Abstract
The Gram-negative bacterium Cronobacter sakazakii is an emerging food-borne pathogen that causes severe invasive infections in neonates. Variation in the O-antigen lipopolysaccharide in the outer membrane provides the basis for Gram-negative bacteria serotyping. The O-antigen serotyping scheme for C. sakazakii, which includes seven serotypes (O1 to O7), has been recently established, and the O-antigen gene clusters and specific primers for three C. sakazakii serotypes (O1, O2, and O3) have been characterized. In this study, the C. sakazakii O4, O5, O6, and O7 O-antigen gene clusters were sequenced, and gene functions were predicted on the basis of homology. C. sakazakii O4 shared a similar O-antigen gene cluster with Escherichia coli O103. The general features and anomalies of all seven C. sakazakii O-antigen gene clusters were evaluated and the relationship between O-antigen structures and their gene clusters were investigated. Serotype-specific genes for O4 to O7 were identified, and a molecular serotyping method for all C. sakazakii O serotypes, a multiplex PCR assay, was developed by screening against 136 strains of C. sakazakii and closely related species. The sensitivity of PCR-based serotyping method was determined to be 0.01 ng of genomic DNA and 103 CFU of each strain/ml. This study completes the elucidation of C. sakazakii O-antigen genetics and provides a molecular method suitable for the identification of C. sakazakii O1 to O7 strains.
INTRODUCTION
Cronobacter (formerly Enterobacter sakazakii) was reclassified as a genus, and now seven named species—Cronobacter sakazakii, C. malonaticus, C. muytjensii, C. dublinensis, C. turicensis, C. condimenti, and C. universalis—have been found (20, 21, 23a). Cronobacter is an emerging opportunistic pathogen causing severe invasive infections in neonates (11, 15, 35). Clinical strains of Cronobacter spp. have been linked to several outbreaks of neonatal meningitis and necrotizing enterocolitis, leading to a high mortality rate (estimated between 33 and 80%) in vulnerable infants (15, 28). C. sakazakii appears to be the most important species in this genus for its dominant isolation frequency and important clinical significance (3, 23, 27, 33).
The O antigen forms part of the lipopolysaccharide (LPS) in the outer membrane of Gram-negative bacteria and is one of the most variable constituents on the cell surface (40). Its variation provides the basis for the serotyping schemes of Gram-negative bacteria. Since it was shown that specific O serotypes were associated with pathogens causing enteritis epidemics in newborn infants between 1945 and 1950, serotyping has been the most widely used method for identifying strains for epidemiological purposes. The O-antigen serotyping scheme for C. sakazakii, which includes seven serotypes (O1 to O7), was recently established for the first time using traditional immunological technologies (46). Several O serotypes were also designed for other Cronobacter species, such as C. muytjensii, C. malonaticus, and C. turicensis (22).
Serotyping is still widely used to characterize isolates for monitoring outbreaks and general bacterial surveillance (17), although other useful molecular typing approaches, such as pulsed-field gel electrophoresis, multilocus sequence typing, and repetitive sequence-based PCR, have been developed (3, 6, 16, 27). Traditional serotyping technology using antisera is specific and still routinely used; however, this approach is labor-intensive, and antisera for all serogroups are difficult to obtain. Therefore, rapid molecular methods for C. sakazakii serotyping are required.
Genes for O-antigen synthesis are normally located on the chromosome as an O-antigen gene cluster, and genetic variation in the gene cluster is the major basis for the diversity of O-antigen forms (40). Similar to Escherichia coli, the C. sakazakii O-antigen gene cluster is located between galF and gnd (34). Furthermore, a conserved 39-bp JUMPStart sequence required for the regulation of downstream genes, is located in the intergenic region between galF and the O-antigen gene cluster (18, 32). Three main classes of genes, including nucleotide sugar synthesis genes, sugar transferase genes, and O-unit processing genes, have been identified in O-antigen gene clusters (40). Particular O-antigen genes, such as O-unit processing genes, are highly specific to individual O serogroups and are used in molecular assay for rapid identification and detection of E. coli, Shigella, and Salmonella relevant strains (7, 8, 14). To date, O-antigen gene clusters and specific primers for three C. sakazakii serotypes (O1, O2, and O3) have been characterized (22, 34). The O-antigen gene clusters of O5 to O7 were referred to in our previously publications but were not analyzed in detail (2, 44).
In the present study, the O-antigen gene clusters of the remaining C. sakazakii O4, O5, O6, and O7 were sequenced and analyzed, providing the possibility of a systematic characterization of the genetics of this pathogen. Specific genes from the four serotypes (O4 to O7) were also identified, thus allowing development of molecular serotyping assay for all seven C. sakazakii serotypes in combination with the previously identified O1 to O3 specific genes.
MATERIALS AND METHODS
Bacterial strains.
Bacterial strains used in the present study are listed in Table 1, and all of these strains have been identified in our previous study (46). C. sakazakii type strains O4, O5, O6, and O7 (laboratory stock numbers G2594, G2706, G2704, and G2592) were isolated from powdered infant formula collected from India, Ireland, China, and France by the Tianjin Entry-Exit Inspection and Quarantine Bureaus of China in 2006 and 2007 (46).
Table 1.
Sources of strains used in this study
| Bacterium | No. of strains and sourcea | No. of strains |
|---|---|---|
| Cronobacter sakazakii | 3A, 2B, 39C,3D,4E, 2F, 2G, 3H, 4I, 4J, 5K, 48L | 119 |
| O1 | 1A, 5C, 2D, 2E, 1F, 1G, 1H, 2I, 2K, 32L | 49 |
| O2 | 2A, 2B, 16C, 1E, 4J, 2K, 7L | 34 |
| O3 | 7C, 1K | 8 |
| O4 | 3C, 1F, 4L | 8 |
| O5 | 3C, 1I | 4 |
| O6 | 1C, 1I, 6L | 8 |
| O7 | 4C, 1D, 1E, 1G, 1H | 8 |
| Other Cronobacter spp. | 1A, 2E, 2I, 1L, 1M | 2 |
| C. malonaticus O1b | 2E | 2 |
| C. muytjensii | 1A | 1 |
| C. dublinensis | 1I, 1L | 2 |
| C. turicensis | 1I, 1M | 2 |
| Enterobacter cloacae | 1A,1N | 2 |
| Enterobacter aerogenes | 2N | 2 |
| Escherichia coli | 1A, 1O | 2 |
| O111 | 1A | 1 |
| O157 | 1O | 1 |
| Salmonella enterica | 1A, 1O | 2 |
| O44 | 1A | 1 |
| D1 | 1O | 1 |
| Shigella flexneri | 1A, 1L | 2 |
| 2a | 1A | 1 |
| 5a | 1L | 1 |
| Total | 136 |
Sources are indicated by superscript letters as follows: A, American Type Culture Collection, USA; B, Czech Collection of Microorganisms (CCM), Czech Republic; C, environmental isolates from food by Tianjin Entry-Exit Inspection and Quarantine Bureau, China; D, environmental isolates from food by Liaoning Entry-Exit Inspection and Quarantine Bureau, China; E, Chinese Academy of Inspection and Quarantine, Beijing, China; F, environmental isolates from food by Jilin Entry-Exit Inspection and Quarantine Bureau, China; G, environmental isolates from food by Neimenggu Entry-Exit Inspection and Quarantine Bureau, China; H, environmental isolates from food by Shenyang Entry-Exit Inspection and Quarantine Bureau, China; I, environmental isolates from food by Xinjiang Entry-Exit Inspection and Quarantine Bureau, China; J, environmental isolates from food by Guangdong Entry-Exit Inspection and Quarantine Bureau, China; K, environmental isolates from food by Hunan Entry-Exit Inspection and Quarantine Bureau, China; L, environmental isolates from food by Beijing Entry-Exit Inspection and Quarantine Bureau, China; M, Environmental isolates from food by Hubei Entry-Exit Inspection and Quarantine Bureau, China; N, China General Microbiological Culture Collection Center (CGMCC), China; and O, National Center for Medical Culture Collection (CMCC), China.
