Abstract
Phakopsora pachyrhizi, the causal agent of Asian soybean rust (ASR), continues to spread across the southeast and midsouth regions of the United States, necessitating the use of fungicides by producers. Our objective in this research was to identify ASR proteins expressed early during infection for the development of immunodiagnostic assays. We have identified and partially characterized a small gene family encoding extracellular proteins in the P. pachyrhizi urediniospore wall, termed PHEPs (for Phakopsora extracellular protein). Two highly expressed protein family members, PHEP 107 and PHEP 369, were selected as ideal immunodiagnostic targets for antibody development, after we detected PHEPs in plants as early as 3 days postinfection (dpi). Monoclonal antibodies (MAbs; 2E8E5-1 and 3G6H7-3) generated against recombinant PHEP 369 were tested for sensitivity against the recombinant protein and extracts from ASR-infected plants and for specificity against a set of common soybean pathogens. These antibodies should prove applicable in immunodiagnostic assays to detect infected soybeans and to identify ASR spores from sentinel surveillance plots.
INTRODUCTION
Phakopsora pachyrhizi, the causal agent of Asian soybean rust (ASR), continues to spread across the southeast and midsouth regions of the United States following introduction in 2004 (25). Due to a lack of resistance to the pathogen in commercial cultivars of soybean, producers must rely on fungicides to combat the disease. In order to make timely, cost-effective decisions regarding the applications of fungicides throughout the growing season, growers, extension agents, and crop consultants need inexpensive, presymptomatic diagnostic decision tools. Our objectives in this research were to identify ASR extracellular proteins expressed early during infection and apply them to the development of P. pachyrhizi-specific immunological reagents for sensitive diagnostic assays.
We recently applied a proteomic approach to identify proteins expressed during germination of P. pachyrhizi urediniospores using two-dimensional gel electrophoresis (2DE) and matrix-assisted laser desorption ionization with tandem time of flight (MALDI-TOF/TOF) mass spectrometry of tryptic peptides (17). In that study, we identified a set of extracellular soluble proteins present in urediniospores, prior to and during germination. The peptides were used to identify corresponding cDNA clones from an expressed sequence tag (EST) sequence database derived from germinating P. pachyrhizi urediniospores (22). The clones were found to be members of a gene family which we have named PHEP (for Phakopsora extracellular protein), containing at least six members. Two highly expressed proteins, PHEP 107 and PHEP 369, were selected for further study as possible targets for immunodiagnostic assays.
We describe here the characterization of the PHEP protein family and development of immunospecific polyclonal (PAb) and monoclonal (MAb) antibodies raised against recombinant PHEP family members with potential as diagnostic reagents for early detection of P. pachyrhizi.
MATERIALS AND METHODS
Rust spores.
Urediniospores of Phakopsora pachyrhizi or Phakopsora meibomiae (Table 1) from the international collection maintained at the Foreign Disease-Weed Science Research Unit were used in this study. Approximately 0.6 g of each individual spore isolate was germinated under sterile conditions for 18 h in a Pyrex dish containing 600 ml of water supplemented with 50 μg/ml of ampicillin and 50 μg/ml of streptomycin.
Table 1.
Phakopsora isolates used in this study
| Species | Isolate | Country (state) or territory/yr | GenBank accession no.a |
|---|---|---|---|
| Phakopsora pachyrhizi | TW 72-1 | Taiwan/1972 | JQ284010 |
| Phakopsora pachyrhizi | TH 02-1 | Thailand/2002 | JQ284011 |
| Phakopsora pachyrhizi | IN 73-1 | India/1973 | JQ284012 |
| Phakopsora pachyrhizi | SA 01-1 | South Africa/2001 | JQ284013 |
| Phakopsora pachyrhizi | PG 01-3 | Paraguay/2001 | JQ284014 |
| Phakopsora pachyrhizi | AL 04-1 | U.S. (AL)/2004 | JQ284015 |
| Phakopsora meibomiae | PR | Puerto Rico/1976 |
GenBank accession numbers for PHEP 369 genomic sequences.
Protein extraction.
For Western blots, soluble protein fractions from fungal spores were extracted as follows: 0.6-g samples were ground in liquid nitrogen, suspended in phosphate-buffered saline (PBS) (24) supplemented with 3 mM dithiothreitol (DTT) and 0.2% sodium dodecyl sulfate (SDS), heated at 95°C for 5 min, and centrifuged at 14,000 × g for 5 min, and supernatant was collected. Aliquots were stored at −20°C. Protein quantification was performed as described by Markwell et al. (19) using bovine serum albumin (BSA) as a standard.
Soybean pathogens.
Extracts from soybean pathogens and infected plants for indirect enzyme-linked immunosorbent assay (ELISA) were generated as described by Baysal-Gurel et al. (3).
