Abstract
A variable-number tandem repeat (VNTR) protocol targeting 10 loci in the Brucella abortus genome was used to assess genetic diversity among 366 field isolates recovered from cattle, bison, and elk in the Greater Yellowstone Area (GYA) and Texas during 1998 to 2011. Minimum spanning tree (MST) and unweighted-pair group method with arithmetic mean (UPGMA) analyses of VNTR data identified 237 different VNTR types, among which 14 prominent clusters of isolates could be identified. Cattle isolates from Texas segregated into three clusters: one comprised of field isolates from 1998 to 2005, one comprised of vaccination-associated infections, and one associated with an outbreak in Starr County in January 2011. An isolate obtained from a feral sow trapped on property adjacent to the Starr County herd in May 2011 clustered with the cattle isolates, suggesting a role for feral swine as B. abortus reservoirs in Starr County. Isolates from a 2005 cattle outbreak in Wyoming displayed VNTR-10 profiles matching those of strains recovered from Wyoming and Idaho elk. Additionally, isolates associated with cattle outbreaks in Idaho in 2002, Montana in 2008 and 2011, and Wyoming in 2010 primarily clustered with isolates recovered from GYA elk. This study indicates that elk play a predominant role in the transmission of B. abortus to cattle located in the GYA.
INTRODUCTION
Brucella abortus continues to be an important veterinary pathogen in the United States, particularly among cattle (Bos primigenius), elk (Cervus elaphus), and bison (Bos bison). The role of elk and bison in the acquisition of B. abortus by cattle in the regions of Idaho, Wyoming, and Montana comprising the Greater Yellowstone Area (GYA) is a politically charged feature of wildlife management and land use programs in that territory (7, 18). Seroprevalence rates among elk herds in the GYA range from 8 to 60%, while in bison herds, seroprevalence ranges from 11 to 75% (8, 9, 12, 21, 22, 23). While the exact means by which cattle may acquire brucellosis from wildlife remains uncertain, it is thought that exposure to aborted fetuses and afterbirth (Fig. 1), feces, or direct contact with infected animals all may constitute routes of infection, an inference supported by observational studies of elk and bison behavior that have documented commingling of these animals with cattle (9).
Fig 1.
Photograph of an aborted bison fetus on pastureland outside Jackson, WY, November 2007. (Photograph courtesy of Jill Randall.)
Apart from the foci of brucellosis associated with the GYA, sporadic cases of brucellosis have been recorded among cattle in Texas; since 2005, these have represented vaccine-associated infections in recipient animals. At present, the vaccine used in the United States is strain RB51, which was licensed for use by the USDA in 1996 (http://www.aphis.usda.gov/animal_health/animal_dis_spec/cattle/downloads/rb51_vaccine.pdf), although apparently some practitioners continued to use their existing stocks of strain 19 after that time. In 2008 the state was declared brucellosis free. However, following detection of seropositive animals at a livestock market, an outbreak was identified in a small beef cattle herd early in 2011 in Starr County, which adjoins the Rio Grande and the Mexican border in southeastern Texas. The herd was depopulated, and 836 cattle located on adjoining premises were tested for seropositivity during the first quarter of 2011. None of these cattle yielded positive test results, and the state continues to hold Class Free status (http://www.tahc.state.tx.us/news/pr/2011/2001Apr_BrucellosisWrapupStarrCounty.pdf).
Since 1934, the USDA and state animal health agencies have partnered in a collaborative effort to reduce the incidence and prevalence of brucellosis in livestock. In February 2008, all 50 states were designated Class Free for the disease in domestic cattle herds (http://www.usda.gov/wps/portal/usda/usdahome?contentidonly=true&contentid=2008/02/0027.xml). In recognition of the success of these efforts, in 2010 the USDA-APHIS Veterinary Services National Slaughter Surveillance Program implemented reduced sampling in states or areas that have been Class Free for five or more years. Alterations to existing surveillance programs are designed to reduce the disruption to slaughter establishments, maintain a surveillance emphasis on relevant geographic areas, and maximize the probability of detection of positive cases in low-risk areas (27).
Characterization of the molecular epidemiology of B. abortus is an important component of efforts by APHIS and state animal health agencies to control the disease among wildlife and livestock. One of the initial protocols used for this purpose was the HOOF-Prints assay of Bricker et al. (5), a variable-number tandem repeat (VNTR) assay (alternately referred to as multilocus variable-number tandem repeat analysis [MLVA]), which exploited the presence of 8-bp tandem repeat sequences at 8 loci in the B. abortus genome. This assay was used to differentiate clusters and groupings among a panel of 97 B. abortus reference strains and field isolates, representing three biovars, collected from different geographic locales in the United States (6).
The study by Beja-Pereira et al. (2) is, to date, the most comprehensive analysis of genetic variation among B. abortus isolates from the GYA. Those authors used an expanded, 10-locus iteration of the HOOF-Prints assay to examine 10 loci among 14 B. abortus cohorts comprised of 25 elk, 10 bison, and 23 cattle from nine locations across Montana, Wyoming, and Idaho. Beja-Pereira et al. concluded that elk, rather than bison, may represent the likely sources of transmission to cattle in the GYA.
Since the publication of those studies, the National Veterinary Services Laboratories (NVSL) and its collaborators in other federal (Centers for Disease Control and Prevention [CDC]) and state (Texas, Idaho, Montana, and Wyoming) agencies have assembled a larger database of B. abortus isolates from the GYA and other locales in the United States. As well, the NVSL has adopted a VNTR-10 protocol for B. abortus that utilizes loci drawn partly from the HOOF-Prints loci and partly from loci associated with a survey of B. abortus isolates of European origin (28). We utilized a genotyping analysis of 366 B. abortus field isolates of veterinary origin to further our understanding of genetic diversity among isolates from different regions of the United States. We also were interested in defining genotypes of B. abortus associated with vaccine-associated, and naturally acquired, infections in cattle in Texas. As well, we used the genotyping analysis to investigate whether lineages of B. abortus associated with cattle in the GYA could also be detected in elk and bison in that region.