Based on previously published data (22).
Genomic DNA extraction and O-antigen gene cluster amplification.
Bacterial genomic DNA was extracted from each strain by using bacterial genomic DNA purification kits (Tiangen Biotech Co., Ltd., Beijing, China) according to the instructions provided by the manufacturer. Long-range PCR of the O-antigen gene cluster was performed with an Expand Long Template PCR system (TaKaRa Biotechnology, Shiga, Japan) using the following primers for amplification of the JUMPStart sequence and gnd gene, respectively: wl-10324 (5′-GCACTGGTAGCTATTGAGCCAGGGGCGGTAGCAT-3′) and wl-2211 (5′-ACTGCCATACCGACGACGCCGATCTGTTGCTTGG-3′). The PCR amplification was performed in 50-μl volumes containing 1× LA buffer (plus MgCl2), 0.4 mM concentrations each dATP, dCTP, dGTP, and dTTP, 0.4 μM concentrations of each primer, 2.5 U of TakaRa LA Taq polymerase, and 2 μl of template DNA (∼500 ng). The following PCR conditions were used for amplification: 32 cycles of denaturation at 94°C for 30 s, annealing at 61°C for 45 s, and extension at 68°C for 15 min.
Sequencing and analysis of O-antigen gene clusters.
The PCR products were gel purified using UNIQ-10 columns (Sangon, Shanghai, China) and were sheared at speed code 9 (20 cycles) to an estimated fragment length of 1,000 to 2,000 bp by HydroShear fragmentation (Digilab, CA). The resulting DNA fragments were repaired and then cloned into the pUC18 vector to produce a shotgun bank. Sequencing was carried out using an ABI 3730 automated DNA sequencer (Applied Biosystems, Foster City, CA). Sequencing data were assembled using the Staden Package software (45) and annotated with the Artemis program (41). BLAST analysis was used to search available databases such as GenBank (5). The Clusters of Orthologous Groups (COG) (48) and Pfam (9) protein motif databases were used to search for conserved protein domains. The program TMHMM 2.0 (25) was used to identify potential transmembrane segments. Sequence alignments and comparisons were performed using the CLUSTAL W program (49).
PCR primer specificity analysis.
Primers were designed based on the wzx and wzy gene sequences using Primer Premier 5.0 software program (Premier Biosoft International, Palo Alto, CA) (Table 2). The sequences of the primers were compared to the sequences in GenBank and the unpublished O-antigen gene sequences in our own laboratory to ensure their specificity. Genomic DNAs extracted from the strains listed in Table 1 were used as templates for amplification. The multiplex PCR was performed by mixing all primers in a final volume of 50 μl containing the following components: 1× PCR buffer; 2.5 mM MgCl2; 400 μM concentrations (each) of dATP, dCTP, dGTP, and dTTP; 0.06 to 0.10 μM primer sets listed in Table 2; and 2.5 U of Taq DNA polymerase and DNA template (50 to 100 ng). The following PCR conditions were used for amplification: an initial denaturation step at 95°C for 5 min, followed by 30 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min. Samples of the PCR products (5 μl) were subjected to agarose gel electrophoresis for examination.
Table 2.
Primers used in this studya
| C. sakazakii serotype | Target gene | Primer | Sequence (5′-3′) | Final concn (μM) | Amplicon size (bp) |
|---|---|---|---|---|---|
| O1 | wzy | wl-35646 | CCCGCTTGTATGGATGTT | 0.10 | 364 |
| wl-35647 | CTTTGGGAGCGTTAGGTT | 0.10 | |||
| O2 | wzy | wl-37256 | ATTGTTTGCGATGGTGAG | 0.06 | 152 |
| wl-37257 | AAAACAATCCAGCAGCAA | 0.06 | |||
| O3 | wzy | wl-37258 | CTCTGTTACTCTCCATAGTGTTC | 0.10 | 704 |
| wl-37259 | GATTAGACCACCATAGCCA | 0.10 | |||
| O4 | wzy | wl-39105 | ACTATGGTTTGGCTATACTCCT | 0.06 | 890 |
| wl-39106 | ATTCATATCCTGCGTGGC | 0.06 | |||
| O5 | wzy | wl-39873 | GATGATTTTGTAAGCGGTCT | 0.10 | 235 |
| wl-39874 | ACCTACTGGCATAGAGGATAA | 0.10 | |||
| O6 | wzy | wl-40041 | ATGGTGAAGGGAACGACT | 0.06 | 424 |
| wl-40042 | ATCCCCGTGCTATGAGAC | 0.06 | |||
| O7 | wzx | wl-40039 | CATTTCCAGATTATTACCTTTC | 0.06 | 615 |
| wl-40040 | ACACTGGCGATTCTACCC | 0.06 |
Annealing temperature, 53°C.
PCR assay sensitivity analysis.
The PCR assay sensitivity was analyzed as follows. Serial dilutions of C. sakazakii O1 to O7 strain (laboratory stock numbers G2539, G2356, G2726, G2594, G2706, G2704, and G2592) genomic DNA (10, 1, 0.1, 0.01, 0.001, 0.0001, and 0.00001 ng) were used as templates to test the sensitivity of the multiplex PCR assay. The sensitivity for isolates in pure culture was also tested. The C. sakazakii O1 to O7 strains were grown in Luria-Bertani (LB) medium to log phase and serially diluted 10-fold in sterile water. Aliquots from the same dilution were removed for planting onto LB agar and for extraction of their genomic DNA to be used as the template for the developed PCR assay. The CFU were counted after overnight growth at 37°C. Amplified products were subjected to agarose gel electrophoresis.
Nucleotide sequence accession numbers.
The DNA sequences of the C. sakazakii O4, O5, O6, and O7 O-antigen gene clusters have been deposited in GenBank under accession numbers JQ674747, JQ674748, JQ674749, and JQ674750, respectively.
RESULTS
Sequence analysis of C. sakazakii O4, O5, O6, and O7 O-antigen gene clusters.