Plant material.
Soybean (Glycine max cv. Williams 82) and Fordhook bush lima bean (Phaseolus lunatus) were grown in potting soil under containment greenhouse conditions (18 to 30°C, 12-h day length, supplemented when needed with lighting from 800-W metal halide lamps at 24 inches). At the 2- to 3-trifoliolate-leaf stage, soybeans and lima beans were spray inoculated with 0.375 mg/ml of P. pachyrhizi isolate TW72-1 and P. meibomiae isolate Puerto Rico, respectively. After inoculation, plants were placed in a dew chamber at 21°C overnight and returned to the greenhouse. One-square-inch infected leaf samples were collected at 0 (preinoculation), 3, 5, 7, and 14 days postinfection (dpi), flash frozen in liquid nitrogen, and stored at −80C. Leaf sample soluble proteins were extracted as described above.
PHEP genomic sequences.
Genomic DNA was extracted from the P. pachyrhizi isolates listed in Table 1, using the DNeasy plant minikit (Qiagen Inc., Valencia, CA) according to the manufacturer's protocol. Full-length PHEP 369 genes were generated by PCR using the primer set 5′-ATG GGA AAA GTT ATC ATC AAT GTG-3′ and 5′-CTT TCC AGC CTT TGC TTT TTC ATC-3′. Fragments were directly sequenced using the BigDye Terminator version 3.1 cycle sequencing kit (ABI, Foster, CA) according to the manufacturer's protocols.
Production of recombinant PHEP (rPHEP) proteins and generation of antibodies against rPHEP 107 and rPHEP 369.
Recombinant proteins were produced under contract (National Cancer Institute—Frederick, Protein Expression Laboratory, Ft. Detrick, MD). Open reading frames (ORFs) for PHEP 107 and PHEP 369 were generated by reverse transcriptase PCR (RT-PCR) with RNA isolated from germinating spores of P. pachyrhizi isolate TW72-1 using the primer sets 5′-ATG GGA AAA GTT ATC ATC AAT GTG-3′ and 5′-CTT TCC AGC CTT TGC TTT TTC ATC-3′ for PHEP 107 and 5′-ATG GGA AAA GTT ATC ATC AAT GTG-3′ and 5′-CTT TCC AGC CTT TGC TTT TTC ATC-3′ for PHEP 369. Both ORFs were cloned into pDest-521 expression vector containing an amino-terminal His-6 tag (Invitrogen, Carlsbad, CA) and overexpressed in Escherichia coli BL21(DE3) cells. Proteins were purified by immobilized metal affinity chromatography using HisTrap Phast columns (GE Healthcare, Piscataway, NJ) according to the manufacturer's protocols. Purified recombinant proteins were used to generate polyclonal antibodies (PAb) in New Zealand rabbits using a commercial provider (Bushover Biologicals, Vassalboro, ME). After the 4th bleed, titer was judged to be appropriate for experiments and production bleeds. Immunoglobulin G (IgG) fractions were purified on immobilized protein A using the manufacturer's protocols, diluted to 1 mg/ml, and stored at −80°C. Recombinant protein PHEP 369 was also used to generate monoclonal antibodies (MAbs) in mouse using a commercial provider (GenScript, Piscataway, NJ) applying standard protocols for murine hybridoma generation, selection, and screening (14). Cell line culture medium samples were screened with rPHEP 369, and IgG fractions were purified by the manufacturer from 1 liter of cell culture medium and stored at −80°C at a dilution of 3 mg/ml. ELISA was used to titer MAbs against rPHEP protein to arrive at working dilutions of 1:5,000 for Western blot analyses.
Western blotting.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 4 to 12% bis-Tris gels (Invitrogen) and transferred to 0.2-μM-pore-size nitrocellulose membranes using a semidry blotter apparatus (Owl Separation Systems, Woburn, MA) according to the manufacturer's guidelines. After transfer, blots were blocked in 3% (wt/vol) dry milk in PBS-0.02% (vol/vol) Tween 20 (PBS-Tw) for 1 h and probed with anti-rPHEP PAbs at 1:5,000 and anti-rPHEP 369 MAbs at 1:5,000 overnight at 4°C (29). Blots were washed 3 times for 5 min in 100 ml PBS-Tw. Blots were then probed with horseradish peroxidase (HRP)-conjugated goat anti-mouse or anti-rabbit IgG (Sigma Chem Co., St. Louis, MO) at 1:10,000 for 1 h, washed 3 times in 100 ml PBS-Tw, and detected using Super Signal West Pico chemiluminescent substrate (Thermo Scientific, Rockford, IL) according to the manufacturer's protocol.
ELISA.
