Abstract
Xyn30D from the xylanolytic strain Paenibacillus barcinonensis has been identified and characterized. The enzyme shows a modular structure comprising a catalytic module family 30 (GH30) and a carbohydrate-binding module family 35 (CBM35). Like GH30 xylanases, recombinant Xyn30D efficiently hydrolyzed glucuronoxylans and methyl-glucuronic acid branched xylooligosaccharides but showed no catalytic activity on arabinose-substituted xylans. Kinetic parameters of Xyn30D were determined on beechwood xylan, showing a Km of 14.72 mg/ml and a kcat value of 1,510 min−1. The multidomain structure of Xyn30D clearly distinguishes it from the GH30 xylanases characterized to date, which are single-domain enzymes. The modules of the enzyme were individually expressed in a recombinant host and characterized. The isolated GH30 catalytic module showed specific activity, mode of action on xylan, and kinetic parameters that were similar to those of the full-length enzyme. Computer modeling of the three-dimensional structure of Xyn30D showed that the catalytic module is comprised of a common (β/α)8 barrel linked to a side-associated β-structure. Several derivatives of the catalytic module with decreasing deletions of this associated structure were constructed. None of them showed catalytic activity, indicating the importance of the side β-structure in the catalysis of Xyn30D. Binding properties of the isolated carbohydrate-binding module were analyzed by affinity gel electrophoresis, which showed that the CBM35 of the enzyme binds to soluble glucuronoxylans and arabinoxylans. Analysis by isothermal titration calorimetry showed that CBM35 binds to glucuronic acid and requires calcium ions for binding. Occurrence of a CBM35 in a glucuronoxylan-specific xylanase is a differential trait of the enzyme characterized.
INTRODUCTION
Biodegradation of xylan, which constitutes approximately one-third of the renewable organic carbon on earth (15), is a critical step in the recycling of carbon in nature and has been targeted as a subject of intense research as a renewable energy resource and for the bioconversion of plant biomass into high-added-value products (34, 46, 48). Catalytic breakdown of xylan is a complex process that requires the coordinated action of several enzymes, among which xylanases (1,4-β-d-xylan xylanohydrolase; EC 3.2.1.8), cleaving internal linkages on the β-1,4-xylose backbone, play a key role. The complex chemical nature and heterogeneity of xylan can account for the multiplicity of xylanases produced by microorganisms (11, 26). The activity of different xylanases with subtle differences in substrate specificities and modes of action contributes to improving the degradation of plant xylan in natural habitats.
Most of the xylanases characterized to date are grouped into glycoside hydrolase families GH10 and GH11 (CAZy) (20). These xylanases do not seem to be specialized for hydrolysis of a particular type of xylan, because they are able to degrade hardwood glucuronoxylans, cereal arabinoxylans, and even algal β-1,4-β-1,3-xylan (2, 25, 32). Recently, three xylanases belonging to family GH30 have been biochemically characterized in detail. The enzymes, XynA from Erwinia chrysanthemi (47), XynC from Bacillus subtilis (38), and Xyn5B from Bacillus sp. BP-7 (18) are specific for glucuronoxylan, as they hydrolyze this polymer but are not active on arabinose branched xylan. Analysis of the three-dimensional (3D) structure of these enzymes suggests that the accommodation of a xylopyranosyl residue substituted with methyl-glucuronic acid in the −2 subsite of the catalytic cleft is a specificity determinant (40, 43). GH30 xylanases must play an important role in complementing the action of GH10 and GH11 enzymes in the depolymerization of glucuronoxylans in plant biomass.
Glycosyl hydrolases frequently display a modular structure comprising catalytic and ancillary modules, such as carbohydrate-binding modules (CBM). Similar to catalytic modules, CBMs are also classified into families based on amino acid sequence homologies (CAZy). The general function of CBMs is to promote the interaction of the enzymes with their target substrates, thereby potentiating their catalytic efficiency (8, 42). Analysis of binding properties of CBMs of family 35 suggests that these modules can direct the enzymes to regions of the plant that are being actively degraded (30). Many xylanases are modular enzymes containing one or more CBM. These noncatalytic modules found in xylanases belong to different families and recognize a diversity of ligands, including xylans, xylose oligomers, and carbohydrates that are not substrates of the xylanases, such as cellulose, in close proximity to xylan in the plant cell wall (7, 8, 17).
Paenibacillus barcinonensis is a recently identified xylanolytic bacteria (36) which produces a set of xylanases, some of which have been successfully evaluated in paper biotechnology (4, 45). Three of these enzymes, Xyn10A, Xyn10B, and Xyn10C, have been cloned and characterized (5, 19, 44). In this study, we describe the identification and cloning of a fourth xylanase from the strain, Xyn30D. Like the few examples of xylanases of family GH30, the enzyme shows specificity for glucuronoxylans, but unlike GH30 xylanases, which are single-domain enzymes, Xyn30D has a modular structure. This xylanase has a carbohydrate-binding module of the CBM35 family which is rarely found in known xylanases, making this enzyme even more unique. These features of Xyn30D suggest that it degrades glucuronoxylan and must therefore have a special role in the bioconversion of xylan-containing biomass into fermentable products.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Paenibacillus barcinonensis was grown as described previously (3). Supernatants of 2-day-old cultures of the strain in 100 ml of BM medium (0.6% Na2HPO4, 0.3% KH2PO4, 0.05% NaCl, 1 mM MgSO4, 1 mM CaCl2, 0.1% yeast extract, 0.25% urea) containing 0.5% glucose or birchwood xylan were cleared by centrifugation and filtered through a 0.22-μm-pore-size membrane (Whatman). Supernatant proteins were collected by precipitation with 10% (wt/vol) trichloroacetic acid at 4°C overnight and centrifugation at 14,000 × g for 2 h at 4°C. Pellets were washed three times with 70% cold ethanol, dried, and resuspended in 400 μl of urea rehydration buffer (8 M urea, 18 mM Tris-HCl [pH 8.0], 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 0.01% blue bromophenol). Samples were then analyzed by two-dimensional electrophoresis in duplicated gels that were stained for proteins or developed as zymograms for enzyme activity. The proteins that showed xylanase activity were excised from the gels, trypsin digested, and analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) using a Voyager DE-PRO (Applied Biosystems) instrument as described elsewhere (23). A peptide with sequence homology to a xylanase from Aeromonas caviae (31) was identified. Degenerated primers deduced from the sequence of the enzyme allowed for the amplification of a portion of a P. barcinonensis gene encoding a xylanase, Xyn30D, whose complete sequence was obtained by gene walking from the DNA fragment amplified using the Genome Walker universal kit (Clontech).