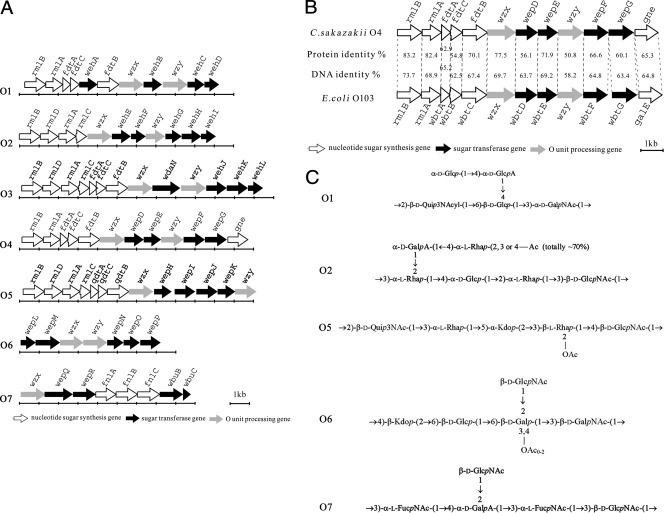
The region between JUMPStart and gnd was sequenced. Sequences of 11,887, 12,299, 7,388, and 8,932 bp were obtained from C. sakazakii O4, O5, O6, and O7 type strains, respectively. A total of 12, 13, 7, and 8 open reading frames (ORFs) (excluding the gnd gene), including orf4-1 to orf4-12 for O4, orf5-1 to orf5-13 for O5, orf6-1 to orf6-7 for O6, and orf7-1 to orf7-8 for O7, were predicted, respectively (Fig. 1A), and functions such as sugar synthesis, sugar transfer, O-antigen processing, and other functions were predicted based on homology with proteins in available databases (Tables 3 to 6). All ORFs had the same transcriptional direction from JUMPStart to gnd, with GC contents ranging from 29.61 to 49.86%, which was lower than that of the C. sakazakii genome (57%) (26).
Fig 1.
(A) Predicted genes of O-antigen gene clusters for C. sakazakii O1 to O7 serotypes. (B) Comparison of the O-antigen gene clusters of C. sakazakii O4 and E. coli O103. (C) O-antigen chemical structures of C. sakazakii O1, O2, O5, O6, and O7.
Table 3.
Characteristics of ORFs in the C. sakazakii O4 O-antigen gene clustera
| ORF | Gene | Gene position (length [aa]) | G+C content (%) | Pfam conserved domain(s) (Pfam no./e value) | Similar protein(s), strain(s) (GenBank accession no.) | % Identical (aa overlap) | Putative function of protein |
|---|---|---|---|---|---|---|---|
| 4-1 | rmlB | 124–1215 (363) | 45.51 | NAD-dependent epimerase/dehydratase family (PF01370/1.7e-78) | dTDP-glucose 4,6-dehydratase, E. coli O103:H2 (YP_003222430) | 84 (356) | Glucose dehydratase |
| 4-2 | rmlA | 1217–2080 (287) | 42.24 | Nucleotidyltransferase (PF00483/6.8e-70) | d-Glucose-1-phosphate thymidylyltransferase, E. coli O103:H2 strain 12009 (YP_003222429) | 84 (286) | Glucose-1-phosphate thymidylyltransferase |
| 4-3 | fdtA | 2082–2483 (133) | 35.57 | WxcM-like, C-terminal (PF05523/1.3e-54) | dTDP-6-deoxy-3,4-keto-hexulose isomerase, E. coli O103:H2 strain 12009 (YP_003222428) | 64 (131) | Hexose isomerase |
| 4-4 | fdhC | 2476–3009 (177) | 32.95 | Putative butyryltransferase, E. coli O103:H2 (EDV84013) | 55 (177) | Butyryltransferase | |
| 4-5 | fdtB | 3014–4120 (368) | 39.74 | DegT/DnrJ/EryC1/StrS aminotransferase family (PF01041/4e-118) | Putative aminotransferase, E. coli (AAS73167) | 69 (372) | Aminotransferase |
| 4-6 | wzx | 4108–5394 (428) | 35.50 | Polysaccharide biosynthesis protein (PF01943/7.3e-13) | O-antigen flippase Wzx, E. coli O103:H2 (YP_003222425) | 79 (419) | Flippase |
| 4-7 | wepD | 5378–6370 (330) | 34.44 | Glycosyltransferase family 2 (PF00535/2.1e-32) | Putative glycosyltransferase, E. coli (EDV84144) | 56 (323) | Glycosyltransferase |
| 4-8 | wepE | 6367–7257 (297) | 34.34 | Glycosyltransferase family 2 (PF00535/7.6e-29) | Putative glycosyltransferase, E. coli O103:H2 (YP_003222423) | 71 (296) | Glycosyltransferase |
| 4-9 | wzy | 7259–8413 (384) | 33.5 | Putative O-antigen polymerase, E. coli (ABK27352) | 51 (378) | Polymerase | |
| 4-10 | wepF | 8410–9507 (365) | 39.7 | Glycosyltransferase group 1 (PF00534/3.5e-37) | Putative glycosyltransferase, E. coli (AAS73172) | 67 (361) | Glycosyltransferase |
| 4-11 | wepG | 9511–10632 (374) | 39.48 | Glycosyltransferase group 1 (PF00534/4.5e-31) | Putative galactosyltransferase, E. coli O103:H2 (ABK27323) | 60 (373) | Galactosyltransferase |
| 4-12 | gne | 10684–11706 (340) | 42.03 | NAD-dependent epimerase/dehydratase family (PF01370/1.7e-58) | UDP-glucose 4-epimerase, H. parainfluenzae ATCC 33392 (ZP_08147973) | 67 (337) | UDP-glucose 4-epimerase |
aa, amino acid(s).
Table 6.
Characteristics of ORFs in the C. sakazakii O7 O-antigen gene clustera
| ORF | Gene | Gene position (length [aa]) | G+C content (%) | Pfam conserved domain(s) (Pfam no./e-value) | Similar protein(s), strain(s) (GenBank accession no.) | % Identical (aa overlap) | Putative function of protein |
|---|---|---|---|---|---|---|---|
| 7-1 | wzx | 69–1358 (430) | 33.02 | Polysaccharide biosynthesis protein (PF01943/8.6e-23) | Polysaccharide biosynthesis protein Wzx, V. vulnificus (ABD38622) | 31 (404) | Flippase |
| 7-2 | wepQ | 1286–2741 (485) | 29.42 | Glycosyltransferase group 1 (PF00534/5.1e-12) | Glycosyltransferase group 1, A. baumannii ABNIH2 (EGT89242.1) | 35 (264) | Glycosyltransferase |
| 7-3 | wepR | 2743–3918 (391) | 31.71 | Glycosyltransferase group 1 (PF00534/4.3e-13) | Glycosyltransferase group 1, Marinomonas sp. (YP_001339674.1) | 28 (392) | Glycosyltransferase |
| 7-4 | fnlA | 3911–4945 (344) | 41.06 | Polysacc_synt_2 (PF02719.8/2.7e-109) | l-Fucosamine synthetase, E. coli (ABM53659) | 93 (344) | Fucosamine synthetase |
| 7-5 | fnlB | 4947–6050 (367) | 41.30 | NAD-dependent epimerase/dehydratase family (PF01370.14/1.2e-13) | FnlB, epimerase, S. dysenteriae (ACA24843) | 78 (367) | Epimerase |
| 7-6 | fnlC | 6050–7180 (376) | 40.93 | UDP-N-acetylglucosamine 2-epimerase (PF02350.12/5.5e-95) | C-2 epimerase, E. coli MS 145-7 (ZP_07692194.1) | 92 (376) | Epimerase |
| 7-7 | wbuB | 7180–8388 (402) | 42.10 | Glycos_transf_1 (PF00534.13/1.6e-09) | l-Fucosamine transferase, S. dysenteriae (ACA24845) | 76 (398) | Glycosyltransferase |
| 7-8 | wbuC | 8379–8783 (135) | 42.71 | Domain of unknown function (DUF1971/0.0009) | WbuC, S. dysenteriae (ACA24846) | 69 (131) | Glycosyltransferase |
aa, amino acid(s).