Samples for indirect ELISA were prepared as described by Baysal-Gurel et al. (3), diluted 1:1 (vol/vol) in carbonate coating buffer, and used at 40% (wt/vol) for plant and 1% (wt/vol) for plant pathogens. One hundred microliters of each sample was added to a polyvinyl chloride plate and incubated overnight at 4°C. Plates were washed 2 times in 200 μl PBS, blocked in 100 μl of 3% dry milk in PBS at room temperature for 1 h, and washed 3 times in 200 μl PBS. One hundred microliters of primary antibody at a 1:5,000 dilution was added per well and incubated at room temperature for 2 h. Plates were washed 3 times in PBS, and alkaline phosphatase (AP)-conjugated goat anti-mouse antibody was added at a concentration of 1:2,000 and incubated at room temperature for 2 h. Plates were then washed 3 times in PBS, 100 μl of AP substrate was added, and absorbance was read at 405 nm in a microtiter plate reader at 15, 30, and 60 min. The results reported in Table 2 are from the 30-min reading.
Table 2.
MAb PHEP 369 ELISA and Western blot reactions with extracts of common soybean pathogens, related rust fungi, and infected plants
| Pathogen | Source | Reactivitya |
|
|---|---|---|---|
| MAb 2E8E5-1 | MAb 3G6H7-3 | ||
| P. pachyrhizi | Ungerminated spores | +++ | +++ |
| P. pachyrhizi | Germinated spores | +++ | +++ |
| P. pachyrhizi | Inoculated soybean | +++ | +++ |
| P. meibomiae | Ungerminated spores | +++ | − |
| P. meibomiae | Germinated spores | − | − |
| P. meibomiae | Inoculated lima bean | + | − |
| Uromyces appendiculatus | Germinated spores | − | − |
| Uromyces appendiculatus | Inoculated common bean | − | − |
| Puccinia punctiformis | Germinated spores | − | − |
| Puccinia graminis | Ungerminated spores | − | − |
| Puccinia graminis tritici | Inoculated plant material | − | − |
| Fusarium graminearum | Inoculated plant material | − | − |
| Peronospora manshurica | Inoculated plant material | − | − |
| Pseudomonas syringae pv. phaseolicola | Inoculated plant material | − | − |
| Xanthomonas campestris pv. glycines | Inoculated plant material | − | − |
| Ustilago tritici | Inoculated plant material | − | − |
| Erysiphe polygoni DC | Inoculated plant material | − | − |
| Septoria lycopersici | Inoculated plant material | − | − |
| Pseudomonas syringae | Inoculated plant material | − | − |
| Cercospora sojina | Inoculated plant material | − | − |
Absorbance readings of less than 0.049 (equivalent to background values) are designated −, readings between 0.050 and 0.300 are designated +, those between 0.310 and 0.800 are designated ++, and those greater than 0.810 are designated +++.
Immunofluorescence localization.
Spores were germinated for 4 to 18 h on glass slides in a moist chamber and incubated in 100 μl primary antibody at 1:500 in PBS-Tw for 1 h at room temperature. Slides were washed 3 times by swirling in 25 ml PBS-Tw in a petri dish on a rocking platform. Secondary antibody (Alexa Fluor 488 goat anti-rabbit IgG [H+L] and Alexa Fluor 514 goat anti-mouse IgG [H+L] [2 mg/ml; Invitrogen, Carlsbad, CA]) was added at a 1:1,000 dilution in PBS-Tw and incubated at room temperature for 1 h. Control slides (no primary antibody) were treated and analyzed as well. Slides were washed 3 times by swirling in 25 ml PBS-Tw in a petri dish on a rocking platform, and fluorescence was visualized on a Nikon E 600 fluorescence microscope using a Nikon B2E/C filter set optimized for fluorescein isothiocyanate (FITC) (excitation of 465 to 495, emission of 515 to 555, barrier of 505 nm) (Nikon, Tokyo, Japan). Image capture was performed with a Diagnostics Instruments real-time monochrome charge-coupled-device (CCD) camera system using SPOT Advanced software version 3.2.4 (Diagnostic Instruments, Sterling Heights, MI).
Inoculation and processing of tissue for transmission electron microscopy.
One set of four pots (one plant per pot) of cultivar Williams soybean plants at the 3- to 4-trifoliolate-leaf stage was spray inoculated with a 105-urediniospore/ml suspension of isolate AL 04-1, applying 15 ml per pot. Plants were placed in a dew chamber at 20° to 22°C. One second trifoliolate leaflet was removed from individual plants 16 h after inoculation. Immediately following the collection of each leaf, the leaves were placed abaxial side down on paper towels to remove excess moisture. The leaves were cut into 1-cm2 pieces with a razor blade and fixed for 3.5 to 8 h in 3% glutaraldehyde at room temperature in 0.05 M phosphate buffer. The glutaraldehyde buffer solution was removed, and leaf pieces were rinsed several times with 0.05 M phosphate buffer and refrigerated in the buffer until processed further. The glutaraldehyde-fixed tissue pieces were cut into 1-mm2 pieces and postfixed in 2% OsO4 in buffer for 2 h. After rinsing in several changes of buffer, samples were dehydrated in an ethanol series and embedded in Spurr's resin (27).