MATERIALS AND METHODS
B. abortus isolates.
Isolates included in this study were obtained from naturally infected samples (primarily tissue, milk/mammary secretions, and vaginal exudate) submitted to the NVSL as part of the USDA-APHIS National Brucellosis Eradication Program, as well as from wildlife surveillance samples submitted by various federal and state agencies. Isolates were collected during an interval that spanned from 1978 to October 2011. The majority of isolates analyzed in this project constituted a subset of the overall isolate collection, with a focus on isolates received after 2000, for which there tended to be more complete documentation. Procedures for the isolation of Brucella bacteria from diagnostic samples, as well as subsequent biochemical identification, were performed by traditional methods (1). In some cases, identification was confirmed by AMOS and/or BASS PCR (4, 13).
A summary of the field isolates used in the study is provided in Table 1 (see also Table S1 in the supplemental material). Concerning multiple isolates from a single animal, those with different VNTR-10 profiles are included in the analysis, while isolates from the same animal, with identical VNTR-10 profiles, are only represented once. Additional B. abortus cultures were used on a routine basis as positive controls for biochemical assays, AMOS PCR, and VNTR-10: two of strain 19, one of biovar 4, one of biovar 2, and one of strain RB51. Unless otherwise noted, the case number assigned to isolates reflects the fiscal year (i.e., October 1 to September 30) in which the isolate was acquired at the National Veterinary Services Laboratories.
Table 1.
Field isolates of Brucella abortus evaluated in this study
| Host species | State | No. of isolates |
|---|---|---|
| Elk (Cervus elaphus) | ID | 19 |
| MT | 23 | |
| WY | 35 | |
| Total for species | 77 | |
| Bison (Bos bison) | SD | 1 |
| MT | 63 | |
| WY | 133 | |
| FL | 1 | |
| WI | 1 | |
| Total for species | 199 | |
| Cattle (Bos primigenius) | ID | 29 |
| MT | 4 | |
| SD | 1 | |
| TX | 38 | |
| WY | 10 | |
| TN | 1 | |
| OK | 1 | |
| NE | 1 | |
| FL | 1 | |
| MS | 1 | |
| Total for species | 87 | |
| Reindeer (Rangifer tarandus) | AK | 1 |
| Llama (Lama glama)a | 1 | |
| Hog (feral) (Sus domestica) | TX | 1 |
The state of origin of the llama isolate was unknown.
Variable-number tandem repeat assay.
Prior to 2008, genotyping of B. abortus isolates (including archived isolates from 1978 onwards) at the NVSL was performed using the HOOF-Prints assay (5). In March 2008, a decision was made to adopt a protocol used at the CDC involving a panel of 21 different VNTR loci (VNTR-21), including loci associated with the HOOF-Prints assay (5, 6) as well as VNTR loci described by Whatmore et al. (28).
A genotyping analysis conducted in mid-2008 on 82 Brucella isolates (including isolates represented in this project) generated VNTR-21 profiles with Simpson's diversity index values ranging from 0 to 86.8%. Based on this analysis, a subset of 10 of the 21 loci, H(OOF)-1, H3, H4, H8, VNTR16, VNTR17, VNTR2, VNTR21, VNTR5A, and VNTR5B, was selected for its ability to differentiate strains into epidemiologically relevant clusters, and this subset was also adopted from late 2008 onwards at the NVSL for VNTR-10 genotyping analyses of B. abortus isolates (B. Harris, unpublished data).
Repository Brucella isolates were stored in Trypticase soy broth with 25% glycerol at −70°C; older isolates were stored as potato agar slants at −70°C. Isolates were subcultured on Trypticase soy agar supplemented with 5% bovine serum and subjected to DNA extraction using InstaGene matrix (Bio-Rad, Hercules, CA). Briefly, a loopful of cells was suspended in 150 to 200 μl InstaGene matrix and incubated at 56°C for 15 min and then incubated at 100°C for 15 min. After heating, the tube was allowed to cool to room temperature and then centrifuged at 2,500 × g for 1 min to pellet the matrix, and a 40-μl aliquot of the supernatant was withdrawn and subjected to viability testing, which consisted of plating the 40 μl of heat-killed cells on Trypticase soy agar with 5% bovine serum and incubation for 5 days. If no growth was observed on the plate, nonviability was established, and 1 μl of the supernatant (equivalent to 5 to 10 ng DNA) was used as template for VNTR PCR. InstaGene-extracted DNA was stored at −70°C, usually for less than 30 days, before being used.
PCR assays were performed using dye-labeled H1 (hexachlorofluoroscein [HEX] conjugated), H3 (6-carboxytetramethyl rhodamine [TAM] conjugated), H4 (6-carboxyfluorescein [FAM]), H8 (TAM), VNTR 16 (FAM), VNTR 17 (HEX), VNTR 2 (HEX), VNTR 21 (TAM), VNTR 5A (HEX), and VNTR 5B (FAM) primers in a multiplex, 10-μl reaction mixture containing 1× PCR buffer, 5% dimethyl sulfoxide, 0.2 mM each deoxynucleoside triphosphate, 400 to 600 nM each primer, 0.25 U FastStart high-fidelity Taq polymerase (Roche, Indianapolis, IN), and 5 to 10 ng B. abortus DNA. Thermal cycling conditions were 5 min at 95°C, then 35 cycles of 30 s at 95°C, 1 min at 55°C, and 1.5 min at 75°C, and a final hold of 5 min at 75°C. All PCRs included as a positive control B. abortus strain 19 (NVSL lot BC-St19) and also no-template controls. This strain 19 isolate is a subculture of the vaccine strain 19 original seed, maintained at the NVSL Brucella and Mycobacterial Reagents Team (BMRT) department in Ames, IA. The subculture is stored as glycerol stocks at −80°C, with aliquots withdrawn from these stocks approximately every 4 to 6 weeks for use as controls for B. abortus biochemical tests and as a source of DNA for use in AMOS PCR and VNTR-10 assays.