Table 4.
Characteristics of ORFs in the C. sakazakii O5 O-antigen gene clustera
| ORF | Gene | Gene position (length [aa]) | G+C content (%) | Pfam conserved domain(s) (Pfam no./e-value) | Similar protein(s), strain(s) (GenBank accession no.) | % Identity (aa overlap) | Putative function of protein |
|---|---|---|---|---|---|---|---|
| 5-1 | rmlB | 183–1265 (360) | 49.86 | NAD-dependent epimerase/dehydratase family (PF01370/3.6e-81) | dTDP-d-glucose-4,6-dehydratase, C. sakazakii (ABX51885.1) | 97 (360) | Glucose dehydratase |
| 5-2 | rmlD | 1268–2164 (298) | 49.72 | RmlD substrate-binding domain (PF04321/2.2e-111) | dTDP-6-deoxy-l-mannose dehydrogenase, C. sakazakii (ABX51886.1) | 96 (295) | Mannose dehydrogenase |
| 5-3 | rmlA | 2214–3092 (292) | 47.55 | Nucleotidyltransferase (PF00483/6.5e-73) | Glucose-1-phosphate thymidylyltransferase, C. sakazakii (ABX51887.1) | 94 (292) | Glucose-1-phosphate thymidylyltransferase |
| 5-4 | rmlC | 3096–3644 (182) | 40.98 | dTDP-4-dehydrorhamnose 3,5-epimerase (PF00908/2.2e-78) | Putative dTDP-4-dehydrorhamnose 3,5-epimerase, Shigella sp. strain D9 (ZP_05431654.1) | 69 (185) | dTDP-4-dehydrorhamnose 3,5-epimerase |
| 5-5 | qdtA | 3647–4066 (139) | 38.57 | dTDP-4-dehydrorhamnose 3,5-epimerase (PF00908/2.2e-78) | QdtA E. coli (ADI77029) | 76 (129) | dTDP-4-oxo-6-deoxy-d-glucose-3,4-oxoisomerase |
| 5-6 | qdtC | 4041–4511 (156) | 38.85 | Transferase hexapeptide (PF00132/0.02) | QdtC E. coli (ADI77035) | 68 (147) | Transacetylase |
| 5-7 | qdtB | 4498–5607 (369) | 41.17 | DegT/DnrJ/EryC1/StrS aminotransferase family (PF01041/1.4e-111) | QdtB E. coli (ADI77030) | 63 (366) | Transaminase |
| 5-8 | wzx | 5604–6854 (416) | 36.45 | Polysaccharide biosynthesis protein (PF01943/2e-10) | O-antigen flippase C. sakazakii (ABX51878) | 47 (413) | Flippase |
| 5-9 | wepH | 6905–7807 (300) | 31.67 | Glycosyltransferase family 2 (PF00535/5.5e-24) | Putative glycosyltransferase P. rettgeri DSM 1131 (EFE51496) | 39 (281) | Glycosyltransferase |
| 5-10 | wepI | 7956–9011 (351) | 33.61 | Glycosyltransferase group 1 (PF00534/8.9e-06) | α-d-GlcNAc α-1,2-l-rhamnosyltransferase V. mimicus VM223 (EEY46453) | 64 (349) | Glycosyltransferase |
| 5-11 | wepJ | 9067–10158 (363) | 32.60 | Hypothetical protein CLOBOL_01216, C. bolteae ATCC BAA-613 (EDP18345) | 26 (255) | Glycosyltransferases | |
| 5-12 | wepK | 10158–11072 (304) | 29.61 | Hypothetical protein glycosyltransferase Eps5K, S. thermophilus | 32 (297) | Glycosyltransferases | |
| 5-13 | wzy | 11056–12153 (365) | 31.23 | Conserved hypothetical protein, Bacteroides sp. strain 1_1_6 (ZP_04850089.1) | 24 (241) | Polymerase |
aa, amino acid(s).
Table 5.
Characteristics of ORFs in the C. sakazakii O6 O-antigen gene clustera
| ORF | Gene | Gene position (length [aa]) | G+C content (%) | Pfam conserved domain(s) (Pfam no./e-value) | Similar protein(s), strain(s) (GenBank accession no.) | % Identity (aa overlap) | Putative function of protein |
|---|---|---|---|---|---|---|---|
| 6-1 | wepL | 47–829 (260) | 31.30 | Glycosyltransferase family 2 (PF00535/4.2e-25) | Glycosyltransferase, F. bacterium BAL38 (ZP_01733772.1) | 38 (118) | Glycosyltransferase |
| 6-2 | wepM | 813–2072 (413) | 30.13 | Capsule polysaccharide biosynthesis protein (PF05159/1.7e-22) | Capsule polysaccharide biosynthesis protein family protein, Marinomonas sp. (ZP_010075937.1) | 34 (334) | Capsule polysaccharide biosynthesis protein family protein |
| 6-3 | wzx | 2065–3270 (401) | 32.18 | Polysaccharide biosynthesis protein (PF01943/2.6e-26) | Polysaccharide biosynthesis protein, B. helcogenes P36-108 (YP_004162007) | 27 (344) | Flippase |
| 6-4 | wzy | 3260–4486 (408) | 30.65 | Wzy, P. mirabilis (ADL32282) | 26 (195) | Polymerase | |
| 6-5 | wepN | 4488–5327 (279) | 32.75 | Core-2/I-branching enzyme (PF02485/2.1e-17) | Glycosyltransferase family 14, M. paludis DSM (ZP_07747376) | 26 (294) | Glycosyltransferase |
| 6-6 | wepO | 5324–6160 (278) | 34.52 | Glycosyltransferase family 2 (PF00535/2e-23) | Glycosyltransferase WceM, Erwinia sp. strain Ejp617 (ADP12985.1) | 50 (267) | Glycosyltransferase |
| 6-7 | wepP | 6185–7225 (346) | 32.76 | Acyltransferase family (PF01757/1.2e-23) | Exopolysaccharide biosynthesis protein, Erwinia sp. strain Ejp617 (ADP10993) | 32 (352) | Acetyltransferase |
aa, amino acid(s).
Nucleotide sugar biosynthesis genes.
