicted to be replaced with 2,159 bp of reporter DNA. This strategy was chosen for two reasons: (i) phoH was not expected to be essential for phage viability or infection, and (ii) replacement of phage DNA with reporter DNA increased the genome size by only ∼1.3 kb and was deemed unlikely to result in the production of defective phage.
Verification of PBSPCA1::luxAB.
To identify the presence of the PBSPCA1::luxAB recombinant phage and to confirm that luxAB integration had occurred at the correct site in the phage genome, cell-free phage supernatants were analyzed by PCR. Primers were designed to detect the presence of lamH (phage DNA; putative tape measure gene), luxB (reporter), and phage DNA that was deleted by the recombination event (Del) or to span the 5′ and 3′ integration junction sites (Fig. 1A). The results of PCR analysis of the recombinant phage were positive for luxB, lamH, 5′-INT, and 3′-INT, as expected, and negative for the PBSPCA1 region predicted to be deleted (Del). PCR analysis performed using the luxB, 5′-INT, and 3′-INT primers generated PCR products of the predicted sizes (Fig. 1B and Table 1), indicating the presence of the reporter and that luxAB had integrated at the correct genome site.
Reporter phage fitness and stability.
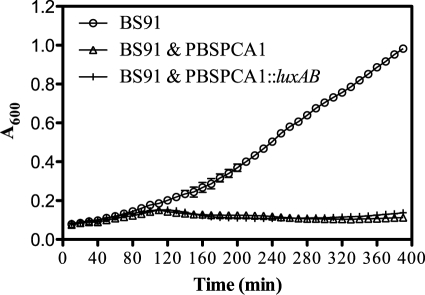
To investigate whether the addition of the heterologous reporter compromised the “fitness” of the recombinant phage, the ability of PBSPCA1::luxAB to lyse BS91 was analyzed. Lysis was assessed by monitoring bacterial growth (culture optical density) in the absence or presence of wild-type PBSPCA1 or PBSPCA1::luxAB phage. Both the wild type and PBSPCA1::luxAB caused a significant drop in OD600, and there were no differences in lysis times (Fig. 2). Recombinant stock titers were also comparable to those of the parental phage (109 to 1010 PFU/ml). These results collectively indicate that (i) a functional PBSPCA1::luxAB reporter phage was generated and (ii) the fitness of the phage was not adversely compromised, at least in rich media, by the introduction of the reporter.
Fig 2.
PBSPCA1-mediated lysis and PBSPCA1::luxAB-mediated lysis of P. cannabina pv. alisalensis showed similar results. P. cannabina pv. alisalensis BS91 was grown in NBY at 28°C until an OD600 0.075 was seen, and the culture was divided into equal portions. Cultures were left untreated (BS91) or infected at a multiplicity of infection of ∼5 with BS91 plus PBSPCA1 or BS91 plus PBSPCA1::luxAB and monitored for OD600 every 10 min (means [n = 3] ± SD).
The stability of the luxAB reporter was tested. The titers of PBSPCA1::luxAB phage that had undergone three rounds of phage amplification were determined using the agar overlay technique. Individual plaques were picked and analyzed by PCR for the presence of the luxB reporter. Of the 23 (96%) plaques from the passaged phage, 22 gave PCR-positive results for luxB (data not shown), indicating that PBSPCA1::luxAB was genetically quite stable.
PBSPCA1::luxAB-mediated detection of P. cannabina pv. alisalensis.
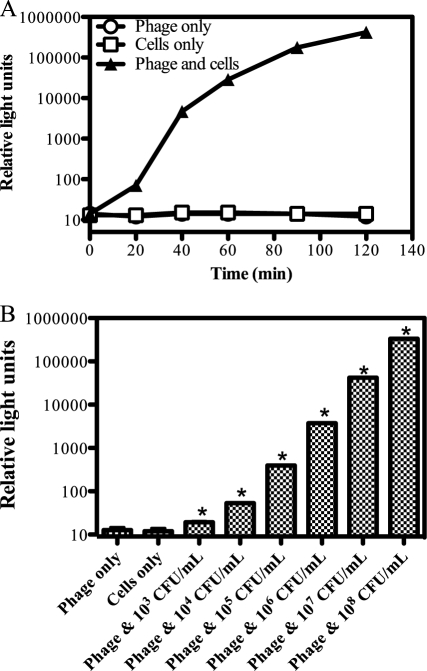
The ability of PBSPCA1::luxAB to transduce a bioluminescent phenotype to P. cannabina pv. alisalensis BS91 was assessed. A steady increase in bioluminescence was seen at 28°C in BS91 phage-infected cells, with a >10,000 increase in signal strength detected within 90 min (Fig. 3A). These data indicate that (i) PBSPCA1::luxAB can rapidly transduce a bioluminescent phenotype to P. cannabina pv. alisalensis and (ii) the LuxAB proteins are functionally stable in P. cannabina pv. alisalensis. Longer incubation (>180 min) of the reporter phage with cells resulted in a gradual decline in signal strength, presumably reflecting phage-mediated cell lysis (data not shown).