PCR products were stored at 4°C in light-safe containers for no more than 48 h before being subjected to electrophoresis on an ABI 3500XL genetic analyzer using the GeneScan 600 LIZ size standard (Applied Biosystems, Carlsbad, CA). Fragment data were analyzed using the GeneMapper 4.1 software package (Applied Biosystems, Carlsbad, CA). VNTR profiles were analyzed using the BioNumerics 6.1 software package. Only isolates that generated unambiguous, well-resolved peaks in the GeneMapper (Applied Biosystems, Carlsbad, CA) electropherograms were included in the database and subsequent statistical analyses (over the past 4 years, fewer than five putative B. abortus isolates were excluded from the project because they failed to generate acceptable GeneMapper profiles). Electropherograms indicative of the absence of a PCR product (i.e., null allele) or zero repeats for a given locus were examined manually, and a confirmatory singelton PCR assay for that locus was performed if the peak profile was considered ambiguous.
Statistical analysis of VNTR-10 data.
Two methods were used to evaluate the performance of the panel of 10 VNTR loci. Calculation of allelic diversity (h) was performed using the following equation: , where n is the number of isolates and xi is the frequency of the ith allele at the locus (19, 24). The Shannon-Wiener index of diversity was calculated using the BioNumerics 6.1 software package (Applied Maths, Saint-Martens-Latem, Belgium). Statistical evaluation of VNTR data was performed using BioNumerics functions for cluster analysis of categorical data using unweighted-pair group method with arithmetic mean (UPGMA) analysis and minimum spanning tree (MST) analysis. The discriminatory power of the VNTR-10 assay was calculated using the website http://insilico.ehu.es/mini_tools/discriminatory_power/index.php (3).
RESULTS
Allelic diversity of VNTR-10 loci.
A total of 366 field isolates and 5 reference strains were used in the genotyping analysis (Table 1; also see Table S1 in the supplemental material). To confirm that the 10 loci analyzed in our assays contained the inferred number of repeats as determined by GeneMapper analysis, PCR amplicons from all 10 loci from eight B. abortus isolates (including four field isolates of biovar 1, a field isolate of strain 19, and laboratory isolates of biovar 4, strain 19, and strain RB51) were sequenced. GeneMapper binning parameters were set according to the number of tandem repeats observed with sequencing (as opposed to inferring these values from in silico data). While we observed isolates with zero repeats for some loci, we did not observe any isolates with null (i.e., absent) loci in our experiment (see Table S1 in the supplemental material).
Using MST analysis, 237 different VNTR-10 types were identified among the 371 isolates (including 5 reference strains) in our collection. The discriminatory power of the VNTR-10 assay was 0.99. The allelic diversity (h) and Shannon-Wiener statistics were calculated for the VNTR-10 data from all isolates (Table 2). For the allelic diversity calculations, three loci, Hoof 8, VNTR 17, and VNTR 21, exhibited little or no diversity (h = 0, 0, and 0.02, respectively). Hoof 3, VNTR 16, and VNTR 5A showed a moderate capacity (i.e., 0.30 ≤ h ≤ 0.70). Four loci (Hoof 1, Hoof 4, VNTR 2, and VNTR 5B) displayed values of h that were >0.75 and thus represented the most discriminatory alleles. Results of the Shannon-Wiener analysis paralleled those observed with allelic diversity; i.e., loci with negligible allelic diversity also exhibited low Shannon-Wiener indices (Table 2).
Table 2.
Shannon-Wiener index of diversity and allelic diversity of 10 Brucella abortus loci used in the VNTR assay of 371 isolates
| Diversity measurea | Diversity score for locus |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hoof 1 | Hoof 3 | Hoof 4 | Hoof 8 | VNTR 16 | VNTR 17 | VNTR 2 | VNTR 21 | VNTR 5A | VNTR 5B | |
| Shannon-Wiener index of diversity | 2.2 | 1.6 | 2.0 | 0 | 0.7 | 0 | 1.7 | 0.06 | 1.2 | 2.2 |
| Allelic diversity (h) | 0.84 | 0.73 | 0.84 | 0 | 0.45 | 0 | 0.79 | 0.02 | 0.70 | 0.87 |
Data were generated from 366 field isolates and 5 reference strains.
Global clustering analysis of all isolates.
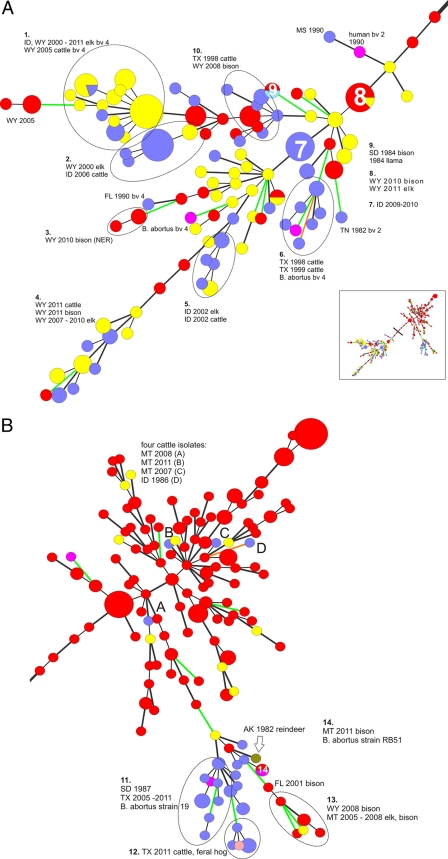
In order to provide a comprehensive, but easily interpretable, gestalt of the relatedness of all 371 isolates, VNTR-10 data were analyzed using MST. The resultant diagram was partitioned into two major portions, referred to for convenience here as the left and right portions; these are reproduced in Fig. 2A and B, respectively.
Fig 2.