To overexpress wild-type xylanase, xyn30D was PCR amplified (KapaHiFi, KAPABiosystems) with the oligonucleotide primers FWwt (5′ CACCATGAAGATGGGATTCA 3′) and BWwt (5′ CGCGGAGGCCGTTACTGTAA 3′) and cloned in pET101/D-TOPO (Invitrogen), giving rise to the recombinant plasmid pET101Xyn30D, which produced the full-length enzyme containing a signal peptide and linked to a C-terminal His6 tag (Xyn30D). A similar strategy was used to clone and overexpress the catalytic module of the enzyme. Oligonucleotide primers FWwt and BWcat (5′ GTTAGCAACAAATGTTGTTACA 3′) were used to amplify and clone the catalytic module in the plasmid or pET101Xyn-CM, which produced the catalytic module linked to a C-terminal His6 tag (Xyn-CM). The DNA encoding the carbohydrate-binding module was amplified with the oligonucleotide primers FWcbm (5′ CATATGGCTAGCCTCTCTGGTGGTAACA 3′) and BWcbm (5′ AAGGATCCTTAGGAGGCCGTTACTGTAA 3′), which contained NheI and BamHI restriction sites, respectively. The amplified fragment was cloned into pET28a doubly digested with NheI and BamHI (Roche) to give rise to the recombinant plasmid pET28aXyn-CBM35 that produced the carbohydrate-binding module with an N-terminal His6 tag (Xyn-CBM35).
Amplified DNA (Expand Hi Fidelity; Roche) encoding Xyn30D, Xyn-CM, Xyn30DΔ414, Xyn30DΔ399, Xyn30DΔ372, or Xyn30DΔ344, was directly cloned in pGEM-T Easy vector (Promega) in Escherichia coli DH5α as the host strain. All the amplifications were performed with the same FWpgem primer (5′ GAGGAGGGTAACGTATGAAG 3′), while the BW primers used were BWwtpgem (5′ TTAGGAGGCCGTTACTGTAA 3′), BWcatpgem (5′ GTTAGCAACAAATGTTGTTACA 3′), BW414 (5′ TTAACTCTGTGCTGGAAGCTGAG 3′), BW399 (5′ TTAAATCGGTGCAAGGCTTGCCA 3′), BW372 (5′ TTAAAACCGCTGACTAGCTGCCG 3′), and BW344 (5′ TTAATTGGTATCCGGGTTTTTG 3′). All DNA constructs were verified by sequencing. Sequence homology was analyzed by BLAST.
Expression and purification of recombinant proteins in E. coli.
Xyn30D, Xyn-CM, and Xyn-CBM35 were purified from E. coli BL21Star(DE3) recombinant clones containing plasmids pET101Xyn30D, pET101Xyn-CM, or pET28aXyn-CBM35, respectively. Exponential-phase cultures (optical density at 600 nm [OD600] of 0.6) were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 37°C for 3 h. Cells were disrupted by using a French press. The recombinant His6 tag proteins were purified from cell extracts by immobilized metal affinity chromatography (IMAC) using HisTrap HP columns of 1 ml (GE Healthcare) and eluted in 20 mM phosphate buffer (pH 7.4) with a 0 to 500 mM imidazole gradient, on a fast protein liquid chromatography system (ÄKTA FPLC; GE Healthcare). Buffer exchange and protein concentration were performed in Centricon centrifugal filter units of 3-kDa molecular mass cutoff (Millipore).
Enzyme assays.
Xylanase activity was assayed by measuring the amount of reducing sugar released from xylan from hardwoods or cereals by the Nelson-Somogyi method (37). The standard assay was performed at 50°C in phosphate buffer (pH 6.5) for 15 min as described previously (44). Birchwood, beechwood, and oat spelt xylans and 4-O-methyl-glucuronoxylan were purchased from Sigma Chemical. Rye and wheat arabinoxylans were purchased from Megazyme. One unit of enzymatic activity was defined as the amount of enzyme that releases 1 μmol of reducing sugar equivalent per min under the assay conditions described. A standard curve of xylose was used to calculate activity units. The Briton-Robinson buffer in a pH range between 3.0 and 12.0 was used to study the optimum pH (10). Protein concentration of the samples was determined using the Bradford method (9). All determinations of enzyme activity were made in triplicate.
Gel electrophoresis and zymograms.
For two-dimensional electrophoresis, proteins were first separated by isoelectric focusing (IEF) on immobilized pH gradient (IPG) strips (7 cm, pH 3 to 10; Amersham Biosciences). The strips were previously rehydrated for 6 h at room temperature. Samples of 50 μg of protein in urea rehydration buffer (8 M urea, 18 mM Tris-HCl [pH 8.0], 4% CHAPS, 0.01% blue bromophenol) were loaded onto the strips, and IEF was performed at 30 V for 6.5 h, 500 V for 1 h, 1,000 V for 1 h, and 5,000 V for 7 h in an IPGphor System (Amersham Biosciences). After being run, the strips were equilibrated for 15 min with a buffer containing 50 mM Tris-HCl (pH 8.8), 6 M urea, 30% glycerol, 2% SDS, and 10 mg/ml dithiothreitol (DTT) followed by a 15-min wash with the same buffer but containing 25 mg/ml iodoacetamide. For the second dimension, the strips were loaded onto SDS-PAGE 12% (wt/vol) polyacrylamide gels and were run at 25 V for 10 min and 120 V for 1 h. For zymogram analysis, gels were soaked in 2.5% (wt/vol) Triton X-100 for 30 min, washed in 50 mM phosphate buffer (pH 6.0) for 30 min, overlaid with agarose gels containing 0.2% birchwood xylan, and incubated at 50°C for 30 min. Agarose gels were then stained with 0.1% (wt/vol) Congo red for 15 min and washed with 1 M NaCl until xylanase spots became visible. For protein staining, SDS-PAGE gels were stained with 0.2% (wt/vol) Coomassie brilliant blue R-350.
To analyze the electrophoretic homogeneity of purified proteins, SDS-PAGE was performed in 15% (wt/vol) polyacrylamide gels essentially as described by Laemmli (27). For detection of xylanase activity, zymogram analysis was performed in SDS-PAGE gels containing 0.2% (wt/vol) birchwood xylan as described previously (44).
Affinity gel electrophoresis (AGE) was performed by following the method of Correia et al. (12). Continuous native polyacrylamide gels containing 7.5% acrylamide in 25 mM Tris-250 mM glycine buffer (pH 8.3) were used. Soluble xylan (2 to 4 mg/ml) was included in gels before polymerization. Gels, with and without xylan, were polymerized at the same time and were run in the same gel tank. About 6 μg of target protein was loaded in each well at room temperature, and gels were run at 10 mA/gel for 2 h. Bovine serum albumin (BSA) was used as a negative noninteracting control.
Binding to insoluble polysaccharides.
Binding activity to insoluble polysaccharides was assessed as described by Hogg et al. with some modifications (21). Briefly, 250 μg of purified Xyn30D, Xyn-CM, or Xyn-CBM35 was mixed with 25 mg of Avicel or insoluble oat spelt xylan in a final volume of 500 μl of 50 mM phosphate buffer (pH 6.5) in 1.5-ml microcentrifuge tubes. The samples were incubated for 1 h at 4°C with gentle orbital mixing. Samples were then centrifuged at 18,000 × g for 20 min, and supernatants, containing unbound protein, were carefully removed. Pellets were washed three times with 400 μl of the same buffer, before being resuspended in 400 μl of 10% SDS and boiled for 10 min to release bound protein. Samples were then analyzed by SDS-PAGE on 10 to 15% (wt/vol) polyacrylamide gels.