Fig 3.
Rapid and sensitive phage-mediated detection of P. cannabina pv. alisalensis BS91. (A) Signal response time. BS91 was grown at 28°C in NBY media, mixed with PBSPCA1::luxAB (time zero; 3.2 × 108 PFU/ml), and incubated at 28°C. Bioluminescence (RLU) was measured over time. Numbers represent means (n = 3) ± SD. (B) Sensitivity limit of detection. BS91 was grown to an OD600 of 0.15 (1.3 × 108 CFU/ml), 10-fold serially diluted, mixed with the reporter phage (3.2 × 108 PFU/ml), and incubated at 28°C for 120 min. Numbers represent means (n = 3) ± SD. *, significant increase (P < 0.05 [Student's t test]) compared to control results.
To investigate assay sensitivity and dose-dependent characteristics, cells serially diluted 10-fold (108 to 103 CFU/ml) were mixed with PBSPCA1::luxAB and analyzed for bioluminescence over time (Fig. 3B). The highest level of CFU per milliliter produced the strongest signal at over 300,000 RLUs within 120 min. As cell numbers decreased, the signal response decreased and the signal response time increased, indicating dose-response characteristics. As few as ∼260 cells (corresponding to 1.3 × 103 CFU/ml) were detectable 120 min after phage addition (P < 0.05 [Student's t test]).
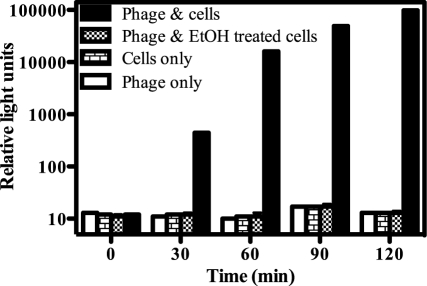
Host cell fitness: detection of viable cells only.
The ability of metabolically active cells to elicit a bioluminescent signal response upon reporter phage infection was compared to the response elicited by compromised cells. Colonies from a freshly grown plate were resuspended in NBY media to an OD600 of 0.08 and either left untreated or treated with 70% (vol/vol) ethanol for 30 min. Ethanol treatment resulted in a 105 to 106 reduction in viable cell levels (to <102 CFU/ml). Following the removal of ethanol, both control and treated cells were incubated with PBSPCA1::luxAB and bioluminescence was assessed over time (Fig. 4). Control cells elicited a rapid and strong bioluminescent response, whereas ethanol-treated cells were unable to elicit a bioluminescence signal. These data indicate that the PBSPCA1::luxAB phage could detect P. cannabina pv. alisalensis directly from colonies without the need for culture outgrowth and that only viable cells were detected.
Fig 4.
Phage-mediated detection of viable cells. P. cannabina pv. alisalensis BS91 colonies were inoculated directly into NBY media until an OD600 of 0.08 was reached. The culture was divided into equal portions and either left untreated or incubated with 70% ethanol for 30 min. Following removal of the ethanol, control and treated cells were incubated with PBSPCA1::luxAB (3.2 × 108 PFU/ml) at 28°C. Bioluminescence (RLU) was measured over time following the addition of 2% n-decanal. Numbers represent means (n = 3) ± SD.
Reporter phage specificity.