For the O4 O-antigen gene cluster, five proteins encoded by orf4-1 to orf4-5 share high-level identity levels (55 to 84%) with known RmlB, RmlA, FdtA, FdtC, and FdtB from E. coli O103. They have been reported to be involved in the synthesis of dTDP-3,6-dideoxy-3-(3-hydroxybutanoylamino)-d-galactose (dTDP-d-Fuc3NHb) in E. coli O103 (10). A protein encoded by orf4-12 shares 67% identity with the Haemophilus parainfluenzae UDP-glucose 4-epimerase (Gne), which catalyzes the conversion of 2-acetamido-2-deoxy-d-glucose (GlcNAc) to 2-acetamido-2-deoxy-d-galactose (GalNAc) (4). Therefore, orf4-1, orf4-2, orf4-3, orf4-4, orf4-5, and orf4-12 were named rmlB, rmlA, fdtA, fdtC, fdtB, and gne, respectively.
The O-antigen structures of O5, O6 and O7 have been defined (2, 44). Five sugars, including one 3-acetamido-3-deoxy-d-quinovose (d-Qui3NAc), two l-rhamnoses (l-Rha), one 3-deoxy-d-manno-2-octulosonic acid (Kdo), and one d-GlcNAc, were found in O5. Five sugars—d-GlcNAc, d-GalNAc, d-galactose (d-Gal), d-glucose (d-Glc), and Kdo—were found in O6, and five sugars, including two N-acetyl-l-fucosamines (l-FucNAc), two d-GlcNAc, and one d-galacturonic acid (d-GalA), were found in O7.
Genes involved in the biosynthesis of common sugar nucleotide precursors (GlcNAc, Glc, and Gal) were not located between the JUMPStart and gnd gene (43). Four proteins encoded by orf5-1, orf5-2, orf5-3, and orf5-4 share high-level identity (69 to 97%) with known RmlB, RmlD, RmlA, and RmlC proteins, which are responsible for l-Rha biosynthesis (12, 13). Three proteins encoded by orf5-5, orf5-6, and orf5-7 share identity (63 to 76%) with QdtA, QdtC, and QdtB, which are involved in the synthesis of d-Qui3NAc (37). Three proteins encoded by orf7-4, orf7-5, and orf7-6 share 78 to 93% identity with the enzymes FnlA, FnlB, and FnlC, which catalyze l-FucNAc biosynthesis (24). Genes responsible for the synthesis of Kdo in O5, Kdo and GalNAc in O6, and GalA in O7 are not present in their O-antigen gene clusters.
Sugar transferase genes.
Four, three, three, and three sugar transferases genes were identified in strains O4, O5, O6 and O7, respectively.
Proteins encoded by orf4-7, orf4-8, orf4-10, and orf4-11 belong to glycosyltransferase families 1 (PF00534) and 2 (PF00535) and share between 56 to 71% identity with E. coli glycosyltransferases. Therefore, they were proposed to encode glycosyltransferases and named wepD, wepE, wepF, and wepG, respectively.
Two proteins encoded by orf5-9 and orf5-10 belong to glycosyltransferase family 2 (PF00535) and 1 (PF00534), respectively, and share identity (39 to 64%) with glycosyltransferases of other genus. Protein encoded by orf5-12 was found to share 32% identity with the Streptococcus thermophilus glycosyltransferase Eps5K. Therefore, orf5-9, orf5-10, and orf5-12 were proposed to encode glycosyltransferases and named wepH, wepI, and wepK, respectively.
Proteins encoded by orf6-1 and orf6-6 belong to glycosyltransferase family 2 (PF00535) and also share 38 and 50% identity with glycosyltransferases of other genus. A protein encoded by orf6-5 shares 26% identity with an Erwinia glycosyltransferase. orf6-1, orf6-5, and orf6-6 were then proposed to be glycosyltransferase genes and named wepL, wepN, and wepO, respectively.
Three proteins encoded by orf7-2, orf7-3, and orf7-7 belong to glycosyltransferase family 1 (PF00534). Two proteins encoded by orf7-2 and orf7-3 share 35 to 28% identity with glycosyltransferases of other genus and are named wepQ and wepR, respectively. A protein encoded by orf7-7 shares high-level identity with WbuB of E. coli, which is an l-fucosamine transferase. Therefore, the protein encoded by orf7-7 is proposed to be responsible for l-FucNAc transfer and named WbuB.
O-antigen-processing genes.
Both wzx and wzy genes were found in O4, O5, and O6 serotypes, whereas only wzx genes were identified in O7. The putative Wzx (O-antigen flippase) from the O4 to O7 serotypes have 11 to 13 well-proportioned transmembrane, which is typical of the topological characteristics of Wzx (31). They also shared 24 to 79% identity with Wzx of other strains. The putative Wzy (O-antigen polymerase) from O4 to O6 serotypes were found to have 10 to 11 predicted transmembrane segments with large periplasmic loops of 46 to 53 amino acid residues, which is typical of the topological character of Wzy (8), and shared 24 to 51% identity values with the Wzy of other strains. Therefore, these genes were proposed to encode the O-antigen flippase and polymerase, respectively.
Additional genes identified.
The function of orf5-11 was not able to be predicted based on searches from the available databases. However, on the basis of the observation that the O antigen for O5 contains five sugars, a glycosyltransferase is absent in the gene cluster (1, 40). orf5-11 was thus proposed as a glycosyltransferase gene and named wepJ. A protein encoded by orf6-7 shares 32% identity with an Erwinia acetyltransferase responsible for the O-acetyl group in the O6 antigen and was named WepP. The protein encoded by orf6-2 shares 34% identity with capsule polysaccharide biosynthesis proteins of other species, although its function in O-antigen synthesis is not known. orf6-2 was named wepM. The protein encoded by orf7-8 shares 69% identity with the WbuC of Shigella, which is a proposed remnant gene product (30). Therefore, orf7-8 was named wbuC.
Selection of target genes.
The wzx and wzy DNA sequences of seven C. sakazakii serotypes were aligned using CLUSTAL W software. The wzx and wzy DNA sequences are highly divergent and serotype specific, with identity levels ranging from 30.1 to 54.6% and 28.9 to 45.0%, respectively (data not shown). Therefore, the wzx and wzy genes represent as suitable targets for the development of a C. sakazakii molecular serotyping scheme.
Primer specificity.
A total of 136 strains, including 49, 34, 8, 8, 4, 8, and 8 C. sakazakii O1 to O7 strains, respectively, 2 C. malonaticus strains, 1 C. muytjensii strain, 2 C. dublinensis strains, 2 C. turicensis strains, and 10 strains of closely related species, were used to test the primer specificity (Table 1). The specificity of each primer pairs was cross-tested with other O serotype strains and the closely related strains. The results showed only the DNA from of corresponding C. sakazakii O serotype strains generated the expected PCR products and no PCR products were amplified from other isolates (see Fig. S1a to g in the supplemental material).
Development of a multiplex PCR assay for identification of all C. sakazakii serotypes.