ITC.
Isothermal titration calorimetry (ITC) was conducted using a VP-ITC (MicroCal) by following the method of Bolam et al. (6). During the titration, Xyn-CBM35 (120 to 180 μM) in 50 mM HEPES buffer (pH 7.0) supplemented or not with 5 mM CaCl2, stirred at 300 rpm in a 1.3586-ml reaction cell maintained at 25°C, was injected with 42 successive aliquots of 7 μl of 3 mM glucuronic acid or polysaccharide at 5 to 25 mg/ml at 200-s intervals. The apo form of Xyn-CBM35, obtained by treatment with Chelex, was titrated against 3 mM CaCl2 to determine whether calcium bound independently to the protein. The results were analyzed by nonlinear regression using a single-site-binding model (Microcal Origin, version 5.0). Thermodynamic parameters were calculated using the standard thermodynamic equation: −RTlnKa = ΔG = ΔH − TΔS, where ΔG, ΔH, and ΔS are the changes in free energy, enthalpy, and entropy of binding, respectively, T is the absolute temperature, and R equals 1.98 cal mol−1 K−1.
Analysis of hydrolysis products from xylan and xylooligosaccharides.
Thin-layer chromatography (TLC) was performed as previously described (19). Neutral xylooligosaccharides and the aldouronic acid mixture standard were purchased from Megazyme. Aldotetraouronic acid 4-O-methyl-α-d-glucuronosyl-(1→2)-β-d-xylopyranosyl-(1→4)-β-d-xylopyranosyl-(1→4)-d-xylose (MeGlcA3Xyl3) and aldopentaouronic acid β-d-xylopyranosyl-(1→4)-[4-O-methyl-α-d-glucuronosyl-(1→2)]-β-d-xylopyranosyl-(1→4)-β-d-xylopyranosyl-(1→4)-d-xylose (MeGlcA3Xyl4) were a gift from Peter Biely. For the analysis of mode of action on glucuronoxylans, Xyn30D (1.7 μM) or Xyn-CM (1.7 μM) was incubated with 1.5% birchwood or beechwood xylan at 50°C in 50 mM phosphate buffer (pH 6.5) for 18 h. The mode of action on branched xylooligosaccharides was analyzed by incubating (1.7 μM) Xyn30D with (7.7 mM) xylooligosaccharides in the same conditions. The reactions were stopped by heating for 15 min at 90°C. All samples and markers were adjusted at pH 6.5 before loading.
For the analysis of xylan hydrolysis products by MALDI-TOF MS, 1 μl of hydrolysates was mixed with 1 μl of matrix solution (10 mg/ml 2,5-dihydroxybenzoic acid dissolved in acetonitrile-water [1:1, vol/vol], 0.1% [wt/vol] trifluoroacetic acid). One microliter of the mixture was spotted onto the MALDI-TOF MS plate and allowed to dry before the analysis. Positive mass spectra were collected with a 4800 Plus MALDI TOF/TOF (ABSciex 2010) spectrometer with an Nd:YAG 200-Hz laser operated at 355 nm.
Bioinformatics tools.
The iCODEHOP version 1.1 tool was used to design PCR primers from protein multiple sequence alignments (http://dbmi-icode-01.dbmi.pitt.edu/i-codehop-context/Welcome). BLAST searches were performed for DNA or protein sequence analysis, including domain classification (1). Putative signal peptide was identified through SignalP 3.0 (16) (http://www.cbs.dtu.dk/services/SignalP/). 3D structure homology models were generated with SwissModel software (http://swissmodel.expasy.org/) based on template gtnA, corresponding to XynC from Bacillus subtilis 168. Pymol software (The PyMOL Molecular Graphics System, version 1.2r3pre; Schrödinger, LLC) was used to visualize the 3D protein models. The ExPASy proteomics server (http://us.expasy.org/tools/protparam.html) was used to analyze the protein physicochemical parameters (ProtParam tool).
Nucleotide sequence accession number.
The DNA sequence of xyn30D gene was submitted to the GenBank database under accession number JQ178343.
RESULTS
Cloning and sequence analysis of Xyn30D.
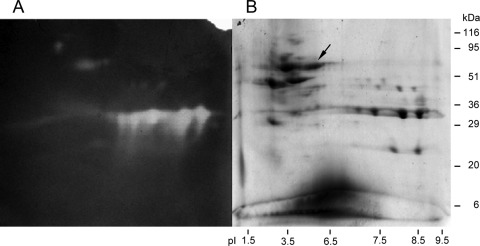
Supernatants of Paenibacillus barcinonensis cultures on BM medium, supplemented or not with 0.5% birchwood xylan, were analyzed in two-dimensional SDS polyacrylamide gels that were also developed for enzyme activity by zymography. Several protein spots were identified that appeared only in the xylan-supplemented cultures (Fig. 1). Those that showed xylanase activity in the zymograms were analyzed by MALDI-TOF MS to determine their amino acid sequence. One of the identified xylanases contained a peptide with sequence homology to a xylanase from Aeromonas caviae (31). Degenerated primers deduced from the sequence of this enzyme allowed amplification of a portion of the gene encoding a new xylanase from P. barcinonensis (Xyn30D), whose complete sequence was obtained by gene walking from the DNA fragment amplified. The open reading frame identified (1,686 bp) encodes a protein of 562 amino acids that shows an N-terminal region of 32 amino acids with the features of a signal peptide. The predicted molecular mass and isoelectric point of the mature protein are 58.5 kDa and 5.9, respectively. Sequence comparison to proteins contained in the NCBI database revealed that the enzyme is a modular xylanase composed of a catalytic module of family 30 of glycosyl hydrolases, GH30, and a carbohydrate-binding module of family 35, CBM35. The modular xylanase was named Xyn30D. It shows high homology to xylanase D from Aeromonas caviae ME-1 (41) and endoxylanase from Aeromonas caviae W-61 (31) (92 and 90% identity, respectively). The catalytic module of P. barcinonensis Xyn30D shows homology to the biochemically characterized GH30 xylanases XynC from Bacillus subtilis (38), Xyn5B from Bacillus sp. BP-7 (18), and XynA from Erwinia chrysanthemi (22, 47) (81, 79, and 41% identity, respectively), while the carbohydrate-binding module of P. barcinonensis Xyn30D shows 41% identity to CBM35 from exo-β-d-glucosaminidase Csxa from Amycolatopsis orientalis (14), 38% identity to rhamnogalacturonan acetyl esterase Rgae12A from Clostridium thermocellum (Che_3141), and 35% identity to CBM35 from Xyn10B from Cellvibrio japonicus (24).
Fig 1.
