Abstract
Sake yeast cells have defective entry into the quiescent state, allowing them to sustain high fermentation rates. To reveal the underlying mechanism, we investigated the PAS kinase Rim15p, which orchestrates initiation of the quiescence program in Saccharomyces cerevisiae. We found that Rim15p is truncated at the carboxyl terminus in modern sake yeast strains as a result of a frameshift mutation. Introduction of this mutation or deletion of the full-length RIM15 gene in a laboratory strain led to a defective stress response, decreased synthesis of the storage carbohydrates trehalose and glycogen, and impaired G1 arrest, which together closely resemble the characteristic phenotypes of sake yeast. Notably, expression of a functional RIM15 gene in a modern sake strain suppressed all of these phenotypes, demonstrating that dysfunction of Rim15p prevents sake yeast cells from entering quiescence. Moreover, loss of Rim15p or its downstream targets Igo1p and Igo2p remarkably improved the fermentation rate in a laboratory strain. This finding verified that Rim15p-mediated entry into quiescence plays pivotal roles in the inhibition of ethanol fermentation. Taken together, our results suggest that the loss-of-function mutation in the RIM15 gene may be the key genetic determinant of the increased ethanol production rates in modern sake yeast strains.
INTRODUCTION
Sake yeast strains, which belong to the budding yeast species Saccharomyces cerevisiae, produce much more ethanol during sake fermentation than any other type of S. cerevisiae strain. Since rapid and high-level ethanol accumulation is desirable for sake brewing in order to shorten fermentation periods and inhibit the growth of undesirable microorganisms, yeast strains with higher fermentation rates have been historically selected as sake yeast. In the past 80 years, a group of genetically closely related modern sake yeast strains (also referred to as the “K7 group” [4]) that exhibit high fermentation rates in sake mash have been isolated. However, the genetic determinants of ethanol production by these modern strains are not completely clear.
In order to elucidate the molecular mechanisms responsible for the excellent fermentation properties of modern sake yeast strains, gene expression profiling has been used to identify the relevant genes and pathways (31, 39, 44). For example, our recent analyses revealed that the stress-responsive transcription factors Msn2p and Msn4p (Msn2/4p) (19, 30) and Hsf1p (32) are significantly inactivated in modern sake yeast cells during fermentation (22, 39). Consistent with these findings, modern sake strains are more sensitive to ethanol stress and heat shock than laboratory strains (35). In spite of the multiple extracellular stresses experienced during fermentation, such as low temperature, low pH, and high ethanol concentrations, increased stress tolerance of yeast cells appears to interfere with rapid ethanol fermentation. In fact, dysfunction of Msn2/4p or Hsf1p positively contributes to fermentation rates (22, 39), at least partly through the modification of glucose metabolism, including the reduced synthesis of storage (e.g., trehalose and glycogen) and structural (e.g., cell wall β-glucans) carbohydrates (D. Watanabe, A. Hirata, Y. Ohya, Y. Fujitani, C. Noguchi, T. Akao, and H. Shimoi, submitted for publication). Although these findings have provided novel insights into the in vivo regulatory mechanisms of ethanol fermentation, how to orchestrate these mechanisms still needs to be elucidated.
Identification of the mutations causing inactivation of Msn2/4p and Hsf1p may help reveal the origins of modern sake yeast strains. Whole-genome sequencing of sake yeast strain Kyokai no. 7 (K7) (1) has facilitated the identification of several sake yeast-specific chromosomal mutations and deletions. Using the available sequence, we recently revealed that the lack of a 2.6-kb DNA region, including the protein phosphatase gene PPT1, is closely associated with the constitutive inhibitory hyperphosphorylation of Hsf1p in modern sake yeast (1, 22). In addition, we also discovered a modern sake strain-specific loss-of-function mutation in the MSN4 gene (msn4C1540T) (39). This mutation, however, does not appear to be solely responsible for the inactivation of Msn2/4p, because K7 still expresses functional Msn2p (40, 41). Furthermore, deletion of the MSN4 gene in S. cerevisiae laboratory strains only modestly increases their fermentation rates (39), indicating that the loss of Msn4p is in itself insufficient to cause the high fermentation rate observed in modern sake yeast. Taken together, these data suggest the existence of an alternative factor that inhibits Msn2/4p activities and contributes to the brewing properties of sake yeast more significantly than the msn4C1540T mutation.
Acquisition of stress tolerance is one of the most outstanding features of yeast cells upon entry into the quiescent state. Quiescent yeast cells display numerous hallmark phenotypes that differentiate them from proliferating cells, including growth arrest as unbudded cells, trehalose and glycogen accumulation, cell wall thickening, increased buoyant density, and elevated resistance to a variety of stresses (2, 10). Notably, sake yeast cells exhibit severe impairments in all of these physiological changes observed in quiescent cells (35, 37; Watanabe et al., submitted), clearly demonstrating that sake yeast strains have defective entry into quiescence during fermentation. Another distinctive feature of quiescent yeast cells is a decreased metabolic rate in conjunction with their decreased proliferation rate. On the basis of a previous report on the decreased glycolytic/fermentation flux of quiescent-like yeast cells during bioethanol production (6), the quiescence deficiency of sake yeast strains appears to be responsible for their increased ethanol production rates.
Recent studies indicate that entry into quiescence is triggered by specialized signaling cascades rather than by passive cellular adaptation (10). In S. cerevisiae, upon nutrient starvation and/or stress conditions, inactivation of nutrient-sensory kinases (i.e., protein kinase A [PKA], TORC1, Sch9p, and the Pho85p/Pho80p complex) leads to activation of the PAS kinase Rim15p, resulting in induction of the quiescent program via Msn2/4p- and Gis1p-mediated gene expression (8, 26, 29, 33, 36). Although the underlying mechanisms by which Rim15p controls these transcription factors remain obscure, Talarek et al. (34) revealed that Rim15p coordinates the transcription of several Msn2/4p- and Gis1p-dependent genes through direct phosphorylation of the α-endosulfine family proteins Igo1p and Igo2p. Therefore, Rim15p acts as the master regulator of initiation of the yeast quiescent program. Accordingly, loss of Rim15p causes a pleiotropic phenotype characterized by defects in several cellular processes, including trehalose and glycogen accumulation, transcriptional derepression of nutrient-regulated genes, induction of thermotolerance and starvation resistance, chronological life span extension, and G1 arrest (8, 26, 29, 42).