Immunocytochemical transmission electron microscopy procedure.
Thin sections obtained using a diamond knife on a Reichert OM2 ultramicrotome were placed on Formvar carbon-coated nickel grids. Sections were etched with 20% hydrogen peroxide for 20 min, blocked with 5% BSA-PBS buffer solution for 20 min, and placed in the primary antibody (dilution 1:100 with 1% BSA-PBS) for 5 h. Sections were rinsed several times in 1% BSA-PBS and placed in gold-labeled (15-nm diameter) goat anti-rabbit IgG antibody (Electron Microscopy Sciences) at a dilution of 1:100 (goat anti-rabbit IgG and 1% BSA-PBS) for 1 h. Sections were rinsed several times in 1% BSA-PBS, air dried, stained in 2% uranyl acetate (pH 5.0) followed by 0.01 M lead citrate (pH 12), and viewed with a Philips 201 transmission electron microscope.
Protein functional analysis.
For analysis of soybean phenotypic responses to PHEPs, recombinant proteins 107 or 369 were suspended in 0.1% PBS-Tw (control sample contained no protein), and 10 μl of 0.1 μg/μl PHEP was applied to leaves as a single microdrop with a micropipettor, vacuum infiltrated, and monitored for a period of 14 days. Protein concentrations of rPHEP proteins were measured as referenced above (19).
For analysis of the effects of antibodies raised against PHEPs on germination of P. pachyrhizi urediniospores, spores were mixed with PAb 107 and 369 or PBS-Tw (control) at a concentration of 10 μg/ml and applied to soybean leaves and glass slides. Spores were observed visually using a compound microscope, and the number of germinated spores was quantitated after 24 h.
RESULTS
Sequence analysis of PHEP family members.
Three low-molecular-weight proteins, termed here PHEP 107 (GenBank accession no. BK008466), PHEP 280 (BK008467), and PHEP 369 (BK008465), were previously identified by MALDI-TOF/TOF mass spectrometry as representing the most visually abundant spots on colloidal blue-stained 2D gels (17). These proteins were identified from searches against translated P. pachyrhizi ESTs, using MASCOT software. All 3 proteins were identified in soluble protein fractions extracted from P. pachyrhizi germinating urediniospores. PHEP 107 and PHEP 369 were present in germination water on which spores were germinated as well and were detected as multiple spots in 2DE. PHEP 107 and PHEP 369 are nearly identical in length (108 and 109 amino acids, respectively) and share 64% amino acid sequence identity (76% amino acid class similarity). tBLASTn searches (2) against the National Center for Biotechnology Information nonredundant (NCBInr) and EST databases revealed three additional ORFs (EH254430, EH26023, and EH238470.1) with high similarity to PHEP 369 (Fig. 1), indicating that the PHEPs represent a small gene family.
Fig 1.
Amino acid alignment of PHEP family proteins, aligned using CLUSTAL (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Asterisks indicate positions which have a single, fully conserved residue. Colons indicate conservation between groups of amino acids with strongly similar properties. Periods indicate conservation between groups of amino acids with weakly similar properties.
We compared the sequences of the PHEP 369 sequences using PCR with rPHEP 369 primers from genomic DNA (gDNA) isolated from ungerminated urediniospores of an international collection of P. pachyrhizi isolates from 6 countries (Table 1). Only two silent mutations were detected within all isolates sequenced (see Fig. S1 in the supplemental material), indicating that the PHEP 369 gene(s) is highly conserved in the species.
Analysis of P. pachyrhizi fosmid and EST sequences with tBLASTn (2) searches against the NCBInr and ESTs revealed four ORFs and one truncated EST with closely related sequences and intron/exon structures to PHEP 369, (EH254430, EH26023, and EH238470.1) (Fig. 1), thus constituting a small gene family. The PHEP 369 sequence was identified in P. pachyrhizi fosmid clone AC188845.2. The 522-bp sequence contained two introns, one 100 bp and the other 98 bp, and three exons. PHEP 369 was encoded with start and stop sites and contained a TATA box 142 bp upstream of the start codon. Conserved domain searches with PHEP family members showed no evidence of sequence similarity or putative functional relationship to any sequences deposited in open source databases. Analysis of the fosmid revealed no other copies for PHEP 369 or any of the related PHEP proteins. Additionally, results of Southern blot analysis (see Fig. S2 in the supplemental material) indicated that PHEP 369 is a single-copy gene.