Secretome analysis of Paenibacillus barcinonensis by two-dimensional electrophoresis. (A) Zymogram of activity on birchwood xylan. (B) Protein-stained two-dimensional SDS polyacrylamide gel. The arrow points to Xyn30D. The positions of molecular mass standards and pI markers are indicated.
Properties of the enzyme.
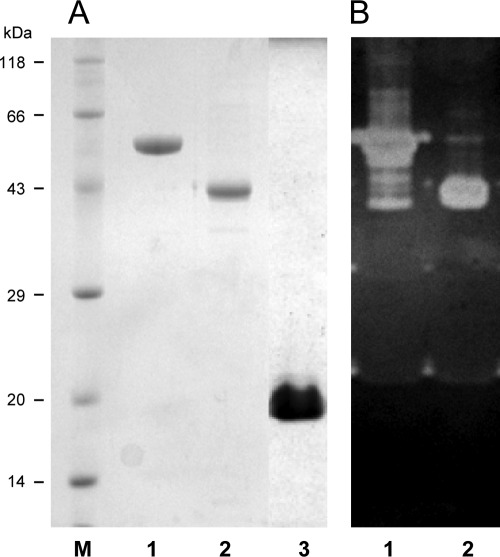
xyn30D was cloned under the control of the high expression T7 promoter from plasmid pET101 in E. coli BL21Star(DE3) to produce a fusion protein of full-length Xyn30D to a C-terminal His tag. A similar construction was performed to clone and overexpress the individual catalytic module GH30 (Xyn-CM). The carbohydrate-binding module CBM35 was cloned in pET28a to produce a fusion protein of this module to an N-terminal His tag (Xyn-CBM35). The three recombinant proteins Xyn30D, Xyn-CM, and Xyn-CBM35 were purified to near homogeneity from cell extracts of the corresponding clones by IMAC (Fig. 2).
Fig 2.
SDS-PAGE analysis of Xyn30D, Xyn-CM, and Xyn-CBM35. (A) Protein staining; (B) zymogram of xylanase activity. Lanes: 1, purified Xyn30D; 2, purified Xyn-CM; 3, purified Xyn-CBM35; M, positions of molecular mass standards.
Purified Xyn30D was analyzed by SDS-PAGE, isoelectric focusing, and zymographic analysis showing an apparent molecular mass of 59 kDa and a pI of 6.0, in accordance with those deduced from the amino acid sequence. Substrate specificity of Xyn30D was determined by evaluating the activity of the enzyme on xylans, other polysaccharides, and aryl-glycosides. The enzyme showed high hydrolytic activity on glucuronic acid branched xylans (glucuronoxylans). Beechwood xylan was the preferred substrate, on which a specific activity of 30.3 xylanase units/mg was found (Table 1). In contrast, the enzyme did not show detectable activity on arabinoxylans from oat spelt, rye, and wheat. Nor did it show activity on any of the other polysaccharides tested, such as crystalline or amorphous celluloses, pNP-β-d-xylopyranoside (pNPX), or other aryl-glycosides, like pNPG and pNPAf. Purified Xyn-CM showed identical substrate specificity to the full-length enzyme.
Table 1.
Substrate specificity of Xyn30D and Xyn-CM
| Substrate | Activity (U/mg)a |
|
|---|---|---|
| Xyn30D | Xyn-CM | |
| Beechwood xylan | 30.3 ± 1.5 | 27.7 ± 2.1 |
| Birchwood xylan | 28.2 ± 2.1 | 27.2 ± 3.2 |
| Oat spelt xylan | ND | ND |
| 4-O-methyl-glucuronoxylan | 19.2 ± 1.8 | 17.5 ± 2.5 |
| Wheat arabinoxylan | ND | ND |
| Rye arabinoxylan | ND | ND |
ND, not detected.
Kinetic parameters of Xyn30D and Xyn-CM on beechwood xylan were determined. The two enzymes showed similar values: Xyn30D showed a Km of 14.72 mg/ml and a kcat value of 1,510 min−1, while Xyn-CM showed a Km of 14.93 mg/ml and a kcat value of 2,024 min−1. The results indicate that the carbohydrate-binding module CBM35 comprised in Xyn30D is not essential for hydrolytic activity of the enzyme under these conditions and on the substrates tested.
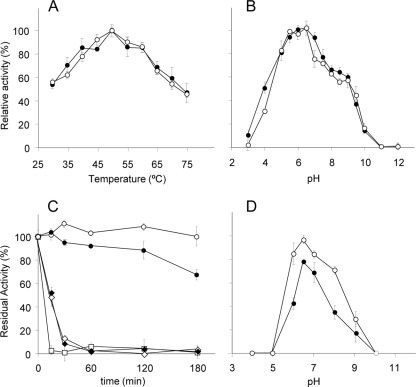
Analysis of the effect of pH and temperature on the hydrolytic activity of Xyn30D and Xyn-CM on beechwood xylan showed similar pH and temperature profiles. Optimal conditions for activity of the two enzymes were 50°C and pH 6.5 (Fig. 3). Thermostability assays showed that Xyn30D remained highly stable up to 50°C after 3 h of incubation at pH 6.5, while Xyn-CM lost 30% of its initial activity after incubation under these conditions. Analysis of the influence of pH on enzyme stability showed that while Xyn30D retained more than 70% of its initial activity after incubation at 50°C for 3 h in buffers ranging from pH 6.0 to 8.0, Xyn-CM was less stable under these conditions, losing more than 50% of its initial activity in buffers at pH lower than 6.0 or higher than 7.0.
Fig 3.
Effect of pH and temperature on activity and stability of Xyn30D and Xyn-CM. (A) Effect of temperature on activity of Xyn30D (○) and Xyn-CM (●); (B) effect of pH on activity of Xyn30D (○) and Xyn-CM (●); (C) effect of temperature on stability of Xyn30D (open symbols) and Xyn-CM (closed symbols). The samples were incubated in 50 mM phosphate buffer (pH 6.5) at 50°C (○, ●), 55°C (♢, ♦), or 60°C (□, ■), and residual activity after different time intervals was determined. (D) Effect of pH on stability of Xyn30D (○) and Xyn-CM (●). The samples were incubated at 50°C in buffers at different pH for 3 h, and residual activity was determined.