Here we investigated the effects of a mutation in the RIM15 gene on the quiescent state of modern sake yeast cells. Our results shed further light on the close relationship between ethanol fermentation and Rim15p-mediated entry into quiescence. The results of this study, together with our previous findings on the defective stress responses of sake yeast, may provide important clues for the genetic improvement of industrial yeast strains toward high-speed ethanol production.
MATERIALS AND METHODS
Strains and plasmids.
S. cerevisiae sake yeast strains Kyokai no. 1 to 15 (K1 to K15) and Kyokai no. 701 (K701; a nonfoaming variant of K7 [24]) were provided by the Brewing Society of Japan. K701 UT-1T (MATa/MATα TRP1/trp1 ura3/ura3) (15) was a kind gift from K. Kitamoto, University of Tokyo. One sake yeast strain (Yabe Kozai), two shochu strains (S-2 and SH-4), two wine strains (Montrachet and OC-2), and four other alcoholic strains (Bu9-7, S-3, RIB1022, and RIB1023) were provided by the National Research Institute of Brewing (Japan). Beer yeast strains (NCYC240, NCYC242, and NCYC1026) were provided by the National Collection of Yeast Cultures (United Kingdom). Laboratory S. cerevisiae strains X2180 and Σ1278b were obtained from the American Type Culture Collection. S. cerevisiae strains BY4741, BY4742, BY4743, and BY4743 Δrim15 were provided by EUROSCARF (Germany). All yeast strains were routinely grown in liquid YPD medium (1% yeast extract, 2% peptone, and 2% glucose) at 25°C unless otherwise stated.
For the construction of the pAUR112-ScRIM15 plasmid, the RIM15 gene was amplified by high-fidelity PCR using KOD Plus DNA polymerase (Toyobo) and X2180 genomic DNA as the template with primers RIM15-F (5′-GGTAGGGATAGCCAAGTTCGAT-3′) and RIM15-R (5′-GGGAGAACTATTCTTCCAGAGG-3′) and was then cloned into the SmaI site of pAUR112 (TaKaRa). Plasmids pAUR112 and pAUR112-ScRIM15 were used to transform K701 UT-1T so as to generate K701 UT-1T(pAUR112) (designated K701 + vector) and K701 UT-1T(pAUR112-ScRIM15) (designated K701 + ScRIM15), respectively. Positive transformants were selected on SD-Ura plates, which consisted of 0.67% yeast nitrogen base without amino acids, 2% glucose, 0.077% complete supplement mixture (CSM)-Ura (Qbiogene), and 2% agar.
The rim155055insA frameshift mutation was introduced into BY4741 and BY4742 by use of a PCR-based mutagenesis method (9) with primers RIM15-REC-DF (5′-GTTAGTAGAGCCACTAGTGGTGTAAGTTTTGACTTAATTATGACAAGCCTTGAAGCTTCCAAAACTTGGTGCTATTGATGACGTACGCTGCAGGTCGAC-3′; the underlined and italicized nucleotides correspond to a single adenine insertion mutation and a double stop codon used to truncate the carboxyl-terminal 75 amino acids of Rim15p, respectively) and RIM15-DR (5′-TTTTTATTCAGTTATTTTTTTTAATTATCTTTATCTTAAAATTTAATCGATGAATTCGAGCTCG-3′) and with plasmid pFA6-kanMX4 (9) as the template to generate strains BY4741 rim155055insA-kanMX and BY4742 rim155055insA-kanMX, respectively. By use of this method, the product of rim155055insA-kanMX had a C terminus identical to that of K7 Rim15p (Fig. 1A). The correct insertions were confirmed by PCR using primers RIM15-REC-F (5′-TGTTTGTGAGCCTATACCGA-3′) and RIM15-R (5′-CTCTAACAAAGGAGAATATATATACG-3′) and sequence analysis. The two strains were mated to generate BY4743 rim155055insA-kanMX/rim155055insA-kanMX (referred to below as BY4743 rim155055insA).
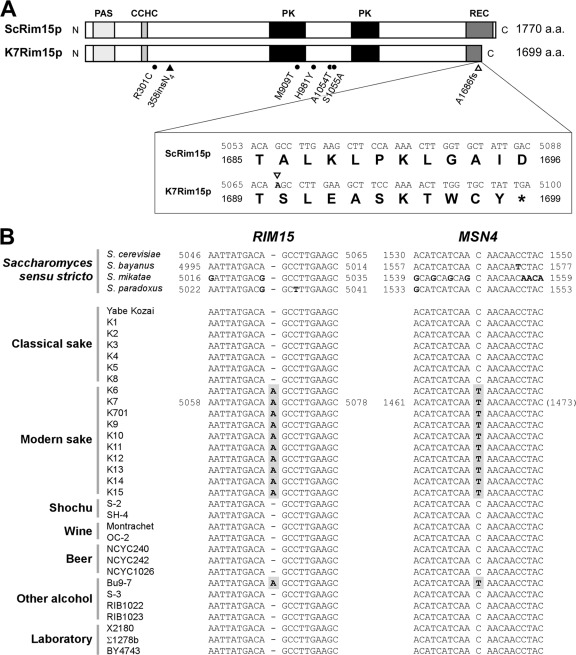
Fig 1.
A frameshift mutation in the RIM15 gene specific to modern sake yeast strains. (A) Comparison between laboratory strain S288C Rim15p (ScRim15p) and the putative RIM15 gene product of sake yeast strain K7 (K7Rim15p). Rectangles with light, medium, and dark shading and filled rectangles represent the PAS, CCHC-type zinc finger, receiver (REC), and protein kinase (PK) catalytic domains, respectively. Nucleotide and amino acid sequences surrounding the frameshift mutation in K7 (A1686fs; corresponding to nucleotides A5053 to C5088 in the ScRIM15 gene) are also shown. Filled circles and filled and open triangles indicate the positions of single amino acid substitutions, insertion of four asparagine residues, and the single-base frameshift mutation, respectively. (B) Distribution of the frameshift mutation in the RIM15 gene (rim155055insA) and the modern sake yeast-specific loss-of-function mutation in the MSN4 gene (msn4C1540T) (39) among Saccharomyces cerevisiae and Saccharomyces sensu stricto strains. Nucleotides that differ from the DNA sequence of S. cerevisiae strain S288C (uppermost) are indicated by boldface. The rim155055insA and msn4C1540T mutations are highlighted in gray.
Thermotolerance test.
Thermotolerance tests were performed as described previously (35). Briefly, yeast cells were precultured overnight in YPD (for BY4743 and its derivative mutants) or SD-Ura (for K701 UT-1T strains carrying pAUR112-based plasmids) medium at 25°C, transferred to YPD medium, grown aerobically at 25°C, and then incubated at a high temperature (50°C for logarithmic-phase cells or 54°C for stationary-phase cells). The number of surviving cells was determined by counting CFU on YPD plates.