Protein profile searches at ExPASy, including PFAM, HAMAP, PROSITE, and ELM (10), indicated no conserved pattern or functional relationship of PHEPs to any well-characterized protein families. Although both proteins contain hydrophobic and charged domains near the N termini, prediction algorithms (4) do not reveal the presence of a canonical signal peptide for either protein, and no signal peptide cleavage site was identified by the prediction algorithms. Additionally, preliminary yeast secretion trap assays, using PHEP107 and PHEP369 clones in a yeast secretion vector (16), were negative (K. Pedley, personal communication; data not shown).
Motif searches applying nonclassical (leaderless) secretion algorithms (5, 6) produced neural network output scores exceeding the normal threshold given for SecretomeP motif searches (12) and also revealed putative carbohydrate and phosphate-binding domains, which may explain why multiple forms of PHEP 369 were identified on 2DE. Lectin blots performed by probing urediniospore proteins separated by 2DE with concanavalin A provided preliminary evidence for mannose-binding domains on PHEP 369 (see Fig. S3 in the supplemental material).
Kyte-Doolittle plots (not shown) displayed clear evidence of hydrophobicity that was evident in regions of the PHEP genes rich in alanine, isoleucine, leucine, and valine (15). Hydrophobic regions were localized to the most conserved regions, while the highest variability was evident in the most hydrophilic domains. Antigenicity plots generated using the ABIE Pro online tool (version 3.0; Chang Biosciences, Castro Valley, CA) revealed numerous peptide sequences as prime candidates for antibody design.
Antibody reactivity and specificity.
Polyclonal antibodies were generated against rPHEP 107 and rPHEP 369 proteins, and IgG fractions were purified for characterization studies. Recombinant proteins could be detected on dot blots by chemiluminescence detection at levels as low as 100 pg at an antibody dilution of 1:5,000. By Western blot analysis, we found that the anti-rPHEP 107 and anti-rPHEP 369 PAbs were not completely specific. Anti-rPHEP 107 PAb was highly reactive with rPHEP 107 and moderately reactive against rPHEP 369. By comparison, anti-rPHEP 369 PAb was highly reactive with rPHEP 369 but only slightly with rPHEP 107 (Fig. 2).
Fig 2.
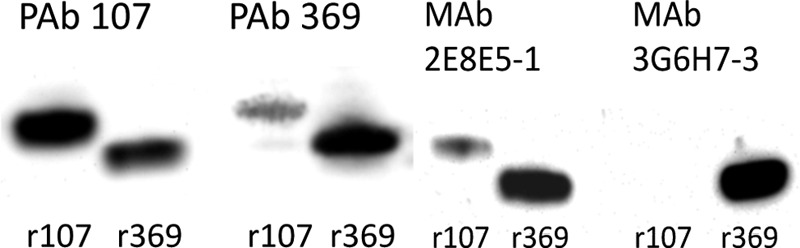
Western blot analysis of rPHEP 107 and rPHEP 369 probed with PAbs and MAbs. Recombinant proteins were loaded at 5 ng total protein per lane.
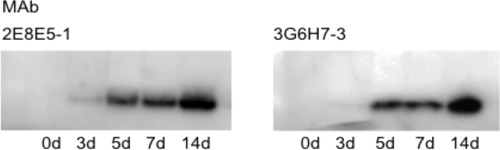
Monoclonal antibodies were then generated against the purified rPHEP 369 protein. Six MAb cell lines were selected after two rounds of screening with rPHEP 369 protein. Two MAb cell lines were specific to rPHEP 369 (3G6H7-3 and 3G6E11) and did not cross-react with rPHEP 107, whereas two MAbs (2E8E5-1 and 2E8H6) reacted with both recombinant proteins. Western blots illustrated the reactivity and specificity for 2 MAbs with rPHEP 107 and 369 (Fig. 2). Using ELISA to determine the sensitivities of MAbs, 300 pg of rPHEP 369 protein was detected at a dilution of 1:3,000 of either 2E8E5-1 (MAb1) or 3G6H7-3 (MAb3) when used as primary antibodies, and 1 ng of rPHEP 369 protein was detected at a dilution of 1:26,000 of either MAb1 or MAb3 when used as primary antibodies. PHEP 369 in leaf soluble protein extracts from soybean inoculated with 0.375 mg/ml of P. pachyrhizi isolate TW72-1 could be detected by Western blotting within 3 dpi using MAb1 and within 5 dpi using MAb3 (Fig. 3).
Fig 3.
Western blot analysis of ASR-infected soybean probed with MAb1 and MAb3. Twenty-four-day-old soybean plants, cultivar Williams 82, were inoculated with isolate TW72-1 and collected at days 0 (uninoculated), 3, 5, 7, and 14 postinoculation. Gels were loaded with 30 μg per lane total protein.