Analysis of the 3D structure of xylanases of family GH30 reveals that they all have a (β/α)8 barrel catalytic module fused to a side β-structure of nine strands, denominated side-associated β-domain (28, 39). Computer modeling of the 3D structure of Xyn30D showed a similar structure of a (β/α)8 barrel with an associated β-domain (see Fig. S1 in the supplemental material). The first strand (βs1) was located at the N terminus of the mature enzyme, amino acids 4 to 7, while the strands βs2 to βs9 were located between the end of the (β/α)8 barrel and the beginning of the CBM35 module, covering Gly331 to Asn421. To evaluate the role of the side β-structure in the catalytic activity of the enzyme, a series of derivatives of the recombinant catalytic module, Xyn-CM, lacking an increasing portion of the β-structure, ranging from only the βs9 strand up to the removal of the seven strands βs3 to βs9, were constructed. Similar to the strategy used to overexpress Xyn30D and Xyn-CM, the truncated derivatives were cloned in E. coli BL21Star(DE3) using the expression vector pET101. All the clones showed high expression of the recombinant proteins, which appeared in the insoluble fraction of cell extracts probably as a result of the formation of inclusion bodies. As different approaches to avoid their formation were unsuccessful, the truncated derivatives, as well as Xyn30D and Xyn-CM, were cloned in a different host strain (E. coli DH5α) and vector (pGEM-T Easy). While Xyn30D and Xyn-CM were expressed in an active form, none of the recombinant clones of the truncated derivatives showed hydrolytic activity on xylan, indicating that a full-length side β-structure is required for enzyme activity of Xyn30D (Table 2).
Table 2.
Enzymatic activity of deleted derivatives of Xyn30D
| Construction | Amino acids | Activitya |
|---|---|---|
| Xyn30D | 1–562 | + |
| Xyn-CM | 1–421 | + |
| Xyn30Δ414 | 1–414 | − |
| Xyn30Δ399 | 1–399 | − |
| Xyn30Δ372 | 1–372 | − |
| Xyn30Δ344 | 1–344 | − |
+, active; −, not active.
Mode of action of Xyn30D.
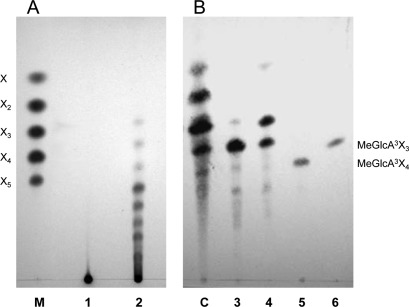
Hydrolysis products released by the enzyme from glucuronoxylans were analyzed by thin-layer chromatography. Birchwood and beechwood xylans were degraded to a mixture of products of intermediate mobility between linear xylooligosaccharides, indicating that they are methyl-glucuronic acid-substituted xylooligomers (aldouronic acids) (Fig. 4). To characterize these products better, the hydrolysate of beechwood xylan was also analyzed by MALDI-TOF MS. The mass spectrum showed the presence of molecular ions of substituted xylooligomers identified as sodium and potassium adducts (see Fig. S2 in the supplemental material and Table 3). The major ions corresponded to substituted xylooligosaccharides consisting of 2 to 8 xylopyranosyl residues and a single MeGlcA residue, indicating that they are aldouronic acids, in accordance with the results of the TLC analysis. Xyn-CM gave a similar pattern of hydrolysis products (see Fig. S2).
Fig 4.
Thin-layer chromatography analysis of the mode of action of Xyn30D. (A) Hydrolysis products from beechwood xylan. Lanes: 1, control, no digested samples; 2, products of xylan hydrolysis by Xyn30D; M, size markers of xylose (X), xylobiose (X2), xylotriose (X3), xylotetraose (X4), and xylopentaose (X5). (B) Hydrolysis products from glucuronic acid-substituted xylooligomers. Lanes: 3, control, no digested aldotetraouronic acid (MeGlcA3Xyl3); 4, products of aldotetraouronic acid hydrolysis by Xyn30D; 5, control, no digested aldopentaouronic acid (MeGlcA3Xyl4); 6, products of aldopentaouronic acid hydrolysis by Xyn30D; C, size markers of a mixture containing aldotetraouronic, aldotriouronic, and aldobiouronic acids and xylose.
Table 3.
Oligosaccharides identified by MALDI-TOF MS after hydrolysis of beechwood xylan by Xyn30D
| Xylooligomer | Mra |
m/z |
||
|---|---|---|---|---|
| Na | K | Na + K | ||
| MeGlcAX2 | 472 | 511.1 | 533.0 | |
| MeGlcAX3 | 604 | 627.1 | 643.1 | 665.1 |
| MeGlcAX4 | 736 | 759.2 | 775.2 | 797.1 |
| MeGlcAX5 | 868 | 891.2 | 907.2 | 929.2 |
| MeGlcAX6 | 1,000 | 1,023.3 | 1,039.2 | 1,061 |
| MeGlcAX7 | 1,132 | 1,155.3 | 1,171.2 | |
| MeGlcAX8 | 1,264 | 1,287.3 | 1,303.3 | |
Mr, molecular weight.
Purified Xyn30D hydrolyzed aldotetraouronic acid (MeGlcA3Xyl3) to xylose and aldotriouronic acid (Fig. 4), while aldopentaouronic (MeGlcA3Xyl4) acid was shortened to aldotetraouronic acid by the enzyme. This is in accordance with the proposed mode of action of GH30 xylanases on glucuronic acid-substituted substrates (38, 47). In contrast to the activity found on substituted oligomers, Xyn30D did not show activity on linear xylooligomers.
Binding to polysaccharides.
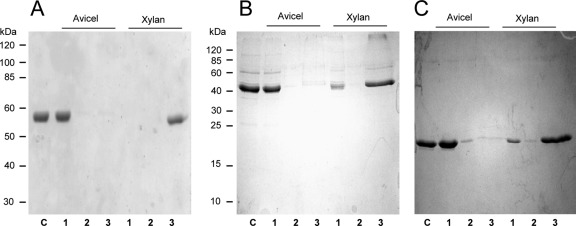
Binding of Xyn30D to insoluble polysaccharides was tested. Avicel and insoluble oat spelt xylan were mixed with Xyn30D, and after 1 h of incubation the fractions bound or unbound to the insoluble polymers were separated by centrifugation and analyzed by SDS-PAGE. Xyn30D bound to insoluble oat spelt xylan, since almost all the enzyme was found adsorbed on the polymer, while none was found in the supernatant and wash fractions (Fig. 5). In contrast, the enzyme did not bind to Avicel, as the protein remained in the supernatants and only a very small amount was detected adsorbed on Avicel. The binding properties of Xyn-CM and Xyn-CBM35 to Avicel and insoluble xylan were also analyzed. As in the case of Xyn30D, purified Xyn-CM and Xyn-CBM35 bound to insoluble oat spelt xylan, although the binding was less efficient. A large amount of Xyn-CM and Xyn-CBM35 adsorbed to the polymer, while a smaller amount remained in the nonadsorbed fraction (Fig. 5). In contrast, Xyn-CM and Xyn-CBM35 did not bind to Avicel.
Fig 5.
SDS-PAGE analysis of binding to insoluble polysaccharides. (A) Binding of Xyn30D. (B) Binding of Xyn-CM. (C) Binding of Xyn-CBM35. Proteins were mixed with Avicel or with the insoluble fraction of oat spelt xylan for 1 h; bound and unbound fractions were separated by centrifugation and analyzed by SDS-PAGE. Lanes: 1, unbound fraction; 2, wash; 3, fraction adsorbed to the polymer; C, control protein. The positions of molecular mass standard proteins are indicated.