Flow cytometric analysis.
Logarithmically growing cells were collected and transferred to liquid YPD medium containing 200 ng/ml rapamycin (Calbiochem). Cell fixation, staining, and flow cytometry analysis were performed as described previously (37).
Determination of intracellular trehalose and glycogen levels.
Intracellular trehalose and glycogen were quantified using previously described methods with minor modifications (25). Briefly, yeast cells (corresponding to an optical density at 660 nm [OD660] of 15 to 30) were frozen in liquid nitrogen and were stored at −80°C until use. Upon thawing, the yeast cells were mixed with glass beads and 250 μl of 250 mM Na2CO3 in a microtube, which was then vigorously agitated for 3 min at 4°C. The resulting lysates were brought to pH 5.2 by the addition of acetic acid and were adjusted to 500 μl by adding an appropriate volume of 200 mM sodium acetate (pH 5.2). Half of the suspension was incubated with trehalase (Roche) at 37°C overnight, and the remainder was incubated with amyloglucosidase (Roche) at 57°C overnight. Glucose in the reaction mixture was directly measured using a glucose CII test kit (Wako). The values for glycogen and trehalose concentrations are reported as glucose equivalents per OD660 unit.
Fermentation monitoring.
Fermentation tests in liquid YPD medium were performed as reported previously (39). Briefly, yeast cells were precultured overnight in YPD medium at 30°C, harvested, inoculated into YPD medium containing 20% glucose at a final OD660 of 0.1, and then further incubated at 30°C for 5 days without shaking. For sake fermentation assays, a single-step sake mash was prepared by mixing 40 g pregelatinized rice, 10 g dried koji (rice cultivated with Aspergillus oryzae), 20 μl 90% lactic acid, and 80 ml water containing yeast cells at a final OD660 of 1.0 and was then incubated at 15°C for 20 days without shaking, as described previously (38). Fermentation was continuously monitored by measuring the volume of evolved carbon dioxide using a Fermograph II apparatus (Atto), as described previously (38).
RESULTS
A frameshift mutation in the RIM15 gene is distributed specifically in modern sake yeast strains.
To determine whether differences in Rim15p exist between S. cerevisiae sake yeast strains and other yeast strains, we compared the nucleotide sequence of RIM15 in the modern sake yeast strain K7 (K7RIM15) to that in laboratory strain S288C (ScRIM15) by using the sequence available in the Sake Yeast Genome Database (SYGD) (http://nribf1.nrib.go.jp/SYGD/) (1). The open reading frame of the K7RIM15 gene is composed of 5,100 nucleotides and contains 34 single nucleotide substitutions, 5 of which are nonsynonymous (R301C, M909T, H981Y, A1054T, and S1055A), an insertion of 4 AAT trinucleotide repeats (358insN4), and the insertion of a single adenine nucleotide immediately after A5067 (corresponding to A5055 in ScRIM15) compared to ScRIM15 (Fig. 1A). The frameshift mutation at nucleotide position 5068 (named A1686fs or 5055insA) is predicted to lead to a premature stop codon that shortens the K7RIM15 gene product by 75 amino acids in the C-terminal region. Since the insertion of an adenine nucleotide at the same site in laboratory strain BY4743 resulted in a rapamycin-sensitive growth phenotype, similar to that observed in the BY4743 Δrim15 disruptant (46) (see Fig. S1 in the supplemental material), we anticipated that the function of Rim15p might be severely impaired or lost in K7 due to the 5055insA mutation.
We next examined the distribution of the 5055insA mutation among 17 sake, 2 shochu, 2 wine, 3 beer, 4 other alcohol, and 3 laboratory yeast strains (Fig. 1B). We had demonstrated previously that the msn4C1540T loss-of-function mutation is specifically localized in genetically closely related modern sake yeast strains, including K6, K7, K701, and K9 to K15 (39). Similarly, all of the modern sake strains examined here were found to contain the rim155055insA mutation. In contrast, nearly all of the other yeast strains tested, including the classical sake yeast strains (K1 to K5, K8, and Yabe Kozai), as well as the shochu, wine, beer, and laboratory yeast strains, did not have this insertional frameshift mutation. Furthermore, sequence analysis using Fungal Sequence Alignment in the Saccharomyces Genome Database (SGD) (http://www.yeastgenome.org/) revealed that the rim155055insA mutation was not found in either Saccharomyces bayanus, S. mikatae, or S. paradoxus. Taken together, these findings indicated that the 5055insA mutation in the K7RIM15 gene represented a novel modern sake yeast-specific mutation.
Expression of functional Rim15p suppresses several physiological traits characteristic of quiescent sake yeast cells.
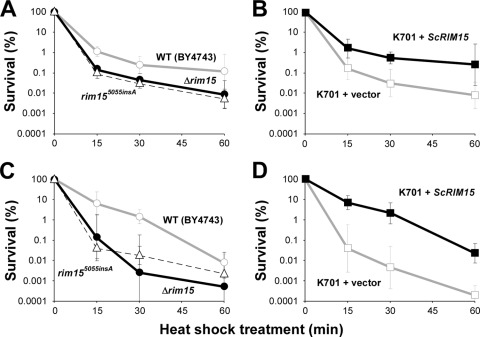
To confirm that the C-terminal truncation of Rim15p is associated with the deficient quiescence-related phenotypes of modern sake yeast, we analyzed the effects of functional RIM15 expression on the acquisition of stress tolerance. In accordance with previous reports (28, 35), a strain BY4743 Δrim15 disruptant and a sake strain (K701 + vector) both exhibited lower thermotolerance than the wild-type strain BY4743 during the logarithmic- and stationary-growth phases (Fig. 2). However, expression of the ScRIM15 gene in the sake yeast strain fully rescued the defective temperature tolerance, as indicated by the increased survival rates of stationary-phase cells after a 15-min heat shock (5.94% for BY4743, 0.13% for BY4743 Δrim15, 0.04% for K701 + vector, and 6.66% for K701 + ScRIM15). This result suggested that the low stress tolerance of sake yeast might be caused primarily by the loss of Rim15p activity.
Fig 2.