(A) Left portion from MST analysis of the VNTR-10 profiles for 371 isolates of Brucella abortus included in this study. Color code: yellow, elk; red, bison; blue, cattle; pink, B. abortus reference strains; turquoise, llama; mauve, feral hog; dark green, reindeer. Each sphere represents a unique VNTR-10 type; smaller spheres contain one isolate, while larger spheres contain multiple isolates with matching VNTR-10 profiles. Thin black lines indicate branches with a difference of 1 locus (of 10 VNTR loci); thick black lines, 2 loci; green lines, 3 loci; orange lines, 4 or more loci. Noteworthy clusters are identified by number. Labels denote the state and year of origin; host animal species are provided in cluster labels when multiple species occupy a cluster. Unless otherwise noted, all isolates are biovar 1; alternate biovars are indicated by abbreviations (i.e., bv 2). The inset depicts the entire MST, with the vertical bar dividing the tree into the left and right portions (depicted in panels A and B, respectively). Total network length, 387; cophenetic correlation coefficient, 63%. (B) Right portion of MST analysis of the VNTR-10 profiles for 371 isolates of Brucella abortus included in this study. Color code: yellow, elk; red, bison; blue, cattle; pink, B. abortus reference strains; purple, llama; mauve, feral hog; dark green, reindeer. Each sphere represents a unique VNTR-10 type; smaller spheres contain one isolate, while larger spheres contain multiple isolates with matching VNTR-10 profiles. Thin black lines indicate branches with a difference of 1 (of 10) VNTR locus; thick black lines, 2 loci; green lines, 3 loci; orange lines, 4 or more loci. Noteworthy clusters are identified by number. Labels denote state and year of origin; host animal species are provided in cluster labels when multiple species occupy a cluster. Unless otherwise noted, all isolates are biovar 1; alternate biovars are indicated by an abbreviation (i.e., bv 2).
The cophenetic correlation coefficient (a statistic that is roughly analogous to the correlation coefficient used for linear regression) for the entire MST was 63%, for a total network length of 383, indicating that the MST was not overly robust in terms of representing the original pairwise distances. Accordingly, the spatial arrangement of the taxa should be interpreted with some degree of caution; as well, it should be noted that there is some element of subjectivity and compromise in deciding which collection of isolates constitutes a “noteworthy” cluster and how best to demarcate such clusters within the confines of a two-dimensional diagram containing a large number of entries.
The left portion (Fig. 2A) contains 100 VNTR-10 types, comprised of biovar 1, biovar 2, and biovar 4 isolates, with 10 prominent clusters (identified by number). Cluster 1 contains strains recovered from GYA elk and cattle, all of biovar 4, from 2000 to 2011, while cluster 2 contains Wyoming elk isolates from 2000 and Idaho cattle isolates from 2006, all of biovar 1. Cluster 3 contains several isolates recovered from Wyoming bison in the National Elk Refuge (NER) in 2010. Clusters 4 and 5 contain GYA elk, cattle, and bison isolates, while cluster 6 is comprised of Texas cattle isolates from two infected herds identified in 1998 and 1999 as well as a reference strain of biovar 4. Cluster 7 is comprised of 2009 to 2010 Idaho cattle isolates, and cluster 8 contains Wyoming elk and bison isolates from 2010 that possess within-cluster identical VNTR-10 profiles. Cluster 9 segregates apparently epidemiologically unrelated isolates: here, a 1984 isolate from a South Dakota bison and an isolate recovered from a llama in 1994. Cluster 10 represents a node into which 1998 isolates from Texas cattle and 2008 Wyoming bison segregate.
The right side (Fig. 2B) of the MST contains 137 VNTR-10 types, which were primarily generated from bison located in the GYA, reflecting the preponderance of these isolates in the database. Four noteworthy clusters are depicted: cluster 11 consists entirely of cattle isolates, including many from Texas from 2005 to 2011, and an archived 1987 strain from a South Dakota cow. Cluster 12 consists of 2011 Texas isolates of bovine and feral pig origins. Cluster 13 contains GYA bison and elk isolates collected from 2005 to 2008. Cluster 14 contains two isolates, both with matching VNTR-10 profiles; these are a reference strain of RB51 and an isolate recovered in 2011 from an aborted bison fetus maintained in a privately owned herd in Montana.
The formation of clusters occupied by epidemiologically unrelated isolates (for example, clusters 10 and 11) indicates that our VNTR-10 typing method is capable of generating homoplasic relationships (i.e., clusters representative of convergent evolution of similar VNTR-10 profiles). Accordingly, we relied on a combination of both epidemiologic data and genotyping data when drawing inferences about possible transmission cycles of B. abortus among animals in Texas and the GYA; expanded analyses of these transmission cycles are presented in the following sections. For assistance in identifying the counties and locales associated with infected animals, readers are directed to the Texas county map at http://quickfacts.census.gov/qfd/maps/texas_map.html. A map of the hunting areas of Wyoming is available at http://gf.state.wy.us/web2011/HUNTING-1000179.aspx. A map of the National Elk Refuge is available at http://www.fws.gov/nationalelkrefuge/Documents/2011_Hunting/2011_Elk_InfoRegs.pdf, and an interactive map of Montana hunting districts is available at http://fwpiis.mt.gov/hunting/planahunt/MapApp/default.aspx?region=1&species=mule.
B. abortus isolates from Texas cattle, 1998 to 2011.
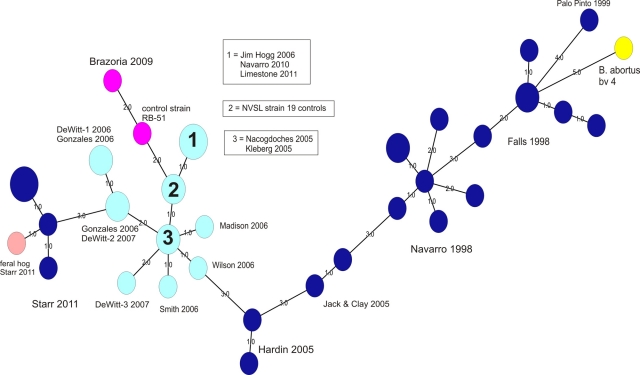
An MST analysis of 38 isolates obtained from cattle from Texas in the years 1998 to 2011 is provided in Fig. 3. These isolates included strains recovered from animals vaccinated with strain 19 and strain RB51. Three prominent clades were noted (going from right to left in Fig. 2): one (dark blue spheres) consists of B. abortus biovar 1 isolates obtained from Palo Pinto, Falls, Navarro, Jack and Clay, and Hardin Counties from 1998 to 2005. The other clade (light blue spheres) consists of field isolates of strain 19, obtained from Gonzales, DeWitt, Smith, Wilson, Madison, Jim Hogg, Limestone, Navarro, Nacogdoches, and Kleberg Counties from 2005 to 2011. A branch of this large “strain 19” cluster contains an isolate recovered from a Brazoria County cow in 2009 and an RB51 reference strain (the two pink nodes in Fig. 3).