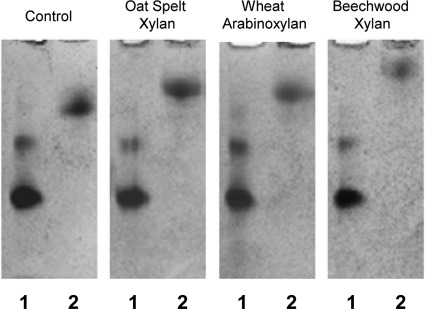
To evaluate the adsorption capacity of the enzyme to soluble polysaccharides, the binding of the purified carbohydrate-binding module, Xyn-CBM35, to soluble fractions of wheat arabinoxylan, oat spelt, and beechwood xylans was analyzed using affinity gel electrophoresis. Migration of Xyn-CBM35 in gels containing soluble xylans was retarded compared with the migration of the protein in gels without xylan (Fig. 6). Beechwood xylan produced the higher retardation of Xyn-CBM35 in the gels, while oat spelt xylan and wheat arabinoxylan retarded migration in similar ways.
Fig 6.
Affinity gel electrophoresis analysis of Xyn-CBM35 binding to soluble polysaccharides. Xyn-CBM35 was analyzed on nondenaturing polyacrylamide gels containing no ligand (control) or soluble xylan from oat spelt, beechwood, or wheat arabinoxylan. Lanes: 1, BSA; 2, Xyn-CBM35.
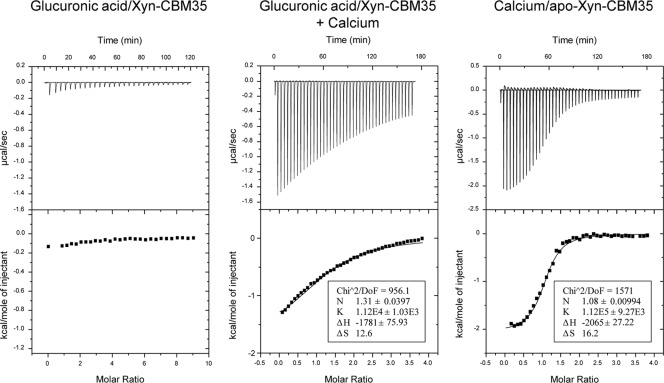
To analyze the binding specificity of Xyn-CBM35 in more detail, isothermal titration calorimetry (ITC) was used to quantify the binding of the protein to soluble xylans and to glucuronic acid, present in variable quantities in glucuronoxylans and arabinoxylans (33). Titration of Xyn-CBM35 with glucuronic acid is shown in Fig. 7. Xyn-CBM35 bound to glucuronic acid with a stoichiometry of 1:1 and an association constant (KA) of 1.12 × 104 M−1. No binding was detected in the absence of calcium added to buffer, indicating that divalent ions were required for binding. To investigate the role of the calcium in ligand binding in more detail, an apo form of Xyn-CBM35 was produced by treating the protein with Chelex. ITC analysis showed that the apo form of Xyn-CBM35 bound to Ca+2 with a stoichiometry of 1:1 and a KA of 1.12 × 105 M−1 (Fig. 7). Binding of Xyn-CBM35 to soluble beech and oat spelt xylans was also analyzed by ITC, but it was not strong enough to be quantified by this technique.
Fig 7.
ITC analysis of Xyn-CBM35 binding to glucuronic acid. The upper part of each panel shows the raw binding heats, and the lower part shows the integrated binding heats minus the dilution control heats fitted to a single-site binding model with MicroCal Origin. Calcium was at 5 mM when included in titrations with glucuronic acid and was at 3 mM (in the syringe) when titrated into apo-Xyn-CBM35.
DISCUSSION
Xylanases are glycosyl hydrolases usually grouped in families GH10 and GH11. However, a few xylanases that exhibit homology to enzymes belonging to other glycoside hydrolase families have also been described (11, 32). Among them, three xylanases of family 30 (GH30), previously classified in family 5, have been biochemically characterized. The enzymes are XynA from Erwinia chrysanthemi (22, 47), XynC from Bacillus subtilis (38), and Xyn5B from Bacillus sp. BP-7 (18). These xylanases are single-domain enzymes whose distinctive trait is their specificity for glucuronoxylans; they show high activity on glucuronoxylans but none on arabinoxylans. These enzymes have an absolute requirement of methyl-glucuronosyl (MeGlcA) side residues for hydrolysis of xylan, releasing as products xylooligosaccharides substituted in the penultimate xylose from the reducing end (38, 47). The crystalline structure of XynA from Erwinia chrysanthemi (43) and XynC from Bacillus subtilis (40) reveals that the accommodation of MeGlcA in the −2 subsite of the catalytic site is a determinant for substrate recognition of these enzymes. GH30 xylanases are targeted to glucuronic acid branched regions of xylans, unlike the majority of xylanases belonging to families GH10 or GH11, which are not active on densely substituted xylan (32). Recently, a GH30 enzyme active on both glucuronoxylans and arabinoxylans from the fungus Bispora sp. MEY-1 has been identified as a xylanase (29). However, the Bispora enzyme has been classified in subgroup G of the GH30 family, while the glucurunoxylan-specific xylanases are classified in subgroup H (39). An arabinoxylan-specific xylanase belonging to the GH5 family has recently been characterized (13). The accommodation of arabinose in subsite −1 of its catalytic groove seems to be a specificity determinant of this new xylanase.
The enzyme we characterized, Paenibacillus barcinonensis Xyn30D, is a glucuronoxylan-specific xylanase. Like xylanases of family GH30, it is highly active on glucuronic acid-decorated xylans and it does not hydrolyze arabinoxylans. Comparison of kinetic constants of Xyn30D and those reported for GH30 xylanases, such as XynC from Bacillus subtilis (38), shows that while the kcat values are in the same range, the Km of Xyn30D is higher than that of the Bacillus subtilis enzyme (14.72 mg/ml versus 1.63 mg/ml). Paenibacillus barcinonensis Xyn30D is a modular enzyme composed of a catalytic module of family 30 (GH30) and a carbohydrate-binding module of family 35 (CBM35). To our knowledge, this is the only example of modular GH30 xylanase characterized to date. Xyn30D shows high sequence identity to xylanase D from Aeromonas caviae ME-1 (41) and endoxylanase from Aeromonas caviae W-61 (31), although the biochemical properties of these enzymes have not been studied in detail. Xylanases belonging to family GH30 share a unique overall structure of the catalytic module comprising a common (β/α)8 barrel linked to a side β-structure consisting of a 9-stranded aligned β-sandwich. This trait has led to the recent reclassification of these xylanases, from family GH5 to family GH30 (39). It has been suggested that the side β-structure has a substrate-targeting role, as a putative carbohydrate-binding module (40). Xyn30D from Paenibacillus barcinonensis shares with GH30 xylanases the overall structure of the catalytic module comprising a (β/α)8 barrel linked to the side β-structure. The role of this side β-structure in the activity of Xyn30D was tested in a series of truncated derivatives of the enzyme that were constructed. None of them retained catalytic activity and was able to hydrolyze glucuronoxylans. The fact that even the derivative with the shortest deletion (which only removed the βs9 strand at the C terminus of the catalytic module) did not show enzymatic activity indicates the essential role of the side β-structure in the catalysis of Xyn30D, as has been previously proposed (39). This is the first reported evidence of the importance of this β-structure associated to the (β/α)8 barrel for the catalytic activity of xylanases of family GH30.