Loss of Rim15p results in reduced thermotolerance of sake yeast. Strains BY4743 (open circles), BY4743 Δrim15 (solid circles), BY4743 rim155055insA (open triangles), and K701 UT-1T carrying an empty vector (open squares) or pAUR112-ScRIM15 (filled squares) were grown aerobically in YPD liquid medium. Yeast cells collected in the logarithmic phase (OD660, 1) (A and B) and stationary phase (7 days after inoculation) (C and D) were subjected to a heat shock for 60 min, and the number of surviving cells was scored on YPD plates. The graphs represent the mean values from two or more independent experiments; error bars indicate standard deviations.
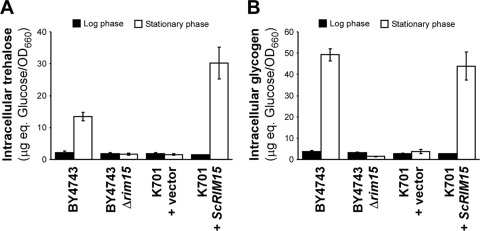
A previous study has shown that RIM15 deletion also causes significant defects in the accumulation of trehalose and glycogen during the stationary phase (26). Similarly, sake yeast cells do not induce trehalose or glycogen synthesis during sake fermentation (Watanabe et al., submitted). Here we also found that the synthesis of both storage carbohydrates was markedly inhibited in the K701 + vector strain in the stationary phase of YPD cultures (Fig. 3). In contrast, the expression of functional ScRIM15 in K701 increased the intracellular quantities of both trehalose and glycogen to levels similar to those observed in wild-type BY4743 cells, demonstrating that the expression of defective Rim15p in sake yeast was also responsible for the decreased accumulation of these storage carbohydrates during stationary phase.
Fig 3.
Loss of Rim15p is responsible for the decreased accumulation of storage carbohydrates in sake yeast. Intracellular trehalose (A) and glycogen (B) levels in the indicated YPD-grown yeast strains were measured during the logarithmic phase (OD660, 1) (filled bars) or stationary phase (7 days after inoculation) (open bars). Data represent the mean values for three independent experiments; error bars indicate standard deviations.
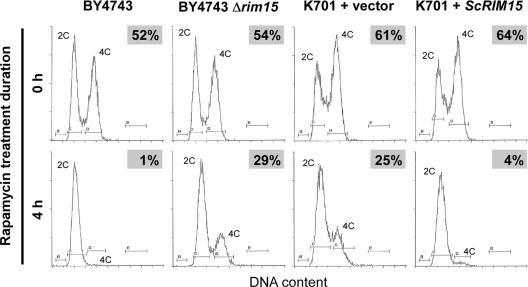
Inactivation of the TORC1 kinase by rapamycin treatment causes multiple quiescent-like phenotypes in S. cerevisiae, including normal G1 arrest, via Rim15p activity. Thus, the deletion of RIM15 inhibits G1 arrest in the presence of rapamycin (26). Consistent with this finding, we have recently shown that sake yeast cells exhibit less-effective G1 arrest during fermentation (37). Together, these studies raise the possibility that defective cell cycle regulation in sake yeast is also triggered by deficient Rim15p functioning. To test this speculation, we assayed rapamycin-induced G1 arrest efficiency in laboratory and sake strains (Fig. 4). Although wild-type BY4743 exhibited almost complete G1 arrest following a 4-h rapamycin treatment, strains BY4743 Δrim15 and K701 + vector both exhibited obvious defects in G1 arrest. Since the K701 + ScRIM15 strain recovered an effective G1 arrest, it appears that the dysfunction of Rim15p in sake yeast likely accounts for the defects in rapamycin-induced G1 arrest.
Fig 4.
Rim15p dysfunction is associated with less-effective G1 arrest in sake yeast cells treated with rapamycin. DNA content frequency histograms determined by flow cytometry are shown for the yeast strains indicated. The relative percentage of cells in the G2/M phase with 4C DNA content is given at the top right of each panel.
Dysfunction of the Rim15p-mediated quiescence program increases ethanol fermentation rates.
Our previous studies showed that defects in stress responses, carbohydrate metabolism, and G1 arrest lead to rapid ethanol fermentation (22, 37, 39; Watanabe et al., submitted). Because all of these physiological defects are closely associated with Rim15p dysfunction, we hypothesized that the loss of Rim15p might facilitate the fermentation properties of sake yeast cells. As shown by the CO2 emission data in Fig. 5A, the BY4743 Δrim15 disruptant exhibited markedly faster fermentation in YPD medium containing 20% glucose than wild-type BY4743 (peak CO2 emission rates, 180.8 ± 11.5 ml/6 h for BY4743 versus 235.3 ± 18.1 ml/6 h for BY4743 Δrim15). Furthermore, the disruption of RIM15 dramatically promoted ethanol fermentation in sake mash (Fig. 5B) (peak CO2 emission rates, 90.8 ± 2.5 ml/6 h for BY4743 versus 142.7 ± 2.8 ml/6 h for BY4743 Δrim15). We also confirmed that the ethanol concentration after 20 days of sake fermentation was significantly higher in the finished sake made from BY4743 Δrim15 (17.03% ± 0.44% by volume) than in that made from wild-type BY4743 (11.19% ± 0.17% by volume) (P = 0.011). In addition, disruption of the IGO1 and IGO2 genes, which encode the only known direct targets of Rim15p (17, 34), raised the fermentation rate in 20% glucose-containing YPD medium (Fig. 5C) (peak CO2 emission rates, 153.5 ± 8.8 ml/6 h for BY4743 versus 189.5 ± 0.3 ml/6 h for BY4743 Δigo1 and 171.5 ± 6.0 ml/6 h for BY4743 Δigo2). Taken together, these results clearly demonstrated that dysfunction of Rim15p and its downstream pathway leads to a striking increase in ethanol fermentation rates.
Fig 5.