The specificities of MAb1 and MAb3 to extracts from plants infected with common U.S. soybean pathogens, extracts of cultured common soybean pathogens, and extracts of spores of related rust fungi were evaluated using ELISA and Western blot analysis and are illustrated in Table 2. MAb1 and MAb3 did not react with extracts from plants infected with common U.S. soybean pathogens, extracts of cultured common soybean pathogens, or extracts of spores of related rust fungi. MAb1 was reactive against P. meibomiae ungerminated spores and slightly reactive against Phaseolus lunatus (lima bean) infected with P. meibomiae collected at 14 dpi during sporulation with numerous pustules (Fig. 4) but did not react with germinated spores of P. meibomiae.
Fig 4.
Western blot analysis of Phakopsora proteins in urediniospores and infected plants probed with MAb1 and MAb3. Urediniospore proteins were loaded at 200 ng/lane (P. pachyrhizi) and 1,000 ng/lane (P. meibomiae) total protein; infected lima bean leaf extracts were loaded at 30 μg/lane total protein. Abbreviations: uPp and gPp, ungerminated and germinated P. pachyrhizi urediniospore proteins, respectively; uP and gPm, ungerminated and germinated P. meibomiae urediniospore proteins; 14d, P. meibomiae-infected lima bean 14 days after infection.
Immunolocalization and immunofluorescent assays (IFA).
An electron microscopy study was undertaken to establish the localization of PHEP proteins in P. pachyrhizi urediniospores. Recombinant PHEP PAbs were chosen for the study because of their reactivity to spores.
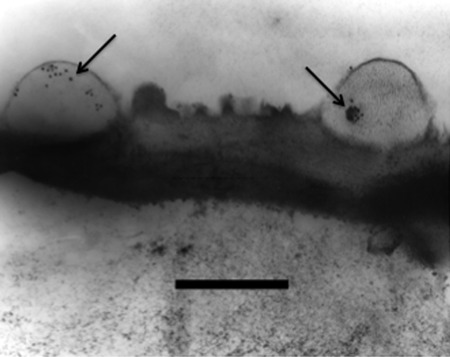
Gold-labeled PAbs localized PHEP 369 to echinulations (spines) on the surface of ungerminated P. pachyrhizi urediniospores (Fig. 5). Gold particles are evident in the spines protruding from the urediniospore wall. Overall, we observed that most spines contained multiple particles.
Fig 5.
Immunocytochemical localization of PHEP 369 protein in echinulations (spines) of the P. pachyrhizi urediniospore wall using MAb1.
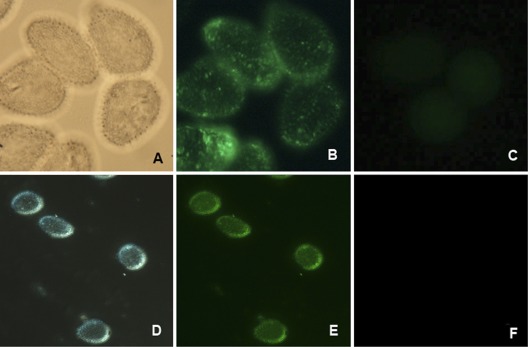
Immunofluorescence assays with urediniospores prior to and during germination confirmed the localization of PHEP 369 to echinulations on the P. pachyrhizi urediniospore surface using PAb 369 (Fig. 6A, B, C) and MAb 2E8E5-1 (Fig. 6D, E, F). Controls without primary antibody showed minimal autofluorescence.
Fig 6.
Immunofluorescence of germinating spores with MAbs and PAbs. (A) MAb1, white light; (B) MAb1, fluorescence; (C) MAb1, control (no primary antibody), fluorescence; (D) MAb3, phase contrast, backlighting; (E) MAb3, phase contrast, fluorescence.; (F) MAb3, control (no primary antibody), phase contrast, fluorescence.
Protein functional analysis.
Considering the sequence analysis of PHEP family members and absence of any known homologues, the functional role of PHEP proteins in fungal development, infection, and interaction with the host is unclear. Attempts were undertaken to establish whether high concentrations of r107 or r369 proteins applied to a soybean leaf surface could induce a phenotypic response. Neither rPHEP 107 nor rPHEP 369 induced a hypersensitive or other visible phenotypic response when infiltrated at quantities up to 1 μg into leaves of susceptible soybean cultivar Williams (see Fig. S4 in the supplemental material). When rPHEP 107 and rPHEP 369 were added to inoculum at the same concentration, they did not prevent subsequent infection (see Fig. S5 in the supplemental material).