The carbohydrate-binding module of Xyn30D belongs to the family CBM35. This is uncommon among xylanases and makes the characterized enzyme more unusual. Only one other xylanase, Xyn10B from Cellvibrio japonicus, has previously been reported to contain a CBM of family 35 (24). ITC analysis showed that the CBM35 from Paenibacillus barcinonensis Xyn30D (Xyn-CBM35) binds glucuronic acid and requires calcium ions for binding. The results are in agreement with the findings for CBM35s of Xyn10B from Cellvibrio japonicus and exo-β-d-glucosaminidase Csxa from Amycolatopsis orientalis but differ from results found with rhamnogalacturonan acetyl esterase Rgae12A from Clostridium thermocellum, which showed only a weak interaction with glucuronic acid (30). Sequence comparison of CBM35s has identified several highly conserved amino acids, among them an asparagine residue which seems to play a key role in ligand orientation (6, 12). The equivalent Asn is found in position 119 of Xyn-CBM35, while most of the conserved amino acids are also found in the CBM. On the basis of protein sequence relatedness, family CBM35s have been grouped into four main clusters which are reported to differ in ligand specificity. Some of the CBM35s, like that of Paenibacillus barcinonensis Xyn30D, bind glucuronic acid decorations, while members of the other clusters bind mannan and galactomannan (12). More CBMs of this family need to be characterized to understand their role in polysaccharide hydrolysis, including their potential role in targeting of the enzymes to areas of the cell wall under biological degradation (30).
ITC was also used to study the binding ability of Xyn-CBM35 to polymeric xylan, but the binding was too weak to be evaluated by this method. Analysis by affinity gel electrophoresis showed that Xyn-CBM35 bound glucuronoxylans and arabinoxylans. Binding of a CBM35 to xylan has previously been reported for the arabinofuranosidase Abf62A from Cellvibrio japonicus (6). The results found show that Xyn-CBM35 binds both xylans and also glucuronic acid. It seems that the CBM35 of Xyn30D targets the enzyme to its substrate, probably by the recognition of glucuronic acid decorations. In fact, the recognition of decorations of hemicelluloses has been proposed as a mechanism of substrate targeting of CBM35 (12). These decorations are also present in arabinoxylans, although steric hindrance of arabinose side chains probably precludes arabinoxylans from catalytic breakage by Xyn30D. The CBM35 of Xyn30D does not seem to be required for hydrolysis of xylan, as the isolated catalytic module shows similar activity to the full-length enzyme on the substrates tested. This indicates that the catalytic and the CBM modules of Xyn30D can function independently, as has frequently been reported for glycosyl hydrolases (21, 35).
Paenibacillus barcinonensis is a powerful xylanolytic microorganism, from which several xylanases of family GH10 have previously been characterized (5, 19, 44). The enzyme identified and characterized in our study, Xyn30D, is a GH30 xylanase with a modular structure comprising a CBM35, which clearly differentiates the enzyme from known xylanases. Xyn30D probably plays a specific role in xylan biodegradation. Its marked preference for glucuronoxylan suggests that it has a distinctive role in the depolymerization of xylan in natural habitats. The biochemical characterization of new GH30 xylanases will be required to compare with the few known examples of these enzymes, in order to understand their role in xylan hydrolysis and their contribution to biomass degradation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Peter Biely for kindly providing aldotetraouronic and aldopentaouronic acids and for scientific advice. We thank the Proteomics Platform of Barcelona Science Park, member of ProteoRed-ISCIII network, for the proteomics study.
The study was supported in part by the Spanish Ministry of Education and Science, grant CTQ2010-20238-C03-02, and AGAUR from Generalitat de Catalunya, grant 2009 SGR 819. Susana Valenzuela was the recipient of an MAE AECI doctorate grant from the Spanish Ministry of Foreign Affairs.
Footnotes
Published ahead of print 23 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Biely P, Vršanska M, Tenkanen M, Kluepfel D. 1997. Endo-β-1,4-xylanase families: differences in catalytic properties. J. Biotechnol. 57:151–166 [DOI] [PubMed] [Google Scholar]
- 3. Blanco A, Pastor FIJ. 1993. Characterization of cellulase-free xylanases from the newly isolated Bacillus sp. strain BP-23. Can. J. Microbiol. 39:1162–1166 [Google Scholar]
- 4. Blanco A, Vidal T, Colom JF, Pastor FIJ. 1995. Purification and properties of xylanase A from alkali-tolerant Bacillus sp. strain BP-23. Appl. Environ. Microbiol. 61:4468–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blanco A, Diaz P, Zueco J, Parascandola P, Pastor FIJ. 1999. A multidomain xylanase from a Bacillus sp. with a region homologous to thermostabilizing domains of thermophilic enzymes. Microbiology 145:2163–2170 [DOI] [PubMed] [Google Scholar]
- 6. Bolam DN, et al. 2004. X4 modules represent a new family of carbohydrate-binding modules that display novel properties. J. Biol. Chem. 279:22953–22963 [DOI] [PubMed] [Google Scholar]
- 7. Boraston AB, et al. 2001. Binding specificity and thermodynamics of a family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A. Biochemistry 40:6240–6247 [DOI] [PubMed] [Google Scholar]
- 8. Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 10. Britton HTS. 1952. Hydrogen ions, 4th ed Chapman and Hall, London, United Kingdom. [Google Scholar]
- 11. Collins T, Gerday C, Feller G. 2005. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 29:3–23 [DOI] [PubMed] [Google Scholar]
- 12. Correia MAS, et al. 2010. Signature active site architectures illuminate the molecular basis for ligand specificity in family 35 carbohydrate binding module. Biochemistry 49:6193–6205 [DOI] [PubMed] [Google Scholar]
- 13. Correia MAS, et al. 2011. Structure and function of an arabinoxylan-specific xylanase. J. Biol. Chem. 286:22510–22520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Côté N, et al. 2006. Two exo-β-d-glucosaminidases/exochitosanases from actinomycetes define a new subfamily within family 2 of glycoside hydrolases. Biochem. J. 394:675–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Vries RP, Visser J. 2001. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 65:497–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emanuelsson O, Brunak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP, and related tools. Nat. Protoc. 2:953–971 [DOI] [PubMed] [Google Scholar]