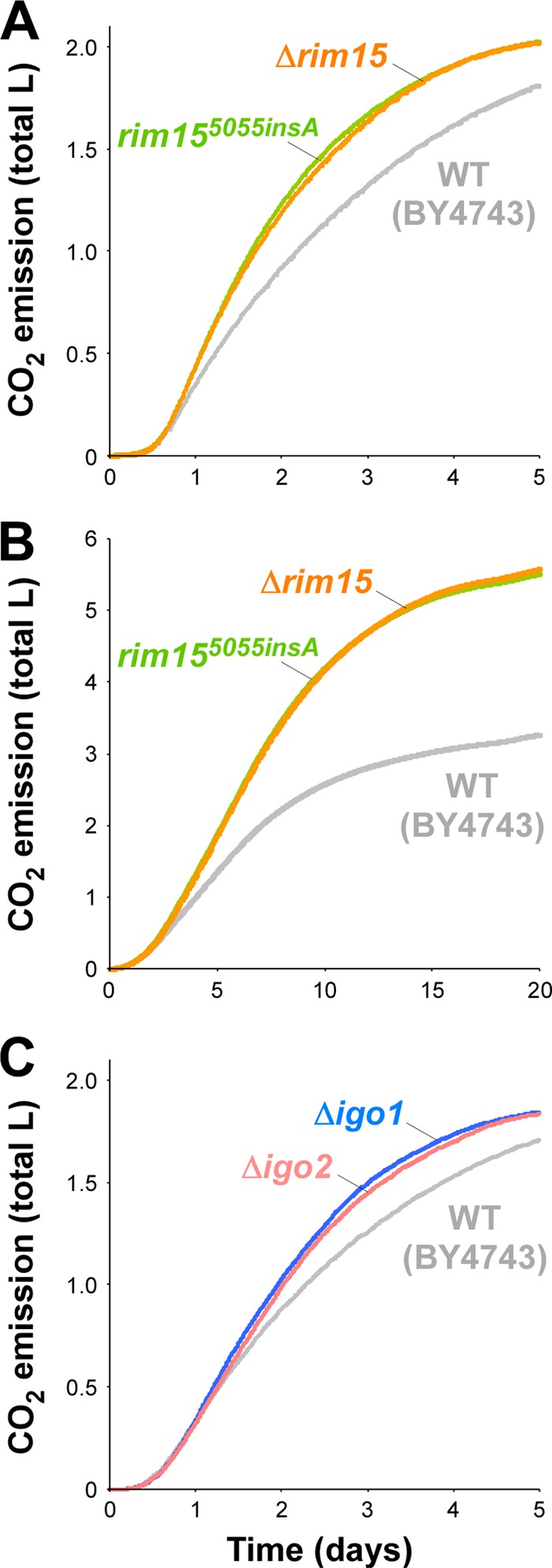
Improvement of the fermentation rate by loss of the Rim15p-mediated quiescence induction pathway. (A) Fermentation tests in YPD medium containing 20% glucose using BY4743 (wild type [WT]) (gray line) and its Δrim15 (orange line) and rim155055insA (green line) mutants. (B) Small-scale sake fermentation tests using BY4743 (WT) and its Δrim15 and rim155055insA mutants. (C) Fermentation tests in YPD medium containing 20% glucose using BY4743 (WT) (gray line) and its Δigo1 (blue line) and Δigo2 (pink line) mutants. Averaged data from two or more independent experiments that yielded similar results are shown.
Notably, the expression of functional RIM15 in K701 did not have a significant effect on ethanol fermentation. As shown in Fig. 6A, expression of a plasmid-borne ScRIM15 gene in K701 only modestly decreased the fermentation rate in YPD medium containing 20% glucose (peak CO2 emission rates, 223.9 ± 7.1 ml/6 h for K701 + vector versus 214.6 ± 3.3 ml/6 h for K701 + ScRIM15). In addition, the viability of the K701 + ScRIM15 cells was significantly higher than that of the K701 + vector cells after a 20-day sake fermentation (Table 1), confirming that the expression of ScRIM15 elevated the stress tolerance of K701 in sake mash, as was observed under heat shock (Fig. 2B and D). Nevertheless, the K701 + ScRIM15 strain exhibited a sake fermentation rate similar to that of the K701 + vector strain (Fig. 6B) (peak CO2 emission rates, 184.0 ± 4.1 ml/6 h for K701 + vector versus 181.9 ± 2.7 ml/6 h for K701 + ScRIM15) and produced almost the same level of ethanol as this control strain (Table 1) (P = 0.074).
Fig 6.
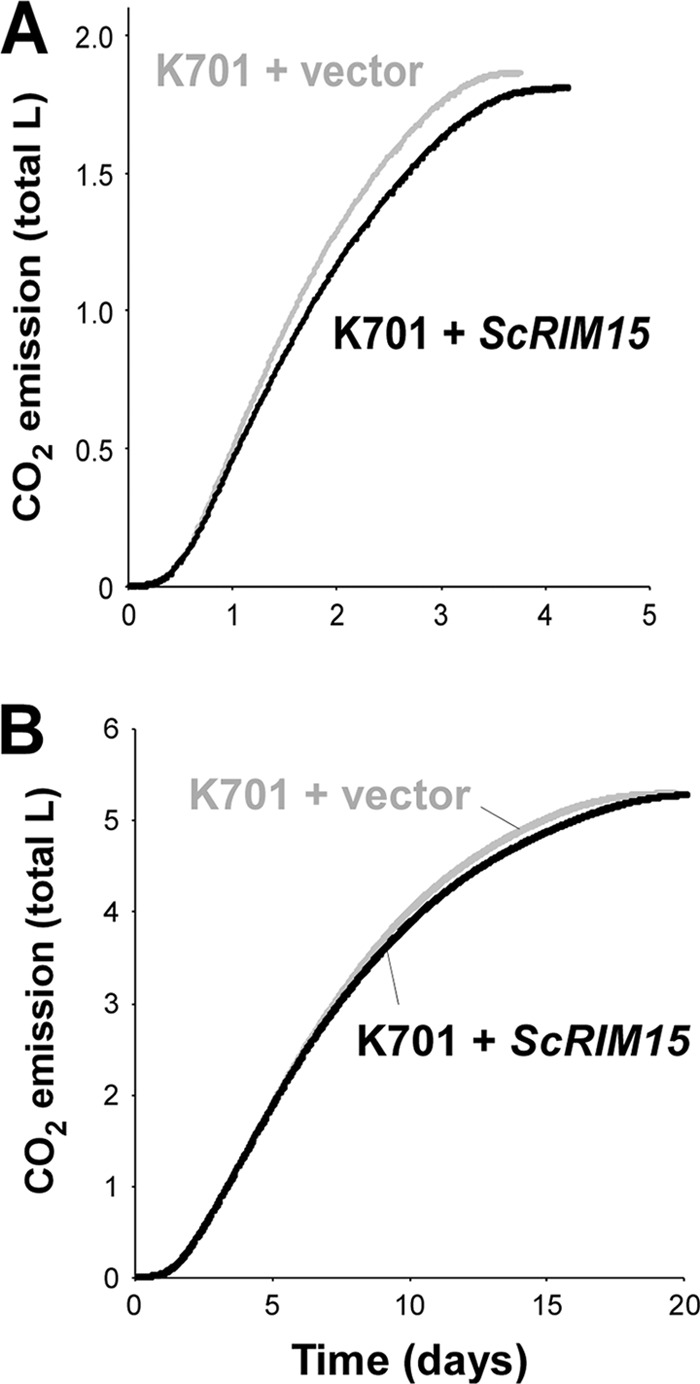
Effects of ScRIM15 expression on the fermentation rate of K701. (A) Fermentation tests in YPD medium containing 20% glucose using the K701 + vector (gray line) and K701 + ScRIM15 (black line) strains. (B) Small-scale sake fermentation tests using the K701 + vector and K701 + ScRIM15 strains. Averaged data from two or more independent experiments that yielded similar results are presented.