Previous studies showed that PBSPCA1 phage infects all strains of P. cannabina pv. alisalensis isolated from widespread geographical locations and from various symptomatic crucifers (5, 6, 11, 17, 18) (data not shown). The activity of PBSPCA1 phage is also specific; none of the 26 strains of P. syringae pv. maculicola tested (data not shown) displayed lysis with the phage by spot tests (9). The ability of PBSPCA1::luxAB to transduce a bioluminescence signal response to P. syringae pv. maculicola, P. marginalis, P. carotovorum, X. campestris pv. campestris, and X. campestris pv. raphani was assessed (Table 2). These species were chosen as important bacterial pathogens of Brassica species. A bioluminescent signal response was not evident above background except for P. syringae pv. maculicola ATCC 51320 and 51322, which produced attenuated signals approximately 100-fold lower than those produced by P. cannabina pv. alisalensis. Thus, even closely related species can be distinguished by the use of this reporter phage.
Table 2.
Reporter phage specificity: inability to detect non-alisalensis Brassica pathogens
| Species | Strain or ATCC no. | Mean no. of relative light units (SD)a |
|---|---|---|
| P. cannabina pv. alisalensis | BS91 | 22,599 (421) |
| P. syringae pv. maculicola | 51320 | 243 (9) |
| P. syringae pv. maculicola | 51321 | 17 (1) |
| P. syringae pv. maculicola | 51322 | 102 (5) |
| P. marginalis | 51281 | 29 (2) |
| P. marginalis | 10844 | 27 (3) |
| P. carotovorum | 495 | 31 (1) |
| P. carotovorum | 138 | 31 (1) |
| X. campestris pv. campestris | 33913 | 25 (1) |
| X. campestris pv. raphani | 49079 | 27 (1) |
Bioluminescence was assessed 60 min following the addition of ∼108 PFU/ml (final concentration) of the reporter phage.
Detection of P. cannabina pv. alisalensis on blight-infected B. rapa.
The ability of PBSPCA1::luxAB to detect P. cannabina pv. alisalensis in planta was tested in a controlled greenhouse environment. At 10 to 14 days postinoculation, the presence of bacterial blight was indicated by large expanding areas of chlorotic (yellow) and necrotic (brown) areas on the leaf surface (Fig. 5A). Leaf tissue from inoculated and noninoculated plants was harvested and assayed with PBSPCA1::luxAB (Fig. 5B). Only background levels of bioluminescence of noninoculated leaves were evident. In comparison, a strong (>100-fold) increase in bioluminescence was obtained from the symptomatic tissues of inoculated plants within 4 h of tissue harvesting. Therefore, PBSPCA1::luxAB can rapidly detect P. cannabina pv. alisalensis in samples from infected Brassica species.
DISCUSSION
The use of phages for the detection of bacterial species is well documented. For example, phage ϕA1122 is used by the CDC for the confirmed identification of Yersinia pestis (10, 13), and γ phage is FDA approved as a standard for the identification of Bacillus anthracis (1). In addition, a phage assay termed FASTPlaqueTB (Biotec Laboratories Ltd., Ipswich, United Kingdom) is marketed for the detection of Mycobacterium tuberculosis, and a phage assay developed by Microphage (Longmont, Colorado) called KeyPath is FDA cleared and marketed for the detection of methicillin-resistant Staphylococcus aureus (MRSA). To reduce the time to detection and to enable detection in complex matrices such as food, environmental, or clinical specimens, reporter phages are being developed as biodetectors of pathogenic bacteria (34). Reporter phages for E. coli, Listeria monocytogenes, Y. pestis, B. anthracis, Salmonella spp., and M. tuberculosis are effective for the detection of bacteria in a range of complex matrices such as milk, ground beef, spinach, soft cheese, and human serum and sputum (2, 3, 22, 23, 27, 29, 32, 33, 36). Although phages had not been previously developed for the detection of plant-pathogenic bacteria, they have received EPA registration for use as biopesticides on tomatoes and peppers to control bacterial spot caused by Xanthomonas campestris pv. vesicatoria and P. syringae pv. tomato (OmniLytics Inc., West Sandy, UT). Phage ϕEa21-4 is currently being investigated as a biopesticide in Canada for use against Erwinia amylovora in order to combat fire blight (20). Therefore, the use of phages (as biocontrol agents) within the agricultural industry is not without precedent.