We then asked whether the presence of PAb r369 during spore germination could disrupt critical stages of the infection process. In experiments incubating P. pachyrhizi urediniospores with PAb 369, we observed no consistent effect of the recombinant protein on urediniospore germination (see Table S1 in the supplemental material) or adhesion to detached leaves (see Table S2 in the supplemental material).
DISCUSSION
We have identified a small gene family of extracellular proteins that are localized to the Phakopsora cell wall. PHEP 107 and 369 are unique low-molecular-weight proteins with no significant similarity to any amino acid or DNA sequence entries in public databases. Evidence reported in this study suggests that these extracellular proteins are associated with P. pachyrhizi urediniospore walls. We found it surprising that no homologues exist in the genomes of other rust fungi.
None of the PHEP family members have RxLR motifs characteristic of some fungal effector peptides (7, 13). No evidence was found for a role in recognition by the host or subsequent virulence during infection, and inhibition assays with antibodies did not indicate a role in germination, growth, or appressorium formation. Although we are not able to assign a function to the two PHEP family members described in this study, we cannot rule out a role in cell wall expansion during growth and development of the germinating urediniospore, as our antibodies may not have penetrated the cell wall during inhibition assays with live, unfixed leaf tissue. The apparently high titers of PHEP 107 and 369 as indicated by spot intensity by 2DE (17) may suggest a possible structural role for these proteins in the echinulations of the spore wall and the spore germling cell wall.
Secreted fungal proteins have largely been studied from the perspective of their roles in adhesion, pathogenesis, and virulence, with early studies focusing on lytic and degradative enzymes with roles in penetration of plant host cuticle and cell wall components (23). Recent progress has been made in genetic and proteomic cataloguing of fungal secretomes (8, 9, 16, 31). Few studies have identified and definitively localized extracellular proteins to the cell wall of obligate fungal pathogens (21) and defined a functional role to the protein(s) (1, 26, 28).
While extracellular proteins in fungal spore and cell walls have been common targets for specific immunochemical diagnostic reagents, few have been directed against surface protein targets in obligate rust, bunts, and smut fungi (11, 18).
Antibodies made to PHEP proteins provide ideal candidates for the development of P. pachyrhizi species-specific immunodiagnostic assays. To our knowledge, rPHEP PAbs and MAbs are the first antibodies designed to react against a set of unique P. pachyrhizi proteins. Antibodies produced in previous studies were generated against complex mixtures of intact urediniospores and pulverized germinated spores (3, 20, 30). Based on our results, MAb 3G687-3 is species specific for P. pachyrhizi, and MAb 2E8ES-1 is genus specific for Phakopsora. While both MAb cell lines detected ASR in presymptomatic leaves, it must be noted that the artificially high infection rates we employed to produce soybean leaf samples for ELISA and Western blot analysis may not represent diagnostic samples typically encountered under field conditions with low to moderate disease pressure.
The antibodies should prove applicable in ELISA and hand-held lateral flow diagnostic assays with infected soybeans, although this may be dependent upon the level of infection. Given the excellent reactivity in immunofluorescence assays, the MAbs should prove as highly specific probes to identify fungal spores trapped in soybean rust sentinel surveillance plots. Both MAbs should also prove useful as tools to quantify fungal biomass in infected leaves, with potential application in screening soybean lines for slow rusting and resistance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Melissa L. Carter, for expert technical assistance and Kerry F. Pedley for performing yeast secretion trap assays with PHEP clones. The authors are grateful for the support provided by the North Central Soybean Research Program and Iowa Soybean Board.
USDA is an equal opportunity provider and employer.
The use of trade, firm, or corporation names in this article is for the information and convenience of the reader. Such use does not constitute an official endorsement of approval by the USDA Agricultural Research Service, NAL, or BIC of any product or service to the exclusion of others that may be suitable.