Table 1.
Effects of expression of a functional RIM15 gene on the sake brewing characteristics of a modern sake yeast straina
| Characteristic | Value (mean ± SD) |
|
|---|---|---|
| K701 + vector | K701 + ScRIM15 | |
| Vol of ethanol (%) | 19.55 ± 0.25 | 19.20 ± 0.28 |
| Sp gr (15°C/4°C) | 1.0086 ± 0.0020 | 1.0083 ± 0.0018 |
| Concn (ppm) of: | ||
| Ethyl acetate | 147.28 ± 4.65 | 147.34 ± 6.33 |
| Ethyl caproate | 0.53 ± 0.05 | 0.52 ± 0.01 |
| Isoamyl acetate | 9.66 ± 0.27 | 8.39 ± 0.29d |
| Acid levelb | 3.10 ± 0.12 | 3.43 ± 0.11e |
| Concn (mg/liter) of: | ||
| Citric acid | 131.7 ± 2.7 | 134.8 ± 7.0 |
| Malic acid | 494.0 ± 17.7 | 732.2 ± 51.8e |
| Succinic acid | 754.5 ± 18.0 | 806.8 ± 27.0e |
| Lactic acid | 553.0 ± 14.3 | 539.9 ± 26.1 |
| Amino acid levelc | 2.29 ± 0.06 | 2.09 ± 0.11d |
| % Dead cells in the sake mash after a 20-day fermentation | 49.5 ± 2.3 | 32.8 ± 3.1d |
Analyzed as reported previously (13).
Sake samples (10 ml) were titrated to pH 7.2 with a 0.1 N NaOH solution. The acid level is defined as the number of milliliters of the 0.1 N NaOH solution required for titration.
After determination of the acid level, the neutralized sake samples were adjusted to pH 8.2 with a 0.1 N NaOH solution and were mixed with 5 ml of a 50% formalin solution (pH 8.2). The mixtures were again titrated to pH 7.2 with a 0.1 N NaOH solution. The amino acid level is defined as the number of milliliters of the 0.1 N NaOH solution required for titration.
Significantly lower than the value for the vector control (P, <0.05 by the Student t test).
Significantly higher than the value for the vector control (P, <0.05 by the Student t test).
The 5055insA mutation abolishes the functions of Rim15p related to stress response and fermentation control.
To identify the mutation responsible for the impaired Rim15p activity in modern sake yeast, we further investigated the effects of the modern sake yeast-specific rim155055insA mutation (Fig. 1B) on several physiological traits. As shown in Fig. 2A and C, the impairment of thermotolerance by rim155055insA was nearly indistinguishable from that by Δrim15 in both the logarithmic and stationary phases. Moreover, the rim155055insA mutant displayed improved fermentation profiles that were identical to those of the Δrim15 disruptant in YPD medium (peak CO2 emission rate, 232.8 ± 14.1 ml/6 h) and sake mash (peak CO2 emission rate, 142.4 ± 1.2 ml/6 h) compared to those of the wild-type strain BY4743 (Fig. 5A and B). The ethanol concentration of the resultant sake (16.76% ± 0.18% by volume) was not significantly different from that of sake made using the Δrim15 mutant (P, 0.269 by the Student t test). These results demonstrate that the 5055insA mutation likely led to a complete loss of Rim15p function related to stress response and fermentation control.
DISCUSSION
Sake strains of S. cerevisiae display fermentation properties superior to those of other types of strains and are considered to have evolved enhanced stress response machineries due to the multiple extracellular stresses experienced during fermentation. In the present study, however, we identified a modern sake yeast-specific loss-of-function mutation (5055insA) in the RIM15 gene (Fig. 1), which encodes a putative upstream activator of the stress-responsive transcription factors Msn2/4p and Gis1p (8, 42). This finding demonstrates that high stress tolerance is not required for efficient ethanol fermentation by sake yeast cells. The dysfunction of Rim15p alone accounted for both the quiescence-related deficient phenotypes (Fig. 2, 3, and 4) and the increased fermentation rates (Fig. 5) that are characteristic of modern sake yeast strains, suggesting that the rim155055insA mutation might have played a pivotal role in establishing the unique core physiological properties of modern sake yeast strains. Our findings, together with a previous report on the higher glycolytic/fermentation flux observed in non-quiescent-like yeast cells (6), indicate that yeast entry into quiescence is a major impediment to rapid ethanol fermentation. Therefore, genetic engineering of the Rim15p-mediated quiescence induction pathway represents a potential strategy for improving the ethanol production rates of other industrial yeast strains.
The identified loss-of-function insertion mutation in Rim15p resides in a C-terminal putative receiver domain (Fig. 1A) whose function is poorly understood. Receiver domains containing a conserved aspartic acid residue typically associate with molecules involved in histidine kinase phosphorelay signaling, which is used by bacteria, slime molds, plants, and fungi to sense and adapt to the extracellular environment (43, 45). For instance, the only known S. cerevisiae histidine kinase, Sln1p, is autophosphorylated under physiological osmotic conditions, resulting in increased phosphorylation of a conserved aspartic acid residue (D554) in the receiver protein Ssk1p. High osmolarity results in dephosphorylation of Sln1p and Ssk1p D554, leading to activation of the mitogen-activated protein kinase (MAPK) Hog1p and induction of target genes whose products protect against osmotic stress (5, 12, 18, 27). Another example of an S. cerevisiae receiver protein is the transcriptional activator Skn7p, whose phosphorylation at a conserved aspartic acid residue (D427) mediates gene expression in response to hypotonic, heat, and cell wall stresses (7, 11, 16, 20). However, phosphorylation of specific threonine residues in the receiver domain is required for its activation under oxidative stress (11, 21), raising the possibility that unidentified serine/threonine protein kinases might also contribute to the regulation of receiver proteins in S. cerevisiae. Because the receiver domain of Rim15p does not contain the aspartic acid residues corresponding to D554 in Ssk1p or D427 in Skn7p, the regulatory mechanism of Rim15p activity via its receiver protein remains unclear. Here we showed clearly that an insertion mutation in the receiver domain of Rim15p causes a complete loss of Rim15p function related to stress response (Fig. 2A and C) and fermentation control (Fig. 5A and B), demonstrating the critical importance of the Rim15p C-terminal domain for the first time. However, it is necessary to identify the specific protein interaction partners and/or phosphorylation sites of the Rim15p receiver domain in order to elucidate the detailed molecular mechanisms by which Rim15p mediates the entry of S. cerevisiae cells into quiescence.