We have generated a P. cannabina pv. alisalensis detection system by integrating the genes encoding bacterial luciferase into the PBSPCA1 genome to create a bioluminescent reporter phage. The recombinant phage is viable and genetically stable and attains titers in lysates similar to those seen with the wild type. PBSPCA1::luxAB phage is able to selectively detect P. cannabina pv. alisalensis by transducing a bioluminescent phenotype to recipient cells. The signal response is rapid (occurring within 20 min), and the level of sensitivity is >103 CFU/ml. Importantly, PBSPCA1::luxAB is able to detect P. cannabina pv. alisalensis on symptomatic plant tissue within 4 h, a significant improvement in time compared to current diagnostics. Greenhouse studies have shown a direct correlation between P. cannabina pv. alisalensis populations in soil and subsequent disease incidence in seedlings (12). P. cannabina pv. alisalensis is closely related to P. syringae pv. maculicola, a pathogen that causes pepper spot of crucifers. Distinguishing between these pathogens is difficult but important, because although the symptoms of pepper spot and bacterial blight are similar, bacterial blight is much more severe and renders crops unmarketable. A bioluminescent signal response was obtained with 2 of the 3 P. syringae pv. maculicola strains tested, but the signal was approximately 100-fold lower than that seen with P. cannabina pv. alisalensis. Consequently, if the reporter phage were to be used for in planta detection, a low bioluminescence signal could reflect nonspecific detection of P. syringae pv. maculicola. Alternatively, since signal strength is dependent on cell density, a low signal response might reflect low bacterial numbers of P. cannabina pv. alisalensis. Therefore, distinguishing between these 2 pathogens might require additional tests in addition to the reporter phage assay.
PBSPCA1::luxAB uses the host's transcriptional and translational machinery to elicit a bioluminescent response. Thus, signal generation is strictly dependent on the host, and only viable cells can elicit a bioluminescent response. This is a valuable attribute when testing contaminated plant debris, soil, and holdover inocula, as it detects the presence of viable cells and thus a potentially infectious pathogen. The reporter phage may also have utility for the detection of contaminated seeds. Plant pathogens that are spread through contaminated seeds cause significant losses in agricultural production each year. There is anecdotal evidence that P. cannabina pv. alisalensis is seed borne, and closely related pathogens are known to be seed borne (14). Antigen and PCR detection technologies generally do not discriminate between viable and dead cells, which limits their utility. Successful pretreatment of a seed lot with bacteriocides still generates a PCR signal and causes rejection of the entire lot even in cases in which the bacteria have been rendered harmless.
This work represents the first successful use of a bioluminescent bacteriophage reporter system to selectively and sensitively detect viable plant-pathogenic bacteria both in pure culture and in planta. The PBSPCA1::luxAB phage can detect the phytopathogenic bacterium P. cannabina pv. alisalensis in infected brassica plants, in addition to differentiating this pathogen from the closely related phytopathogenic P. syringae pv. maculicola. The ability to selectively identify viable (living) bacteria affords a significant advantage over other tests that generate signals whether the tested cells are viable or not. This is extremely important in diagnostics where infectivity of the pathogen is critical, such as in seed testing and field monitoring.
ACKNOWLEDGMENTS
This research was supported by the National Science Foundation Small Business Innovative Research (grant 1012059) and in part by the U.S. Department of Education and the U.S. Department of Agriculture.
Any opinions, findings, conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the funding agencies.
We thank Darren Wray for technical assistance and the Medical University of South Carolina Biotechnology Resource laboratory for sequencing data.