Fig 3.
Minimum spanning tree analysis of the VNTR-10 profiles for 38 B. abortus isolates recovered from Texas cattle and a feral hog, 1998 to 2011. Each sphere represents a unique VNTR-10 type; smaller spheres contain one isolate, while larger spheres contain multiple isolates with matching VNTR-10 profiles. Labels denote county and year of origin; multiple isolates from the same county are designated by a numerical suffix attached to the county name. Color code: dark blue, B. abortus biovar 1; turquoise, B. abortus strain 19; pink, B. abortus strain RB51; yellow, B. abortus biovar 4. Smaller-font numbers assigned to branches indicate the number of loci (of a total of 10 loci) that were different between the linked isolates. Total network length, 55; cophenetic correlation coefficient, 80%.
The final clade (dark blue spheres) consists of 5 B. abortus biovar 1 isolates recovered from cattle from an owner located in Starr County in 2011; these isolates display limited similarity (∼ 60%) with all other isolates associated with Texas cattle. This cluster also contains an isolate from a feral pig (sow) recovered by the State-Federal Diagnostic Laboratory in Austin; the animal was 1 of a group of 12 pigs trapped in May 2011 on Starr County property adjacent to the location of the infected cattle herd. Interestingly, serologic testing performed at the State-Federal Diagnostic Laboratory indicated that all 12 feral pigs were seronegative for Brucella (M. Hamelwright, personal communication). The feral pig isolate displayed 83% similarity in its VNTR-10 profile with those from the Starr County cattle; it differed from one cattle isolate, case B11-0206, at only one locus, H1, by only 2 repeats.
Because of the limited similarity that the Starr County B. abortus isolates displayed with our other Texas isolates, we were interested in querying genotype profiles associated with clinical (i.e., human) isolates. When profiled using an MLVA-15 assay (14) performed at the Zoonoses and Select Agent Laboratory at the Centers for Disease Control and Prevention, the Starr County cattle isolates did not exactly match any entries in the CDC database of human B. abortus strains; the isolate displaying nearest-neighbor clustering with the Starr County strains was a 2007 clinical isolate provided by the Arizona State Health Department, and it displayed ∼76% similarity in its MLVA-15 profile (see Fig. S1 in the supplemental material).
B. abortus isolates associated with GYA cattle and wildlife, 1999 to 2011.
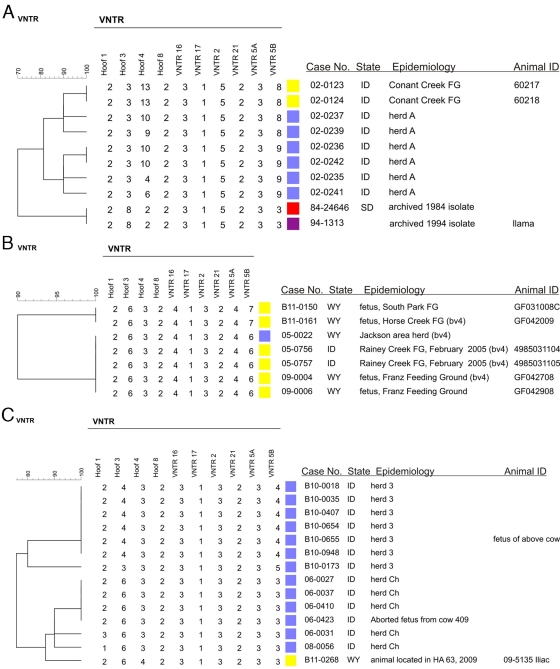
MST analysis identified a number of clusters containing GYA elk, bison, and cattle, including clusters 1, 2, 4, 5, and 13 (Fig. 2A and B). In order to provide readers with a more detailed examination of these and other cattle-related outbreaks, we constructed UPGMA dendrograms (which display the VNTR-10 numerical values for all loci for all isolates) for the strains involved in these clusters. These UPGMA dendrograms are presented in Fig. 4A to F.
Fig 4.
Dendrograms generated from UPGMA analyses of VNTR-10 profiles of B. abortus isolates associated with GYA cattle and bison brucellosis outbreaks. Color key: cattle, blue; bison, red; elk, yellow; pink, reference strain; isolates from other animal sources are indicated in the text of their labels. All isolates were biovar 1 unless otherwise indicated. FG, feeding ground; HA, hunt area; NER, National Elk Refuge.
Isolates associated with a 2002 outbreak among a herd of cattle in Idaho are depicted in Fig. 4A. None of the cattle isolates (herd A) matched VNTR-10 profiles with other isolates in the database; nearest neighbors to these cattle isolates included two isolates recovered from elk in the Conant Creek Feeding Ground in Idaho. Also clustering with the Idaho cattle isolates were two archived strains of B. abortus, one recovered from a South Dakota bison in 1984 and a 1994 llama isolate (its state of origin is unknown).
Figure 4B depicts a B. abortus biovar 4 isolate recovered from a cow in Jackson, WY, in 2005. This cattle isolate exactly matched the VNTR-10 profiles of other isolates in our database. These matches included two biovar 4 isolates obtained in 2005 from elk in the Rainey Creek Feeding Ground in Idaho and biovar 4 and biovar 1 isolates obtained in 2008 from two elk fetuses, ID numbers GF042708 and GF042908, respectively, located on the Franz Feeding Ground in Wyoming. Nonmatching nearest-neighbor isolates to these included other strains obtained from aborted elk fetuses located at the South Park Feeding Ground in 2011.
Figure 4C summarizes B. abortus strains associated with brucellosis among two cattle herds (herd Ch and herd 3) in Idaho in 2006 and 2010. Isolates from both outbreaks segregated with only one other strain from our collection, a 2009 isolate recovered from an elk in Hunt Area 63 (HA 63) in Wyoming.