Primer concentrations of 0.05, 0.06, 0.08, 0.10, and 0.20 μM were analyzed to identify the optimal PCR conditions, respectively, and the best combinations (0.10 μM for C. sakazakii O1, O3, and O5 serotypes and 0.06 μM for C. sakazakii O2, O4, O6, and O7 serotypes) are used and listed in Table 2. Using the optimized specific primer concentrations, all of the seven primer pairs were mixed and used to screen DNA isolated from 136 strains. The results demonstrated all of the C. sakazakii strains produced the expected PCR products and non-sakazakii isolates gave no PCR products. Representative PCR results using C. sakazakii O1 to O7 genomic template DNA are shown in Fig. 2a. Finally, coded DNA samples from 20 strains of Cronobacter spp. were randomly selected and used as templates to further evaluate the specificity and sensitivity of the multiplex PCR assay using a double-blind test. The PCR results identified 2, 2, 1, 3, 1, 2, and 2 strains as C. sakazakii O1 to O7, respectively, and 7 strains as other Cronobacter spp. These results were consistent with conventional antisera serotyping methods (46).
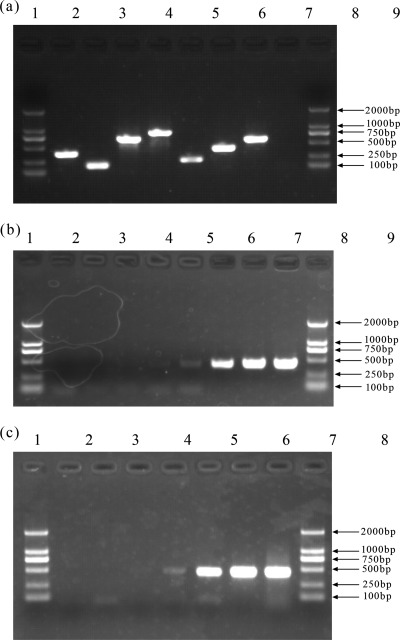
Fig 2.
Specificity and sensitivity analysis of PCR assay for C. sakazakii O1 to O7. (a) Agarose gel electrophoresis of the PCR products obtained from respective C. sakazakii O1 to O7 strains by using the multiplex PCR assay. Lanes 1 and 10, DL2000 DNA markers; lane 2, O1; lane 3, O2; lane 4, O3; lane 5, O4; lane 6, O5; lane 7, O6; lane 8, O7; lane 9, negative control. (b) Agarose gel electrophoresis of the PCR products obtained from serially diluted chromosomal DNA of C. sakazakii O6 strain G2704. Lanes 1 and 10, DL2000 DNA markers; lane 2, negative control; lane 3, 0.00001 ng; lane 4, 0.0001 ng; lane 5, 0.001 ng; lane 6, 0.01 ng; lane 7, 0.1 ng; lane 8, 1 ng; lane 9, 10 ng. (c) Agarose gel electrophoresis of the PCR products obtained from serially diluted culture of C. sakazakii O6 strain G2704. Lanes 1 and 9, DL2000 DNA markers; lane 2, negative control; lane 3, 101 CFU/ml; lane 4, 102 CFU/ml; lane 5, 103 CFU/ml; lane 6, 104 CFU/ml; lane 7, 105 CFU/ml; lane 8, 106 CFU/ml.
Sensitivity of serotyping using genomic DNA and pure culture.
The sensitivity of the multiplex PCR assay for the serotyping of O1 to O7 was evaluated using a serial 10-fold dilution ranging from 0.00001 to 10 ng of genomic DNA from C. sakazakii O serotypes O1 to O7 as templates. Positive signals were generated using 0.01 ng of DNA for all seven serotypes. Representative PCR results using C. sakazakii O6 diluted genomic template DNA are shown in Fig. 2b. Similarly, serially diluted (101 to 108 CFU/ml) pure cultures of C. sakazakii O serotypes O1 to O7 were used as templates, and positive signals were obtained using 103 CFU/ml for all seven serotypes. Representative PCR results using C. sakazakii O6 genomic template DNA exacted from diluted pure cultures are shown in Fig. 2c.
DISCUSSION
O-antigen gene clusters of C. sakazakii O4 to O7 were sequenced and analyzed in the present study. The O1 to O3 gene clusters have been identified previously. Therefore, in combination, these data complete identification of all O-antigen gene clusters among the seven serotypes, thus providing the potential for systematic analysis of all C. sakazakii O-antigen gene clusters.
C. sakazakii O-antigen gene clusters of all seven serotypes are located on the chromosome between the housekeeping genes galF and gnd, as are the O-antigen gene clusters of E. coli, Shigella sp., Salmonella sp., and Citrobacter freundii (52). Based on the elucidated O-antigen structures of C. sakazakii O1, O2, O5 to O7, and O1 to O7 antigen gene clusters, it was determined that these O antigens contain rare sugars, including d-GalNAc, d-ClcA, d-GalA, d-Qui3NAc, 3-acylamido-3,6-didesoxy-d-glucose (d-Qui3NAcyl), d-Fuc3NHb, l-Rha, Kdo, and l-FucNAc (Fig. 1C). Genes responsible for GalNAc, Kdo, and GalA are not present in O-antigen gene clusters. Kdo is the obligatory component of the LPS core, and two genes, ugd and gla, responsible for the synthesis of GalA are involved in colonic acid synthesis. Therefore, it is reasonable that the genes responsible for the synthesis Kdo and GalA are located outside of the O-antigen gene cluster. In fact, these genes could be found in the genome sequences of C. sakazakii (NC_009778) and C. turicensis (NC_013282) (the locus tags are ESA-01502, ESA-02426, ESA-3591, ESA3590, ESA-02586, ESA-01534, and ESA-01535 in C. sakazakii and CTU-24190, CTU-15240, CTU-03800, CTU-03810, CTU-13590, CTU-23880, and CTU-23870 in C. turicensis). The gene for the synthesis of GalNAc is gne, which could be located inside or outside of the O-antigen gene cluster. In C. sakazakii O4, gne was found, while in C. sakazakii O1 and O6, gne is absent in their O-antigen gene clusters. By amplifying gne using the primer pair targeting the gene (5-ACAGATTGGTGATGTTCG and 5-ATCAAAGCAATATCCACC), we found the gne could be located elsewhere in the chromosomes of C. sakazakii O1 and O6 (data not shown). Similar cases have been also been identified in E. coli (12, 13).
It has been reported in E. coli that the gene (wecA) responsible for the transfer of GalNAc-1-phosphate or GlcNAc-1-phosphate to the carrier lipid undecaprenol phosphate (UndP) for initiation of O unit synthesis is located outside the O-antigen gene cluster (1, 40). Based on the known O-antigen structures and gene clusters, five or six sugars are known to be present in C. sakazakii O antigens, and one fewer glycosyltransferase than sugars in O units has been noted. Therefore, a similar arrangement is proposed for C. sakazakii. For O5, no acetyltransferase gene (necessary for the O-acetyl modification of the O antigen) was found in its O-antigen gene cluster, indicating that this gene may be located on the prophage in the chromosome, similar to Shigella (19, 29, 38).
Three different pathways (the Wzx/Wzy, the ABC transporter, and the synthase pathways) have been described for the assembly and processing of O antigens (39). Most O antigens, particularly heteropolymeric O antigens, appear to be synthesized by the Wzx/Wzy pathway. The seven C. sakazakii O serotypes express a heteropolymeric O antigen and are synthesized by the Wzy/Wzx pathway (22, 34). The wzy gene is not present in its O-antigen gene cluster of C. sakazakii O7. However, the LPS of C. sakazakii O7 was extracted and separated by SDS–15% PAGE (data not shown). The results showed that C. sakazakii O7 produced normal LPS, which indicated that this strain has a functional wzy gene (36). Similar arrangements occur in Salmonella serogroups A, B, and D, which have a functional wzy gene outside the O-antigen gene cluster (50).