Footnotes
Published ahead of print 16 March 2012
REFERENCES
- 1. Abshire TG, Brown JE, Ezzell JW. 2005. Production and validation of the use of gamma phage for identification of Bacillus anthracis. J. Clin. Microbiol. 43:4780–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Banaiee N, et al. 2001. Luciferase reporter mycobacteriophages for detection, identification, and antibiotic susceptibility testing of Mycobacterium tuberculosis in Mexico. J. Clin. Microbiol. 39:3883–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brigati JR, et al. 2007. Bacteriophage-based bioluminescent bioreporter for the detection of Escherichia coli O157:H7. J. Food Prot. 70:1386–1392 [DOI] [PubMed] [Google Scholar]
- 4. Bull CT, Du Toit L. 2009. First report of bacterial blight on conventionally and organically grown arugula in Nevada caused by Pseudomonas syringae pv. alisalensis. Plant Dis. 93:109. [DOI] [PubMed] [Google Scholar]
- 5. Bull CT, Goldman PH, Koike ST. 2004. Bacterial blight on arugula, a new disease caused by Pseudomonas syringae pv. alisalensis in California. Plant Dis. 88:1384. [DOI] [PubMed] [Google Scholar]
- 6. Bull CT, Goldman PH, Smith RF, Koike ST. 2004. Pseudomonas syringae pv. alisalensis and Pseudomonas syringae pv. maculicola cause disease on crucifers used in cover crop mixtures. Phytopathology 94:S150 [Google Scholar]
- 7. Bull CT, Mauzey SJ, Koike ST. 2010. First report of bacterial blight on Brussels sprouts (Brassica oleracea L. var. gemmifera) caused by Pseudomonas cannabina pv. alisalensis in California. Plant Dis. 94:1375. [DOI] [PubMed] [Google Scholar]
- 8. Bull CT, Rubio I. 2011. First report of bacterial blight of crucifers caused by Pseudomonas cannabina pv. alisalensis in Australia. Plant Dis. 95:1027. [DOI] [PubMed] [Google Scholar]
- 9. Carlson K. 2005. Working with bacteriophages: common techniques and methodological approaches, p 437–494 In Kutter E, Sulakvelidze A. (ed), Bacteriophages: biology and applications. CRC Press, Boca Raton, FL [Google Scholar]
- 10. Chu MC. 2000. Laboratory manual of plague diagnostic tests. Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 11. Cintas NA, Koike ST, Bull CT. 2002. A new pathovar, Pseudomonas syringae pv. alisalensis pv. nov., proposed for the casual agent of bacterial blight of broccoli and broccoli raab. Plant Dis. 86:992–998 [DOI] [PubMed] [Google Scholar]
- 12. Cintas NA, Koike ST, Bunch RA, Bull CT. 2006. Holdover inoculum of Pseudomonas syringae pv. alisalensis from broccoli raab causes disease in subsequent plantings. Plant Dis. 90:1077–1084 [DOI] [PubMed] [Google Scholar]
- 13. Dennis DT, Gage KL, Gratz N, Poland JD, Tikhomirov E. 1999. Plague manual: epidemiology, distribution, surveillance, and control. World Health Organization, Geneva, Switzerland [Google Scholar]
- 14. Gitaitis R, Walcott R. 2007. The epidemiology and management of seedborne bacterial diseases. Annu. Rev. Phytopathol. 45:371–397 [DOI] [PubMed] [Google Scholar]
- 15. Gross DC, Vidaver AK. 1990. Transformation of Pseudomonas syringae pv. syringae, p 501 In Klement Z, Rudolph K, Sands DC. (ed), Methods in phytobacteriology. Akademiai Kiado, Budapest, Hungary [Google Scholar]
- 16. Keinath AP, Wechter WP, Smith JP. 2006. First report of bacterial leaf spot on leafy Brassica caused by Pseudomona syringae pv. maculicola in South Carolina. Plant Dis. 90:683. [DOI] [PubMed] [Google Scholar]
- 17. Koike ST, Kammeijer K, Bull CT, O'Brien D. 2006. First report of bacterial blight of romanesco cauliflower (Brassica oleracea var. botrytis) caused by Pseudomonas syringae pv. alisalensis in California. Plant Dis. 90:1551. [DOI] [PubMed] [Google Scholar]
- 18. Koike ST, Kammeijer K, Bull CT, O'Brien D. 2007. First report of bacterial blight of rutabaga (Brassica napus var. napobrassica) caused by Pseudomonas syringae pv. alisalensis in California. Plant Dis. 91:112. [DOI] [PubMed] [Google Scholar]
- 19. Kuhn J, et al. 2002. Detection of bacteria using foreign DNA: the development of a bacteriophage reagent for Salmonella. Int. J. Food Microbiol. 74:229–238 [DOI] [PubMed] [Google Scholar]