Figure 4D depicts the isolates clustered based on a diagnosis of brucellosis in two Montana cattle herds in 2008 (ranch A) and September 2011 (ranch B). The ranches are adjacent to each other, and a shared ridgeline runs through both properties; elk frequently descend this ridgeline to explore the pastures. The closest match for the 2008/ranch A isolate was a B. abortus isolate obtained in 2010 from a Montana elk; the only difference in VNTR-10 profile between these two isolates was the presence of 5 repeats for the bovine isolate, versus 6 repeats for the elk isolate for locus VNTR 2. The 2011/ranch B isolate was obtained from a heifer; this isolate possessed a unique VNTR-10 profile. Nearest neighbors to the 2011 isolate included B. abortus isolates recovered from bison from the GYA during 2005 to 2008.
Figure 4E provides results from the largest recent outbreak of brucellosis among cattle in the GYA, an outbreak taking place in Meeteetse, Park County, WY, in November and December 2010. Cattle at two ranches (designated herd A and herd B) were diagnosed with brucellosis; the four isolates representing these two cohorts segregated into separate clusters, with no exact matches among other strains in our database (Fig. 4E). For herd A, the B. abortus isolates from two infected animals displayed heterogeneity in their VNTR-10 profiles, with differences present at Hoof 3; for herd B, isolates from two different animals displayed the same VNTR-10 profile. Nearest neighbors to the herd A and herd B strains included isolates obtained in 2007 and 2010 from Wyoming elk in HA 61.
A 2-year-old female bison kept on a private ranch (herd C) in the Park County area was diagnosed with brucellosis in November 2010; the B. abortus isolate from this animal (case number B11-0053) occupied a singleton branch and showed limited similarity in its VNTR-10 profile with the cattle isolates from Meeteetse and the isolates obtained from elk in HA 61 (Fig. 4E).
Figure 4F depicts the relationships between three bison cases originating from the same (privately owned) herd in the Gallatin Gateway region of Montana and associated elk isolates. Isolate B11-0011 was recovered in October 2010 from the dam of an aborted calf (no isolation was made from the calf). The isolate possessed a VNTR-10 profile identical to that of an isolate obtained in 2009 from a hunter-killed elk in Montana (animal ID BR090731).
A second isolate, case number B11-0280, was recovered from a fetus aborted by a dam vaccinated (with strain RB51) in the fall of 2010; this isolate displayed a VNTR-10 profile equivalent to that of our in-house strain, the RB51 control. The third isolate (case number B11-0473) originating in the herd was recovered from a 2-year-old male reactor sampled in April 2011; this isolate of B. abortus exhibited a difference of one tandem repeat at locus VNTR 2 with an isolate obtained in 2009 from a Montana elk (animal ID BR081200) (Fig. 4F).
DISCUSSION
The analysis of B. abortus isolates based on the VNTR-10 panel was implemented at the NVSL early in 2008 in order to aid efforts by APHIS staff, state veterinarians, epidemiologists, and wildlife managers to define foci of transmission and inform control strategies. In general, our VNTR-10 assay proved useful for evaluating genetic diversity among B. abortus isolates, with only three loci (Hoof 8, VNTR 17, and VNTR 21) displaying negligible indices of diversity. We did observe geographically and temporally disparate isolates possessing exactly the same profiles, a phenomenon known as homoplasy (in which unrelated isolates independently evolve matching genotypes); accordingly, we relied on a combination of genetic and epidemiologic data in order to make conclusions about the involvement of various lineages of B. abortus in the cattle outbreaks described in this report.
Our study has some weaknesses. First, the inclusion of migratory animals, such as elk and bison, reflects the site where the animal was sampled and not necessarily where it acquired the infection; accordingly, inferences about the geographic distributions of selected clusters or groupings of elk and bison strains of B. abortus should be made with some degree of caution. Second, by querying 10 loci, instead of the panels of 15 or more loci used in other Brucella VNTR assays (14, 15, 16, 17, 26, 28), we may have restricted our ability to generate finer partitioning among isolates with highly similar VNTR-10 profiles. Third, we did not perform multiple-locus sequence typing (MLST) on our isolates; while this technique may not necessarily be more advantageous for partitioning among otherwise-closely grouped isolates, it can reveal the existence of single-nucleotide polymorphisms associated with noteworthy features of the Brucella genome (11, 29). Fourth, our knowledge of the background histories and epidemiologic data of submissions is limited to the information provided on the sample submission form.
Our analysis of genetic variability among B. abortus isolates in Texas incorporates isolates of strain 19 and strain RB51 recovered from vaccinated cattle and reflects ongoing surveillance for such infections on the part of animal health practitioners in that state. Examination of VNTR-10 profiles for these clinical isolates indicated that alterations in tandem repeat number occur, compared to the profiles we have observed for strain 19 and strain RB51 isolates used as laboratory controls.
It is unclear if the alterations in VNTR-10 profiles we observed in vaccine-derived clinical isolates were a result of genetic alterations associated with host-mediated selection for particular lineages of B. abortus following inoculation. In the 4 years (i.e., 2008 to 2012) during which we have employed the VNTR-10 and VNTR-21 assays (the latter used for analysis of Brucella suis isolates), we have routinely used a passaged isolate of strain 19 as a positive control, and we have not observed alterations in its VNTR-10 or VNTR-21 profile during that time. In their description of the VNTR-21 assay, Whatmore et al. (28) examined serially passaged (14 passages over 270 days) isolates of B. abortus, B. suis, and Brucella melitensis; alterations in the VNTR-21 profile were restricted to the B. abortus isolate, to locus VNTR 12B, to an increase in tandem repeat number of +1 (28). In their study of multiply passaged vaccine strains of B. abortus, Kulakov et al. (16) observed variation in 4 of 12 VNTR loci, indicating that within-strain variability in genotype profiles was a feature of these particular lineages of B. abortus. This phenomenon may explain the variation observed in clinical isolates obtained from vaccinated cattle from Texas; however, more-detailed investigations are necessary before definitive conclusions can be drawn regarding postinoculation genetic variability in vaccine strains.