How does the dysfunction of Rim15p in modern sake yeast strains result in increased ethanol production rates? The most plausible explanation is that the loss of Rim15p activity leads to elevated fermentation rates through inactivation of Msn2/4p. A previous study of the genomewide transcriptional profiles of Δrim15, Δmsn2/4, and Δgis1 cells revealed that the entire set of genes that require Rim15p for induction during diauxic shift is found within larger combined sets of Msn2/4p- and Gis1p-dependent genes (8), indicating that these transcription factors act as putative downstream effectors of Rim15p. Furthermore, we have reported that impaired Msn2/4p activities are partly responsible for the high fermentation rates of modern sake yeast strains (39). Inactivation of Msn2/4p, in concert with a defective Hsf1p-mediated ethanol stress response (22), contributes to the repression of genes related to stress-responsive carbohydrate metabolism, which diversifies the cellular glucose flux (Watanabe et al., submitted). In agreement with this phenomenon, we showed that the expression of a functional RIM15 gene in a modern sake yeast strain appears to recover the synthesis of the storage carbohydrates trehalose and glycogen (Fig. 3). We recognize, however, that inactivation of Msn2/4p alone is insufficient to cause the high fermentation rates in Rim15p-deficient strains, since a Δrim15 mutant strain displayed fermentation properties superior to that of a Δmsn2 Δmsn4 double disruptant (Fig. 5A and B) (39). We also found that Rim15p dysfunction also affects cell cycle regulation under rapamycin treatment in the present study (Fig. 4). Since G1 progression is involved in the regulation of fermentation rates (37), Rim15p dysfunction might partly contribute to rapid ethanol production through a decrease in G1 arrest efficiency during fermentation. In addition, we revealed that the only known targets of Rim15p, namely, Igo1p and Igo2p (17, 34), also inhibit fermentation (Fig. 5C), albeit by unknown mechanisms. To elucidate the complete regulatory mechanisms underlying Rim15p-mediated quiescence entry and ethanol fermentation, the downstream effectors of Rim15p and their roles in fermentation control should be investigated comprehensively.
Expression of a functional RIM15 gene fully suppressed all of the quiescence-related deficient phenotypes of sake yeast (Fig. 2, 3, and 4) but did not significantly affect fermentation rates (Fig. 6). We also confirmed that the coexpression of functional RIM15 and MSN4 genes in strain K701 had only a minor effect on the fermentation profile (data not shown). Together, these data suggest that RIM15, MSN4, and their yet unidentified target(s) responsible for the shutdown of fermentation activity in response to stress signals might have become pseudogenes during the sake yeast domestication process. Our established K701 + ScRIM15 strain thus exhibited enhanced stress tolerance and remarkable sake-brewing characteristics (Table 1). Notably, the expression of ScRIM15 did not alter the levels of ethanol or ethyl esters, which are the main contributors to sake aromas. We also found that the use of K701 + ScRIM15 decreased the proportion of dead cells in sake mash containing >19% ethanol by volume, and thus, the amino acid content derived from yeast cell lysates, which often affects the quality of sake negatively, was significantly reduced. Moreover, ScRIM15 expression contributed to the increased acid level in the finished sake, primarily through elevation of the malic acid level, which may impart a pleasant sourness to sake. The breeding of high-malic-acid-producing yeast strains is one of the central issues in the sake industry (3, 14, 23). Considering the close association between malic acid production and stress responses (23), Rim15p-mediated stress signaling might be responsible for the observed enhancement of malic acid productivity. Based on the data presented in this study, we propose a novel and potentially effective method for sake yeast breeding, in which a functional RIM15 gene is stably integrated into the chromosomes of modern sake strains. Further elucidation of the mechanisms underlying Rim15p-mediated metabolic control is expected to provide the technical basis for improving the industrial production of ethanol and other useful compounds using yeast.
Supplementary Material
ACKNOWLEDGMENTS
We greatly appreciate the gift of the K701 UT-1T yeast strain from K. Kitamoto (University of Tokyo, Tokyo, Japan).
This work was supported by the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry and by a Noda Institute for Scientific Research (NISR) Young Investigator Grant.
