Abstract
Eukaryotic marine microbes play pivotal roles in biogeochemical nutrient cycling and ecosystem function, but studies that focus on the protistan biogeography and genetic diversity lag-behind studies of other microbes. 18S rRNA PCR amplification and clone library sequencing are commonly used to assess diversity that is culture independent. However, molecular methods are not without potential biases and artifacts. In this study, we compare the community composition of clone libraries generated from the same water sample collected at the San Pedro Ocean Time Series (SPOTs) station in the northwest Pacific Ocean. Community composition was assessed using different cell lysis methods (chemical and mechanical) and the extraction of different nucleic acids (DNA and RNA reverse transcribed to cDNA) to build Sanger ABI clone libraries. We describe specific biases for ecologically important phylogenetic groups resulting from differences in nucleic acid extraction methods that will inform future designs of eukaryotic diversity studies, regardless of the target sequencing platform planned.
INTRODUCTION
Biologists who sample natural microbial communities face challenges in estimating a community's true diversity through population subsamples. Since the late 1980s, culture-independent PCR-amplified clone libraries that target the small subunit (SSU) 16S rRNA taxonomic genes have served as a proxy for the diversity and composition of natural microbial bacterial and archaeal populations (15, 16, 33a). However, studies of microbial eukaryotic populations have lagged behind those of their generally smaller prokaryote counterparts (7, 25). More recently, natural assemblages of microbial eukaryotes have been assessed and compared using the SSU 18S rRNA gene. These culture-independent gene surveys have revealed extensive microbial diversity that was previously undetected with culture-dependent methods and morphological identification (10, 13, 28, 33, 44).
The sampling of the 18S rRNA gene or gene transcript is commonplace for studies of microbial eukaryotes, but several factors can complicate or bias diversity assessments (1, 37, 40, 42). The resulting analysis of community composition can be dependent on the filter size fraction, lysis method, whether DNA or RNA is targeted, the PCR primers used, and the PCR thermal cycling regimen (2). Each of these different steps can affect library diversity and skew results away from the true community composition. For example, a comparison of the contribution of five marine stramenopile (MAST) groups to the total sample using a PCR-amplified library versus fluorescence in situ hybridization (FISH) showed that the PCR primers that targeted the full 18S rRNA gene overestimated two groups and underestimate one group, whereas the primer set that only targeted a portion of the gene gave a more accurate representation of the actual abundance of each group (29). Different primer sets designed to target a particular group of organisms can also retrieve sequences with very little overlap (40). Additionally, both chimeric sequences (4) and intraindividual ribosomal RNA polymorphisms (reviewed by Richards and Bass [37]) can artificially increase diversity.
While PCR bias has received some attention and has been studied quantitatively, there are still other unknown biases in sequencing libraries that have not been fully evaluated, especially for microbial eukaryotes. The purpose of this study is to tease out the biasing effects on community composition specifically due to the use of different extraction methods (mechanical versus chemical) and the extraction of different nucleic acids (DNA versus RNA). Methodological decisions for extraction methodologies take place prior to library construction, so results should be informative no matter what type of downstream sequencing platform is planned.
MATERIALS AND METHODS
Sample collection.
Seawater was collected at the San Pedro Ocean Time Series (SPOTs) station off the coast of southern California (33°33′N, 118°24′W) on 17 April 2009. This site is the focus of a long-term USC Microbial Observatory study and is well characterized (10). Five hundred liters of seawater was collected from the surface (5 m) and prefiltered using a 20-μm Nitex mesh into acid-washed and preconditioned carboys. The seawater sample was sequentially filtered through 142-mm 3.0-μm Versapor and 0.8- and 0.1-μm Supor impact membrane filters (Pall Life Sciences). Filters were frozen at −80°C immediately after filtration.
Nucleic acid extraction.
We used two nucleic extraction methods for both the 3.0- and 0.8-μm-size fraction filters. Replicate filters were either extracted by mechanical lysis (ML) or by a gentler chemical lysis (CL).
Mechanical lysis.