In the present study, O-antigen gene clusters of C. sakazakii O4 and E. coli O103 were observed to share an overall DNA identity of 60 to 74% (Fig. 1B), which is lower than the identities between housekeeping genes of bacteria (79 to 92%) (data not shown). The O antigen is exposed on the cell surface and undergo higher selection pressure for better adaptation to the situation; therefore, the genes in the O-antigen gene cluster were found to diverge at a higher rate than the housekeeping genes (42). If the overall levels of similarity in O-antigen gene clusters are higher than that for housekeeping genes, the O antigen may arise as a result of acquisition of the gene cluster by interspecies lateral transfer since species divergence (42). However, in the present study, the O-antigen gene clusters had a lower level of identity than did the housekeeping genes. Two possible explanations were proposed for this case. One is that the O-antigen gene cluster was present in the common ancestral species, and the O-antigen genes diverged at a high rate than the housekeeping genes. The other is that the O-antigen gene cluster was acquired by one species from the other for a long time and was undergoing higher selection pressure than were the housekeeping genes. Most O antigens are distinct among different genus. Therefore, it is proposed that this O-antigen form is highly adaptive in C. sakazakii and E. coli, which occupy different niches, and may be important for the pathogenicity of these strains.
The O-antigen gene clusters of C. sakazakii O6 and C. malonaticus O2 were found to share high-level identity (99.3%), which may imply recent lateral transfer of the respective O-antigen gene cluster between these species. Similar cases have also been reported in E. coli strains (53). The O-antigen gene clusters shared by C. sakazakii O3 and C. muytjensii O1, C. turicensis O1, and C. malonaticus O1 have also been recently reported (18). However, these pairs of O-antigen gene clusters exhibit a lower identity of 91%, in the range of or lower than the corresponding housekeeping gene identities (89 to 91% and 93 to 94%, respectively) (data not shown). These data indicate that the O-antigen gene clusters could not be evolved from recent lateral transfer. It is likely that these two O-antigen forms have the same case with that for C. sakazakii O4 and E. coli O103 discussed above.
Development of molecular serotyping methods is important for epidemiological surveillance of C. sakazakii, which is becoming an emerging food-borne pathogen infecting humans, especially infants. Some O-antigen genes, such as wzx and wzy, are highly specific to individual O serotypes, thus providing the potential for use in molecular assay for rapid identification and serotyping of relevant strains (51), which has been proved in the identification of C. sakazakii serotypes O1, O2, and O3 (22, 34). Many PCR-based typing methods targeting these O-antigen-specific genes have proved to be reliable, rapid, and sensitive for identifying isolates of E. coli, Shigella, and Salmonella from clinical, food, and environmental samples (7, 8, 14). In the present study, C. sakazakii O4 to O7 specific genes were identified, and O1 to O7 specific primers based on the targeted genes were designed, synthesized, and screened. The specific gene-based PCR serotyping method used here has been shown to be accurate and sensitive (22) and therefore provides a potential method that is suitable for serotyping C. sakazakii O1 to O7 strains.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National 973 Program of China grants 2012CB721001, 2012CB721101, and 2011CB504900; National Natural Science Foundation of China (NSFC) key program grant 31030002; NSFC General Program grants 31000044 and 81171524; National 863 Program of China grant 2011BAK10B02; Fundamental Research Funds for the Central Universities (65010721, 65020121, and 65020061); Research Fund for the Doctoral Program of Higher Education of China (grant 20090031120023); and National Key Programs for Infectious Diseases of China (2009ZX10004-108).
Footnotes
Published ahead of print 23 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alexander DC, Valvano MA. 1994. Role of the rfe gene in the biosynthesis of the Escherichia coli O7-specific lipopolysaccharide and other O-specific polysaccharides containing N-acetylglucosamine. J. Bacteriol. 176:7079–7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arbatsky NP, et al. 2011. Elucidation of the structure and characterization of the gene cluster of the O-antigen of Cronobacter sakazakii G2592, the reference strain of C. sakazakii O7 serotype. Carbohydr. Res. 346:1169–1172 [DOI] [PubMed] [Google Scholar]
- 3. Baldwin A, et al. 2009. Multilocus sequence typing of Cronobacter sakazakii and Cronobacter malonaticus reveals stable clonal structures with clinical significance which do not correlate with biotypes. BMC Microbiol. 9:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bengoechea JA, et al. 2002. Functional characterization of Gne (UDP-N-acetylglucosamine-4-epimerase), Wzz (chain length determinant), and Wzy (O-antigen polymerase) of Yersinia enterocolitica serotype O:8. J. Bacteriol. 184:4277–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benson DA, et al. 2000. GenBank. Nucleic Acids Res. 28:15–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caubilla-Barron J, et al. 2007. Genotypic and phenotypic analysis of Enterobacter sakazakii strains from an outbreak resulting in fatalities in a neonatal intensive care unit in France. J. Clin. Microbiol. 45:3979–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DebRoy C, et al. 2004. Detection of Escherichia coli serogroups O26 and O113 by PCR amplification of the wzx and wzy genes. Appl. Environ. Microbiol. 70:1830–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Feng L, et al. 2004. Identification of Escherichia coli O114 O-antigen gene cluster and development of an O114 serogroup-specific PCR assay. J. Clin. Microbiol. 42:3799–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Finn RD, et al. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247–D251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fratamico PM, DebRoy C, Strobaugh TP, Jr, Chen CY. 2005. DNA sequence of the Escherichia coli O103 O antigen gene cluster and detection of enterohemorrhagic E. coli O103 by PCR amplification of the wzx and wzy genes. Can. J. Microbiol. 51:515–522 [DOI] [PubMed] [Google Scholar]
- 11. Giovannini M, et al. 2008. Enterobacter sakazakii: an emerging problem in paediatric nutrition. J. Int. Med. Res. 36:394–399 [DOI] [PubMed] [Google Scholar]
- 12. Giraud MF, et al. 1999. Purification, crystallization, and preliminary structural studies of dTDP-6-deoxy-d-xylo-4-hexulose 3,5-epimerase (RmlC), the third enzyme of the dTDP-l-rhamnose synthesis pathway, from Salmonella enterica serovar Typhimurium. Acta Crystallogr. D Biol. Crystallogr. 55:706–708 [DOI] [PubMed] [Google Scholar]
- 13. Giraud MF, et al. 1999. Overexpression, purification, crystallization and preliminary structural study of dTDP-6-deoxy-l-lyxo-4-hexulose reductase (RmlD), the fourth enzyme of the dTDP-l-rhamnose synthesis pathway, from Salmonella enterica serovar Typhimurium. Acta Crystallogr. D Biol. Crystallogr. 55:2043–2046 [DOI] [PubMed] [Google Scholar]