We also cannot definitively rule out a possible contribution of culture-induced alterations in VNTR profile to our interpretation of typing-based associations among field isolates. Detection of such contributions would require that each field isolate undergo several passages, followed by VNTR typing of some or all of these passaged cultures, which is impractical from a logistical standpoint (particularly given the obligations imposed by working with select agents). However, because the locus (VNTR 12B) reported by Whatmore et al. (28) as displaying passage-induced alterations for an isolate of B. abortus is not employed in our VNTR-10 assay, we do not believe that our interpretation of our data has been biased due to this phenomenon.
The outbreak of B. abortus infection among a small herd of cattle in Starr County in January 2011 was the first outbreak of brucellosis in Texas since the state was designated by APHIS as Class Free of the disease in 2008. Since the isolates displayed a unique VNTR-10 profile not previously observed in the NVSL database, we queried the CDC B. abortus database for possible matches. The sole isolate clustering with the Starr County strains, an isolate recovered from a patient from Arizona in 2007, displayed 76% similarity in its MLVA-15 profile. Interestingly, a feral pig trapped in May 2011 on property adjacent to the farm where the outbreak took place yielded an isolate that displayed 83% similarity in VNTR-10 profile with the Starr County cattle B. abortus strains. While genotyping alone cannot determine if the feral pig was a source of the infection in the cattle, or vice versa, the data affirm conclusions made in a previous study regarding feral pigs as potential reservoirs for B. abortus and indicate that further analyses of this population of animals, as well as other potential reservoir hosts, in the Starr County area are warranted (10, 25).
Our analysis of outbreaks in the GYA from 2002 to 2011 indicated that only one (biovar 4) strain of B. abortus originating in cattle possessed a VNTR-10 profile exactly matching that of a wildlife isolate; this was strain 05-0022 recovered in 2005 from a Jackson, WY, cow that matched 2005 isolates from elk located at the Rainey Creek Feeding Ground in eastern Idaho, as well as isolates recovered from aborted fetuses in 2009 at the Franz Feeding Ground in Wyoming (Fig. 4B). The exact matching of the VNTR-10 profiles and the fact that these areas are in close enough proximity to enable contact between Rainey Creek-associated elk and the affected cattle herd provide our strongest evidence for transmission of B. abortus between these two species.
Based on UPGMA-mediated comparisons of VNTR-10 profiles, GYA cattle isolates possessed VNTR-10 profiles with 70 to 95% similarity to isolates from elk. While bison isolates displayed ≤80% similarity to cattle isolates, bison isolates possessed VNTR-10 profiles with >80% similarity to those observed in elk (Fig. 4A to F). These observations suggest a greater degree of genetic propinquity between GYA elk isolates and those from cattle and a lesser degree of genetic propinquity between GYA bison isolates and those from cattle.
A November 2010 outbreak among cattle and privately owned bison in the Meeteetse, Park County, WY, area represents the largest outbreak of brucellosis in the GYA since 2009 (Fig. 4E). As best can be determined from the investigation of the outbreak, there were no links (i.e., commingling of animals) between either of the two affected cattle herds, nor were there any links with the affected bison herd. Serologic testing of the two cattle herds immediately following the discovery of the infected cattle indicated a prevalence of ∼1%. Our existing observations regarding seroprevalence rates among cattle herds in the GYA indicate that seroprevalence rates of 2.5% or greater are associated with exposure windows to infection of greater than 18 months (J. Belfrage, personal communication). Accordingly, we hypothesize, based on genotyping and epidemiologic data, that the two cattle herds and the bison herd in Meeteetse were exposed to infected elk within a 12-month interval preceding the outbreak.
Indications of a complex epidemiology involving bison located on a private ranch in Montana surfaced in October 2010 and in March and April 2011. The genotyping data generated from isolates present in this herd indicate that both vaccine-related infections, as well as infections with a field strain of B. abortus, may take place among bison within the same herd. These observations demonstrate the utility of VNTR-10 analysis in distinguishing between the two sources of infection.
Our genotyping data reinforce earlier conclusions that elk constitute the most likely reservoir for this pathogen among GYA wildlife, an observation with implications for management of the numerous winter feed grounds for wildlife in the region, as well as control measures, such as selective culling of bison herds and vaccination of elk and bison (9). Although these feed grounds were originally established to provide supplemental nutrition for wildlife, they have evolved to become a primary disease management tool by providing separation of elk and bison from domestic cattle. Unfortunately, feed grounds also provide conditions that allow brucellosis to proliferate and maintain itself in these wild elk and bison populations, by forcing high concentrations of animals on small areas of land during peak transmission periods (8). Feed grounds and other management techniques (hazing) are not able to prevent all commingling events (and, presumably, transmission) between domestic livestock and free-ranging elk and bison (23). We recognize that our genotyping data cannot definitively indicate the direction of transmission (i.e., elk to bison to cattle or vice versa), but they can inform the decision-making process for management of wildlife populations in the GYA.
We have demonstrated that the VNTR-10 assay can be useful in supporting inferences about disease transmission among comparatively circumscribed populations of cattle and wild animals in the GYA and Texas. However, improved tools for B. abortus genotyping are needed. Projects to acquire genomic sequences for as many as 300 Brucella spp. isolates are under way in the United States and Europe (http://www.broadinstitute.org/annotation/genome/brucella_group/News.html). These projects promise to generate improved diagnostic assays, based on characteristics (such as canonical single-nucleotide polymorphisms) capable of robustly differentiating isolates at the subspecies level (20).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dale Preston, Neil Anderson, Ryan Clarke, Mike O'Brien, Gordon Luikart, John Treanor, Mike McDole, Deb Dufficy, Barbara Martin, Deb Donch, Michael Hamelwright, and John Belfrage for providing isolates and epidemiologic information. Ben Rosenthal of the USDA-ARS assisted with the UPGMA analyses of B. abortus genotypes, and Monica Reising of the USDA-APHIS Center for Veterinary Biologics assisted with the statistical analyses.