Footnotes
Published ahead of print 23 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Akao T, et al. 2011. Whole-genome sequencing of sake yeast Saccharomyces cerevisiae Kyokai no. 7. DNA Res. 18:423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allen C, et al. 2006. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 174:89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Asano T, Kurose N, Tarumi S. 2001. Isolation of high-malate-producing sake yeasts from low-maltose-assimilating mutants. J. Biosci. Bioeng. 92:429–433 [DOI] [PubMed] [Google Scholar]
- 4. Azumi M, Goto-Yamamoto N. 2001. AFLP analysis of type strains and laboratory and industrial strains of Saccharomyces sensu stricto and its application to phenetic clustering. Yeast 18:1145–1154 [DOI] [PubMed] [Google Scholar]
- 5. Bahn YS. 2008. Master and commander in fungal pathogens: the two-component system and the HOG signaling pathway. Eukaryot. Cell 7:2017–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benbadis L, Cot M, Rigoulet M, François J. 2009. Isolation of two cell populations from yeast during high-level alcoholic fermentation that resemble quiescent and nonquiescent cells from the stationary phase on glucose. FEMS Yeast Res. 9:1172–1186 [DOI] [PubMed] [Google Scholar]
- 7. Brown JL, Bussey H, Stewart RC. 1994. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 13:5186–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cameroni E, Hulo N, Roosen J, Winderickx J, De Virgilio C. 2004. The novel yeast PAS kinase Rim15 orchestrates G0-associated antioxidant defense mechanisms. Cell Cycle 3:462–468 [PubMed] [Google Scholar]
- 9. Goldstein AL, McCusker JH. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541–1553 [DOI] [PubMed] [Google Scholar]
- 10. Gray JV, et al. 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68:187–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He XJ, Mulford KE, Fassler JS. 2009. Oxidative stress function of the Saccharomyces cerevisiae Skn7 receiver domain. Eukaryot. Cell 8:768–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaserer AO, Andi B, Cook PF, West AH. 2009. Effects of osmolytes on the SLN1-YPD1-SSK1 phosphorelay system from Saccharomyces cerevisiae. Biochemistry 48:8044–8050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Katou T, Kitagaki H, Akao T, Shimoi H. 2008. Brewing characteristics of haploid strains isolated from sake yeast Kyokai no. 7. Yeast 25:799–807 [DOI] [PubMed] [Google Scholar]
- 14. Kitagaki H, Kato T, Isogai A, Mikami S, Shimoi H. 2008. Inhibition of mitochondrial fragmentation during sake brewing causes high malate production in sake yeast. J. Biosci. Bioeng. 105:675–678 [DOI] [PubMed] [Google Scholar]
- 15. Kitamoto K, Oda K, Gomi K, Takahashi K. 1990. Construction of uracil and tryptophan auxotrophic mutants from sake yeasts by disruption of URA3 and TRP1 genes. Agric. Biol. Chem. 54:2979–2987 [Google Scholar]
- 16. Li S, et al. 2002. The eukaryotic two-component histidine kinase Sln1p regulates OCH1 via the transcription factor, Skn7p. Mol. Biol. Cell 13:412–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luo X, Talarek N, De Virgilio C. 2011. Initiation of the yeast G0 program requires Igo1 and Igo2, which antagonize activation of decapping of specific nutrient-regulated mRNAs. RNA Biol. 8:14–17 [DOI] [PubMed] [Google Scholar]
- 18. Maeda T, Wurgler-Murphy SM, Saito H. 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369:242–245 [DOI] [PubMed] [Google Scholar]
- 19. Martínez-Pastor MT, et al. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15:2227–2235 [PMC free article] [PubMed] [Google Scholar]
- 20. Morgan BA, Bouquin N, Merrill GF, Johnston LH. 1995. A yeast transcription factor bypassing the requirement for SBF and DSC1/MBF in budding yeast has homology to bacterial signal transduction proteins. EMBO J. 14:5679–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morgan BA, et al. 1997. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16:1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Noguchi C, Watanabe D, Zhou Y, Akao T, Shimoi H. 2012. Association of constitutive hyperphosphorylation of Hsf1p with a defective ethanol stress response in Saccharomyces cerevisiae sake yeast strains. Appl. Environ. Microbiol. 78:385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oba T, et al. 2011. Properties of high malic acid-producing strains of Saccharomyces cerevisiae isolated from sake mash. Biosci. Biotechnol. Biochem. 75:2025–2029 [DOI] [PubMed] [Google Scholar]
- 24. Ouchi K, Akiyama H. 1971. Non-foaming mutants of sake yeast: selection by cell agglutination method and by froth flotation method. Agric. Biol. Chem. 35:1024–1032 [Google Scholar]
- 25. Parrou JL, François J. 1997. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal. Biochem. 248:186–188 [DOI] [PubMed] [Google Scholar]
- 26. Pedruzzi I, et al. 2003. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell 12:1607–1613 [DOI] [PubMed] [Google Scholar]
- 27. Posas F, et al. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865–875 [DOI] [PubMed] [Google Scholar]
- 28. Reinders A, Bürckert N, Boller T, Wiemken A, De Virgilio C. 1998. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev. 12:2943–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roosen J, et al. 2005. PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Mol. Microbiol. 55:862–880 [DOI] [PubMed] [Google Scholar]
- 30. Schmitt AP, McEntee K. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 93:5777–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shobayashi M, Ukena E, Fujii T, Iefuji H. 2007. Genome-wide expression profile of sake brewing yeast under shaking and static conditions. Biosci. Biotechnol. Biochem. 71:323–335 [DOI] [PubMed] [Google Scholar]
- 32. Sorger PK, Pelham HR. 1987. Purification and characterization of a heat-shock element binding protein from yeast. EMBO J. 6:3035–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Swinnen E, et al. 2006. Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae. Cell Div. 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Talarek N, et al. 2010. Initiation of the TORC1-regulated G0 program requires Igo1/2, which license specific mRNAs to evade degradation via the 5′-3′ mRNA decay pathway. Mol. Cell 38:345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Urbanczyk H, et al. 2011. Sake yeast strains have difficulty in entering a quiescent state after cell growth cessation. J. Biosci. Bioeng. 112:44–48 [DOI] [PubMed] [Google Scholar]
- 36. Wanke V, Pedruzzi I, Cameroni E, Dubouloz F, De Virgilio C. 2005. Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. EMBO J. 24:4271–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watanabe D, et al. 2011. Ethanol fermentation driven by elevated expression of the G1 cyclin gene CLN3 in sake yeast. J. Biosci. Bioeng. 112:577–582 [DOI] [PubMed] [Google Scholar]
- 38. Watanabe D, Ota T, Nitta F, Akao T, Shimoi H. 2011. Automatic measurement of sake fermentation kinetics using a multi-channel gas monitor system. J. Biosci. Bioeng. 112:54–57 [DOI] [PubMed] [Google Scholar]
- 39. Watanabe D, et al. 2011. Enhancement of the initial rate of ethanol fermentation due to dysfunction of yeast stress response components Msn2p and/or Msn4p. Appl. Environ. Microbiol. 77:934–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Watanabe M, et al. 2007. Elevated expression of genes under the control of stress response element (STRE) and Msn2p in an ethanol-tolerance sake yeast Kyokai no. 11. J. Biosci. Bioeng. 104:163–170 [DOI] [PubMed] [Google Scholar]
- 41. Watanabe M, Watanabe D, Akao T, Shimoi H. 2009. Overexpression of MSN2 in a sake yeast strain promotes ethanol tolerance and increases ethanol production in sake brewing. J. Biosci. Bioeng. 107:516–518 [DOI] [PubMed] [Google Scholar]
- 42. Wei M, et al. 2008. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 4:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wolanin PM, Thomason PA, Stock JB. 2002. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 3:REVIEWS3013.1–3013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu H, et al. 2006. Global gene expression analysis of yeast cells during sake brewing. Appl. Environ. Microbiol. 72:7353–7358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wuichet K, Cantwell BJ, Zhulin IB. 2010. Evolution and phyletic distribution of two-component signal transduction systems. Curr. Opin. Microbiol. 13:219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xie MW, et al. 2005. Insights into TOR function and rapamycin response: chemical genomic profiling by using a high-density cell array method. Proc. Natl. Acad. Sci. U. S. A. 102:7215–7220 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.