- 17. Fujimoto Z, et al. 2002. Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module. J. Mol. Biol. 316:65–78 [DOI] [PubMed] [Google Scholar]
- 18. Gallardo O, et al. 2010. Characterization of a family GH5 xylanase with activity on neutral oligosaccharides and evaluation as a pulp bleaching aid. Appl. Environ. Microbiol. 76:6290–6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gallardo O, et al. 2010. Structural insights into the specificity of Xyn10B from Paenibacillus barcinonensis and its improved stability by forced protein evolution. J. Biol. Chem. 285:2721–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Henrissat B, Davies G. 1997. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7:637–644 [DOI] [PubMed] [Google Scholar]
- 21. Hogg D, et al. 2003. The modular architecture of Cellvibrio japonicus mannanases in glycoside hydrolase families 5 and 26 points to differences in their role in mannan degradation. Biochem. J. 371:1027–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hurlbert JC, Preston JF. 2001. Functional characterization of a novel xylanase from a corn strain of Erwinia chrysanthemi. J. Bacteriol. 183:2093–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Irar S, Oliveira E, Pagès M, Goday A. 2006. Towards the identification of late-embryogenic abundant phosphoproteome in Arabidopsis by 2-DE and MS. Proteomics 6:S175–S185 [DOI] [PubMed] [Google Scholar]
- 24. Kellett LE, et al. 1990. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem. J. 272:369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kolenová K, Vršanská M, Biely P. 2006. Mode of action of endo-β-1,4-xylanases of families 10 and 11 on acidic xylooligosaccharides. J. Biotechnol. 121:338–345 [DOI] [PubMed] [Google Scholar]
- 26. Kulkarni N, Shendye A, Rao M. 1999. Molecular and biotechnological aspects of xylanases. FEMS Microbiol. Rev. 23:411–456 [DOI] [PubMed] [Google Scholar]
- 27. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 28. Larson SB, Day J, Barba dela Rosa AP, Keen NT, McPherson A. 2003. First crystallographic structure of a xylanase from glycoside hydrolase family 5: implications for catalysis. Biochemistry 42:8411–8422 [DOI] [PubMed] [Google Scholar]
- 29. Luo H, et al. 2010. Molecular cloning and characterization of the novel acidic xylanase XYLD from Bispora sp. MEY-1 that is homologous to family 30 glycosyl hydrolases. Appl. Microbiol. Biotechnol. 86:1829–1839 [DOI] [PubMed] [Google Scholar]
- 30. Montanier C, et al. 2009. Evidence that family 35 carbohydrate binding modules display conserved specificity but divergent function. Proc. Natl. Acad. Sci. U. S. A. 106:3065–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Okai N, et al. 1998. Molecular properties and activity of a carboxyl-terminal truncated form of xylanase 3 from Aeromonas caviae W-61. Biosci. Biotechnol. Biochem. 62:1560–1567 [DOI] [PubMed] [Google Scholar]
- 32. Pollet A, Delcour JA, Courtin CM. 2010. Structural determinants of the substrate specificities of xylanases from different glycoside hydrolase families. Crit. Rev. Biotechnol. 30:176–191 [DOI] [PubMed] [Google Scholar]
- 33. Puls J, Schuseil J. 1993. Enzymological aspects of microbial hemicellulases with emphasis on fungal systems, p 1–27. In Coughlan MP, Hazlewood GP. (ed), Hemicelluloses and hemicellulases. Portland Press, London, United Kingdom [Google Scholar]
- 34. Ragauskas AJ, et al. 2006. The path forward for biofuels and biomaterials. Science 311:484–489 [DOI] [PubMed] [Google Scholar]
- 35. Sakka M, Higashi Y, Kimura T, Ratanakhanokchai K, Sakka K. 2011. Characterization of Paenibacillus curdlanolyticus B-6 Xyn10D, a xylanase that contains a family 3 carbohydrate-binding module. Appl. Environ. Microbiol. 77:4260–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sánchez MM, et al. 2005. Paenibacillus barcinonensis sp. nov., a xylanase-producing bacterium isolated from a rice field in the Ebro River delta. Int. J. Syst. Evol. Microbiol. 55:935–939 [DOI] [PubMed] [Google Scholar]
- 37. Spiro RG. 1966. The Nelson-Somogyi copper reduction method. Analysis of sugars found in glycoprotein. Methods Enzymol. 8:3–26 [Google Scholar]
- 38. St. John FJ, Rice JD, Preston JF. 2006. Characterization of XynC from Bacillus subtilis subsp. subtilis strain 168 and analysis of its role in depolymerization of glucuronoxylan. J. Bacteriol. 188:8617–8626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. St. John FJ, González JM, Pozharski E. 2010. Consolidation of glycosyl hydrolase family 30: a dual domain 4/7 hydrolase family consisting of two structurally distinct groups. FEBS Lett. 584:4435–4441 [DOI] [PubMed] [Google Scholar]
- 40. St. John FJ, Hurlbert JC, Rice JD, Preston JF, Pozharski E. 2011. Ligand bound structures of a glycosyl hydrolase family 30 glucuronoxylan xylanohydrolase. J. Mol. Biol. 407:92–109 [DOI] [PubMed] [Google Scholar]
- 41. Suzuki T, Ibata K, Hatsu M, Takamizawa K, Kawai K. 1997. Cloning and expression of a 58-kDa xylanase VI gene (xynD) of Aeromonas caviae ME-1 in Escherichia coli which is not categorized as a family F or family G xylanase. J. Ferment. Bioeng. 84:86–89 [Google Scholar]
- 42. Tomme P, et al. 1998. Characterization and affinity applications of cellulose-binding domains. J. Chromatogr. B. 715:283–296 [DOI] [PubMed] [Google Scholar]
- 43. Urbániková L, Vršanská M, Mørkeberg Krogh KBR, Hoff T, Biely P. 2011. Structural basis for substrate recognition by Erwinia chrysanthemi GH30 glucuronoxylanase. FEBS J. 278:2105–2116 [DOI] [PubMed] [Google Scholar]
- 44. Valenzuela SV, Diaz P, Pastor FIJ. 2010. Recombinant expression of an alkali stable GH10 xylanase from Paenibacillus barcinonensis. J. Agric. Food Chem. 58:4814–4818 [DOI] [PubMed] [Google Scholar]
- 45. Valls C, et al. 2010. Obtaining low-HexA-content cellulose from eucalypt fibres: which glycosyl hydrolase family is more efficient? Carbohydr. Polym. 80:154–160 [Google Scholar]
- 46. Van Vleet JH, Jeffries TW. 2009. Yeast metabolic engineering for hemicellulosic ethanol production. Curr. Opin. Biotech. 20:300–306 [DOI] [PubMed] [Google Scholar]
- 47. Vršanská M, Kolenová K, Puchart V, Biely P. 2007. Mode of action of glycoside hydrolase family 5 glucuronoxylan xylanohydrolase from Erwinia chrysanthemi. FEBS J. 274:1666–1677 [DOI] [PubMed] [Google Scholar]
- 48. Wang J, Sun B, Cao Y, Tian Y, Wang C. 2009. Enzymatic preparation of wheat bran xylooligosaccharides and their stability during pasteurization and autoclave sterilization at low pH. Carbohydr. Polym. 77:816–821 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.