Footnotes
Published ahead of print 23 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ahn N, Kim S, Choi W, Im KH, Lee YH. 2004. Extracellular matrix protein gene, EMP1, is required for appressorium formation and pathogenicity of the rice blast fungus, Magnaporthe grisea. Mol. Cells 17:166–173 [PubMed] [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Baysal-Gurel F, et al. 2008. An immunofluorescence assay to detect urediniospores of Phakopsora pachyrhizi. Plant Dis. 92:1387–1393 [DOI] [PubMed] [Google Scholar]
- 4. Bendtsen JD, Nielsen H, von Heijine G, Brunak S. 2004. Improved predictions of signal peptides: Signal P 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 5. Bendtsen JD, Juhl Jensen L, Blom N, von Heijne G, Brunak S. 2004. Feature based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17:349–356 [DOI] [PubMed] [Google Scholar]
- 6. Bendtsen JD, Kiemer L, Fausboll A, Brunak S. 2005. Non-classical protein secretion in bacteria. BMC Microbiol. 5:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chisholm ST, Coaker G, Day B, Staskawicz BJ. 2006. Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124:803–814 [DOI] [PubMed] [Google Scholar]
- 8. de Groot PWJ, Ram AF, Klis FK. 2005. Features and functions of covalently linked protein in fungal cell walls. Fungal Genet. Biol. 42:657–675 [DOI] [PubMed] [Google Scholar]
- 9. de Groot PWJ, et al. 2009. Comprehensive genomic analysis of cell wall genes in Aspergillus nidulans. Fungal Genet. Biol. 46:S72–S81 [DOI] [PubMed] [Google Scholar]
- 10. Gasteiger E, Hoogland GAC, Ivanyi I, Appel RD, Bairoch A. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gupta MK, Narwal S, Umpathy V, Mall R, Kumar A. 2009. Characterisation of potential antigen(s) of Tilletia indica teliospore walls to develop a specific immunoassay for Karnal bunt detection. Food Agric. Immunol. 20:79–94 [Google Scholar]
- 12. Horton P, Park K-J, Obayashi T, Hakai K. 2006. Protein subcellular localization prediction with WoLF PSORT. Proc. 4th Annu. Asia Pacific Bioinform. Conf. 6:39–48 [Google Scholar]
- 13. Jones JDG, Dangl JL. 2006. The plant immune system. Nature 444:323–329 [DOI] [PubMed] [Google Scholar]
- 14. Kohler G, Milstein C. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256:495–497 [DOI] [PubMed] [Google Scholar]
- 15. Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132 [DOI] [PubMed] [Google Scholar]
- 16. Lee S-J, et al. 2006. A functional screen to characterize the secretomes of eukaryotic pathogens and their hosts in planta. MPMI 19:1368–1377 [DOI] [PubMed] [Google Scholar]
- 17. Luster DG, McMahon MB, Carter ML, Fortis LL, Nunez A. 2010. Proteomic analysis of germinating urediniospores of Phakopsora pachyrhizi. Proteomics 10:3549–3557 [DOI] [PubMed] [Google Scholar]
- 18. Luster DG, Bonde MR, Peterson GL, Hack MA, Schaad NW. 1998. Antigenic glycoproteins in the teliospore wall of Tilletia indica. Mycologia 90:180–188 [Google Scholar]
- 19. Markwell MA, Haas SM, Bieber LL, Tolbert NE. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87:206–210 [DOI] [PubMed] [Google Scholar]
- 20. Mendesa RK, Carvalhala RF, Stach-Machadob DR, Kubotaa LT. 2009. Surface plasmon resonance immunosensor for early diagnosis of Asian rust on soybean leaves. Biosens. Bioelectron. 24:2483–2487 [DOI] [PubMed] [Google Scholar]
- 21. Mueller O, et al. 2008. The secreome of the maize pathogen Ustilago maydis. Fungal Genet. Biol. 45:S63–S70 [DOI] [PubMed] [Google Scholar]
- 22. Posada-Buitrago ML, Frederick R. 2005. Expressed sequence tag analysis of the soybean rust pathogen Phakopsora pachyrhizi. Fungal Genet. Biol. 42:949–962 [DOI] [PubMed] [Google Scholar]
- 23. Rast DM, Baumgartner D, Mayer C, Hollenstein O. 2003. Cell wall-associated enzymes in fungi. Phytochemistry 64:339–366 [DOI] [PubMed] [Google Scholar]
- 24. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, second ed, vol 3 Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 25. Schneider RW, Hollier CA, Whitam HK. 2005. First report of soybean rust caused by Phakopsora pachyrhizi in the continental United States. Plant Dis. 89:774. [DOI] [PubMed] [Google Scholar]
- 26. Schoffelmeer EAM, Vossen JH, van Doorn AA, Cornelissen BJC, Haring MA. 2001. FEM1, a Fusarium oxysporum glycoprotein that is covalently linked to the cell wall matrix and is conserved in filamentous fungi. Mol. Genet. Genomics 265:143–152 [DOI] [PubMed] [Google Scholar]
- 27. Spurr AR. 1969. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26:31–43 [DOI] [PubMed] [Google Scholar]
- 28. Stringer MA, Timberlake WE. 1995. dewA encodes a fungal hydrophobin component of the Aspergillus spore wall. Mol. Microbiol. 16:33–44 [DOI] [PubMed] [Google Scholar]
- 29. Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose shsets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vittal R, Haudenshield JS, Hartman GL. 2010. Detection of viable Phakopsora pachyrhizi spores by indirect immunofluorescence and propidium iodide staining. Phytopathology 100:S131 [Google Scholar]
- 31. Wu Z, et al. 2008. Proteomic analysis of spore wall proteins and identification of two spore wall proteins from Nosema bombycis (Microsporidia). Proteomics 8:2447–2461 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.