- 20. Lehman SM, Kropinski AM, Castle AJ, Svircev AM. 2009. Complete genome of the broad-host-range Erwinia amylovora phage ΦEa21-4 and its relationship to Salmonella phage Felix O1. Appl. Environ. Microbiol. 75:2139–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lelliott RA. 1988. Bacteria I—Pseudomonas and Xanthomonas, p 143–145 In Smith IM, Dunez J, Lelliott RA, Phillips DH, Archer SA. (ed), European handbook of plant diseases. Blackwell Scientific, Oxford, United Kingdom [Google Scholar]
- 22. Loessner MJ, Rees CE, Stewart GS, Scherer S. 1996. Construction of luciferase reporter bacteriophage A511::luxAB for rapid and sensitive detection of viable Listeria cells. Appl. Environ. Microbiol. 62:1133–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loessner MJ, Rudolf M, Scherer S. 1997. Evaluation of luciferase reporter bacteriophage A511::luxAB for detection of Listeria monocytogenes in contaminated foods. Appl. Environ. Microbiol. 63:2961–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mauzey SJ, Koike ST, Bull CT. 2010. First report of bacterial blight on cabbage (Brassica oleracea var. capitata L.) caused by Pseudomonas cannabina pv. alisalensis in California. Plant Dis. 95:71. [DOI] [PubMed] [Google Scholar]
- 25. McCarter SM, Jones JB, Gitaitis RD, Smitley DR. 1983. Survival of Pseudomonas syringae pv. tomato in association with tomato seed, soil, host tissue, and epiphytic weed hosts in Georgia. Phytopathology 73:1393–1398 [Google Scholar]
- 26. Murillo G, Mehta RG. 2001. Cruciferous vegetables and cancer prevention. Nutr. Cancer 41:17–28 [DOI] [PubMed] [Google Scholar]
- 27. Oda M, Morita M, Unno H, Tanji Y. 2004. Rapid detection of Escherichia coli O157:H7 by using green fluorescent protein-labeled PP01 bacteriophage. Appl. Environ. Microbiol. 70:527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rimmer SR. 2007. Diseases caused by bacteria—bacterial leaf spot, p 58–59 In Rimmer SR, Shattuck VI, Buchwaldt L. (ed), Compendium of Brassica diseases. APS Press, St. Paul, MN [Google Scholar]
- 29. Ripp S, Jegier P, Johnson CM, Brigati JR, Sayler GS. 2008. Bacteriophage-amplified bioluminescent sensing of Escherichia coli O157:H7. Anal. Bioanal. Chem. 391:507–514 [DOI] [PubMed] [Google Scholar]
- 30. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 31. Sarris PF, Karri IV, Goumas DE. 2010. First report of Pseudomonas syringae pv. alisalensis causing bacterial blight of arugula (Eruca vesicaria subsp. savita) in Greece. New Dis. Rep. 22:22 [Google Scholar]
- 32. Schofield DA, Molineux IJ, Westwater C. 2009. Diagnostic bioluminescent phage for detection of Yersinia pestis. J. Clin. Microbiol. 47:3887–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schofield DA, Westwater C. 2009. Phage-mediated bioluminescent detection of Bacillus anthracis. J. Appl. Microbiol. 107:1468–1478 [DOI] [PubMed] [Google Scholar]
- 34. Smartt AE, Ripp S. 2011. Bacteriophage reporter technology for sensing and detecting microbial targets. Anal. Bioanal. Chem. 400:991–1007 [DOI] [PubMed] [Google Scholar]
- 35. Tang L, et al. 2008. Consumption of raw cruciferous vegetables is inversely associated with bladder cancer risk. Cancer Epidemiol. Biomark. Prev. 17:938–944 [DOI] [PubMed] [Google Scholar]
- 36. Thouand G, Vachon P, Liu S, Dayre M, Griffiths MW. 2008. Optimization and validation of a simple method using P22::luxAB bacteriophage for rapid detection of Salmonella enterica serotypes A, B, and D in poultry samples. J. Food Prot. 71:380–385 [DOI] [PubMed] [Google Scholar]
- 37. Wechter WP, Farnham MW, Smith JP, Kinaith AP. 2007. Identification of resistance to peppery leaf spot among Brassica juncea and Brassica rapa plant introductions. HortScience 42:1140–1143 [Google Scholar]
- 38. Wechter WP, Keinath AP, Farnham MW, Smith JP. 2010. First report of bacterial leaf blight on broccoli and cabbage caused by Pseudomonas syringae pv. alisalensis in South Carolina. Plant Dis. 94:132. [DOI] [PubMed] [Google Scholar]