Footnotes
Published ahead of print 16 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alton GG, Jones LM, Angus RD, Verger JM. 1988. Techniques for the brucellosis laboratory, p 34–61 INRA Publications, Paris, France [Google Scholar]
- 2. Beja-Pereira A, et al. 2009. DNA genotyping suggests that recent brucellosis outbreaks in the Greater Yellowstone Area originated from elk. J. Wildl. Dis. 45:1174–1177 [DOI] [PubMed] [Google Scholar]
- 3. Bikandi J, San Millan R, Rementeria A, Garaizar J. 2004. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR, and endonuclease restriction. Bioinformatics 22:798–799 [DOI] [PubMed] [Google Scholar]
- 4. Bricker BJ, Ewalt DR, Olsen SC, Jensen AE. 2003. Evaluation of the Brucella abortus species-specific polymerase chain reaction assay, an improved version of the Brucella AMOS polymerase chain reaction assay for cattle. J. Vet. Diagn. Invest. 15:374–378 [DOI] [PubMed] [Google Scholar]
- 5. Bricker BJ, Ewalt DR, Halling SM. 2003. Brucella ‘HOOF-Prints’: strain typing by multi-locus analysis of variable number tandem repeats (VNTRs). BMC Microbiol. 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bricker BJ, Ewalt DR. 2005. Evaluation of the HOOF-Print assay for typing Brucella abortus strains isolated from cattle in the United States: results with four performance criteria. BMC Microbiol. 5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheville NF, McCullough DR. 1998. Brucellosis in the Greater Yellowstone Area, p 16–88 National Research Council Press, Washington, DC [Google Scholar]
- 8. Cross PC, Edwards WH, Scurlock BM, Maichak EJ, Rogerson JD. 2007. Effects of management and climate on elk brucellosis in the Greater Yellowstone ecosystem. Ecol. Appl. 17:957–964 [DOI] [PubMed] [Google Scholar]
- 9. Cross PC, et al. 2010. Probable causes of increasing brucellosis in free-ranging elk of the Greater Yellowstone ecosystem. Ecol. Appl. 20:278–288 [DOI] [PubMed] [Google Scholar]
- 10. Davis DS, Boeer WJ, Mims JP, Heck FC, Adams LG. 1979. Brucella abortus in coyotes. I. Serologic and bacteriologic surveys in eastern Texas. J. Wildl. Dis. 151:367–372 [DOI] [PubMed] [Google Scholar]
- 11. De BK, et al. 2008. Novel Brucella strain (BO1) associated with a prosthetic breast implant infection. J. Clin. Microbiol. 46:43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Etter RP, Drew ML. 2006. Brucellosis in elk of eastern Idaho. J. Wildl. Dis. 42:271–278 [DOI] [PubMed] [Google Scholar]
- 13. Ewalt DR, Bricker BJ. 2003. Identification and differentiation of Brucella abortus field and vaccine strains by BASS-PCR. Methods Mol. Biol. 216:97–108 [DOI] [PubMed] [Google Scholar]
- 14. Huynh LY, et al. 2008. Multiple locus variable number tandem repeat (VNTR) analysis (MLVA) of Brucella spp. identifies species specific markers and insights into phylogenetic relationships, p 47–54 In Georgiev VS, Western K, McGowan JJ. (ed), National Institute of Allergy and Infectious Diseases, NIH: frontiers in research, vol 1 Humana Press, Totowa, NJ [Google Scholar]
- 15. Kilic S, et al. 2011. Multiple-locus variable-number tandem-repeat analysis genotyping of human Brucella isolates from Turkey. J. Clin. Microbiol. 49:3276–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kulakov Y, Zheludkov MM, Sclyarov OD. 2010. Variable-number tandem repeat markers for identification of Brucella abortus 82 and 75/79-AV vaccine strains. Vaccine 28S:F41–F45 [DOI] [PubMed] [Google Scholar]
- 17. Le Fleche P, et al. 2006. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meyer ME, Meahger M. 1995. Brucellosis in free-ranging bison in Yellowstone, Grand Teton, and Wood Buffalo National Parks: a review. J. Wildl. Dis. 31:579–598 [DOI] [PubMed] [Google Scholar]
- 19. Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small sample of individuals. Genetics 89:583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. O'Callaghan D, Whatmore A. 2011. Brucella genomics as we enter the multi-genome era. Brief. Funct. Genomics 10:334–341 [DOI] [PubMed] [Google Scholar]
- 21. Rhyan JC, et al. 2009. Pathogenesis and epidemiology of brucellosis in Yellowstone bison: serologic and culture results from adult females and their progeny. J. Wildl. Dis. 45:729–739 [DOI] [PubMed] [Google Scholar]
- 22. Roffe TJ, et al. 1999. Brucellosis in Yellowstone National Park bison: quantitative serology and infection. J. Wildl. Manage. 63:1132–1137 [Google Scholar]
- 23. Scurlock BM, Edwards WH. 2010. Status of brucellosis in free-ranging elk and bison in Wyoming. J. Wildl. Dis. 46:442–449 [DOI] [PubMed] [Google Scholar]
- 24. Selander RK, et al. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stoffregen W, et al. 2007. Diagnostic characterization of a feral swine herd enzootically infected with Brucella. J. Vet. Diagn. Invest. 19:227–237 [DOI] [PubMed] [Google Scholar]
- 26. Tiller RV, et al. 2009. Comparison of two multiple-locus variable-number tandem repeat (VNTR) analysis (MLVA) methods for molecular strain typing of human Brucella melitensis isolates from the Middle East. J. Clin. Microbiol. 47:2226–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. USDA-APHIS 2011. National brucellosis slaughter surveillance plan. USDA-APHIS, Washington, DC: http://www.aphis.usda.gov/animal_health/animal_diseases/brucellosis/downloads/nat_bruc_slaughter_surv_plan.pdf [Google Scholar]
- 28. Whatmore AM, et al. 2006. Identification and characterization of variable-number tandem-repeat markers for typing of Brucella spp. J. Clin. Microbiol. 44:1982–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Whatmore AM, Perrett L, MacMillan AP. 2007. Characterisation of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol. 7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.