- 14. Han W, et al. 2007. DNA microarray-based identification of serogroups and virulence gene patterns of Escherichia coli isolates associated with porcine postweaning diarrhea and edema disease. Appl. Environ. Microbiol. 73:4082–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Healy B, et al. 2010. Cronobacter (Enterobacter sakazakii): an opportunistic foodborne pathogen. Foodborne Pathog. Dis. 7:339–350 [DOI] [PubMed] [Google Scholar]
- 16. Healy B, et al. 2008. Evaluation of an automated repetitive sequence-based PCR system for subtyping Enterobacter sakazakii. J. Food Prot. 71:1372–1378 [DOI] [PubMed] [Google Scholar]
- 17. Herikstad H, Motarjemi Y, Tauxe RV. 2002. Salmonella surveillance: a global survey of public health serotyping. Epidemiol. Infect. 129:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hobbs M, Reeves PR. 1994. The JUMPstart sequence: a 39-bp element common to several polysaccharide gene clusters. Mol. Microbiol. 12:855–856 [DOI] [PubMed] [Google Scholar]
- 19. Huan PT, Whittle BL, Bastin DA, Lindberg AA, Verma NK. 1997. Shigella flexneri type-specific antigen V: cloning, sequencing and characterization of the glucosyl transferase gene of temperate bacteriophage SfV. Gene 195:207–216 [DOI] [PubMed] [Google Scholar]
- 20. Iversen C, et al. 2007. The taxonomy of Enterobacter sakazakii: proposal of a new genus Cronobacter gen. nov. and descriptions of Cronobacter sakazakii comb. nov. Cronobacter sakazakii subsp. sakazakii, comb. nov., Cronobacter sakazakii subsp. malonaticus subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., and Cronobacter genomospecies 1. BMC Evol. Biol. 7:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iversen C, et al. 2008. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov., and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int. J. Syst. Evol. Microbiol. 58:1442–1447 [DOI] [PubMed] [Google Scholar]
- 22. Jarvis KG, et al. 2011. Molecular characterization of Cronobacter lipopolysaccharide O-antigen gene clusters and development of serotype-specific PCR assays. Appl. Environ. Microbiol. 77:4017–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Joseph S, Forsythe SJ. 2011. Predominance of Cronobacter sakazakii sequence type 4 in neonatal infections. Emerg. Infect. Dis. 17:1713–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a. Joseph S, et al. 2011. Cronobacter condimenti sp. nov., isolated from spiced meat and Cronobacter universalis sp. nov., a novel species designation for Cronobacter sp. genomospecies 1, recovered from a leg infection, water, and food ingredients. Int. J. Syst. Evol. Microbiol. 9:225. [DOI] [PubMed] [Google Scholar]
- 24. Kneidinger B, et al. 2003. Three highly conserved proteins catalyze the conversion of UDP-N-acetyl-d-glucosamine to precursors for the biosynthesis of O antigen in Pseudomonas aeruginosa O11 and capsule in Staphylococcus aureus type 5: implications for the UDP-N-acetyl-l-fucosamine biosynthetic pathway. J. Biol. Chem. 278:3615–3627 [DOI] [PubMed] [Google Scholar]
- 25. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 26. Kucerova E, et al. 2010. Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS One 5:e9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuhnert P, Korczak BM, Stephan R, Joosten H, Iversen C. 2009. Phylogeny and prediction of genetic similarity of Cronobacter and related taxa by multilocus sequence analysis (MLSA). Int. J. Food Microbiol. 136:152–158 [DOI] [PubMed] [Google Scholar]
- 28. Lai KK. 2001. Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine (Baltimore) 80:113–122 [DOI] [PubMed] [Google Scholar]
- 29. Lindberg AA, Wollin R, Gemski P, Wohlhieter JA. 1978. Interaction between bacteriophage Sf6 and Shigella flexneri. J. Virol. 27:38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu B, et al. 2008. Structure and genetics of Shigella O antigens. FEMS Microbiol. Rev. 32:627–653 [DOI] [PubMed] [Google Scholar]
- 31. Liu D, Cole RA, Reeves PR. 1996. An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J. Bacteriol. 178:2102–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marolda CL, Valvano MA. 1998. Promoter region of the Escherichia coli O7-specific lipopolysaccharide gene cluster: structural and functional characterization of an upstream untranslated mRNA sequence. J. Bacteriol. 180:3070–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miled-Bennour R, et al. 2010. Genotypic and phenotypic characterisation of a collection of Cronobacter (Enterobacter sakazakii) isolates. Int. J. Food Microbiol. 139:116–125 [DOI] [PubMed] [Google Scholar]
- 34. Mullane N, et al. 2008. Molecular analysis of the Enterobacter sakazakii O-antigen gene locus. Appl. Environ. Microbiol. 74:3783–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mullane NR, et al. 2007. Enterobacter sakazakii an emerging bacterial pathogen with implications for infant health. Minerva Pediatr. 59:137–148 [PubMed] [Google Scholar]
- 36. Nakhamchik A, Wilde C, Rowe-Magnus DA. 2007. Identification of a Wzy polymerase required for group IV capsular polysaccharide and lipopolysaccharide biosynthesis in Vibrio vulnificus. Infect. Immun. 75:5550–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pfostl A, et al. 2008. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-α-d-glucose. Biochem. J. 410:187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pupo GM, Lan R, Reeves PR. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. U. S. A. 97:10567–10572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reeves PP, Wang L. 2002. Genomic organization of LPS-specific loci. Curr. Top. Microbiol. Immunol. 264:109–135 [PubMed] [Google Scholar]
- 41. Rutherford K, et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 42. Samuel G, Hogbin JP, Wang L, Reeves PR. 2004. Relationships of the Escherichia coli O157, O111, and O55 O-antigen gene clusters with those of Salmonella enterica and Citrobacter freundii, which express identical O antigens. J. Bacteriol. 186:6536–6543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Samuel G, Reeves P. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338:2503–2519 [DOI] [PubMed] [Google Scholar]
- 44. Shashkov AS, Arbatsky NP, Knirel YA. 2011. Structures and genetics of Kdo-containing O-antigens of Cronobacter sakazakii G2706 and G2704, the reference strains of serotypes O5 and O6. Carbohydr. Res. 346:1924–1929 [DOI] [PubMed] [Google Scholar]
- 45. Staden R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5:233–241 [DOI] [PubMed] [Google Scholar]
- 46. Sun Y, et al. 2011. Development of an O-antigen serotyping scheme for Cronobacter sakazakii. Appl. Environ. Microbiol. 77:2209–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reference deleted.
- 48. Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637 [DOI] [PubMed] [Google Scholar]
- 49. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang L, Andrianopoulos K, Liu D, Popoff MY, Reeves PR. 2002. Extensive variation in the O-antigen gene cluster within one Salmonella enterica serogroup reveals an unexpected complex history. J. Bacteriol. 184:1669–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang L, Reeves PR. 1998. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect. Immun. 66:3545–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang L, Wang Q, Reeves PR. 2010. The variation of O antigens in gram-negative bacteria. Subcell. Biochem. 53:123–152 [DOI] [PubMed] [Google Scholar]
- 53. Wang Q, et al. 2009. Genetic and structural analyses of Escherichia coli O107 and O117 O-antigens. FEMS Immunol. Med. Microbiol. 55:47–54 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.