DNA and RNA were extracted using the PowerSoil total RNA extraction kit and the DNA elution accessory kit (MoBio, Carlsbad, CA) according to the manufacturer's protocol. Eluted ML DNA was treated with RNase One (Promega, Madison, WI) in a 10-μl reaction mixture comprised of 1 μl enzyme (10 U), 1 μl reaction buffer, and 8 μl extracted DNA. The reaction mixture was incubated at 37°C for 10 min. The enzyme, salts, and oligonucleotides then were removed using the DNA Clean and Concentrator-5 kit (Zymo, Orange, CA), and the eluted DNA was used in subsequent PCRs.
Chemical lysis.
Total nucleic acids were extracted by Amplicon Express using a method based on Miller et al. (32) and Howe (http://hg.wustl.edu/hdk_lab_manual/yeast/yeast4.html), with additional phenol-chloroform extractions and ethanol washes. RNA samples derived from both ML and CL were treated in a 10-μl reaction mixture containing 1 μl RQ1 RNase-free DNase (1U) (Promega, Madison, WI), 1 μl reaction buffer, and 8 μl of extracted RNA and incubated at 37°C for 10 min. After adding 1 μl of stop solution, the mixture was incubated for an additional 10 min at 65°C to inactivate the enzyme. The treated RNA was purified using the RNeasy MinElute cleanup kit (Qiagen, Valencia, CA). This procedure was repeated until a PCR to check for the presence of 18S DNA was negative. The RNA sample was then reverse transcribed into cDNA using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) using random decamers (Integrated DNA Technologies) as the primer. The resulting cDNA was used in the PCR amplification reaction.
DNA and cDNA library construction and sequencing. (i) Mechanical lysis library.
18S SSU rRNA genes were amplified from DNA and cDNA using universal eukaryotic primers Euk-A (5′-AACCTGGTTGATCCTGCCAGT-3′) and Euk-B (5′-GATCCTTCTGCAGGTTCACCTAC-3′) (31). The manufacturer's protocol for GoTaq polymerase (Promega) was modified in the following way to obtain optimal amplifications. The final concentrations in each 50-μl PCR were 0.5 mM each primer, 1× Promega buffer B, 2.5 mM Promega MgCl2, 250 mM Promega deoxynucleoside triphosphates (dNTPs), 2.5 U of Promega Taq in buffer B, and 1 to 2 μl of DNA extract in 50-ml reaction volumes. The thermal cycling protocol consisted of an initial denaturation step for 2 min at 95°C, followed by 25 to 35 cycles of 30 s at 95°C, 30 s at 55°C, 2 min at 72°C, and then a final extension step for 7 min at 72°C. Four to five replicate PCRs were pooled for the subsequent cloning reaction.
PCR products were separated on a 1.5% agarose gel, and the products of the expected size (∼1,800 bp) were excised from the gel. DNA was purified from the gel slices using the QIAquick gel purification kit (Qiagen) according to the manufacturer's protocol. The eluted DNA was further cleaned and concentrated using the Clean and Concentrator-5 kit (Zymo). The purified PCR products were ligated into the pCR2.1-TOPO vector (Invitrogen). Products were then purified again, mixed with TOP10-competent cells (Invitrogen), and then electroporated in a cuvette using a BTX ECM 399 electroporator (Holliston, MA). After shocking, sterile SOC medium (MP Biomedicals, Salon, OH) was added to the cell suspension. The mixture was transferred to a 14-ml cell culture tube and incubated at 37°C for 1 h with shaking at 250 rpm. Subsequently, the bacterial clones were plated and picked according to a standard protocol. Two 96-well plates from each library were sequenced using the Euk-570F primer (5′-GTAATTCCAGCTCCAATAGC-3′) (43) on an ABI 377 Sanger sequencer.
(ii) Chemical lysis library.
The CL DNA libraries were constructed by Amplicon Express with the following deviations from the methods described above. The libraries were transformed into Invitrogen DH10B T1r electrocompetent cells. Clones (n = 768) were picked from each library and arrayed into 3,384-well plates using a Genetix Qpix. DNA was extracted for 192 of these clones from each library and sequenced using an ABI 377 Sanger sequencer with the Euk-570F primer.
Community richness and diversity analyses. (i) Sequence preprocessing.
Raw Sanger sequences were trimmed using Phred (14, 15) with the default error probability cutoff of 0.05. Trimmed sequences were screened for chimeras using a local implementation of pintail (3) that used a subset of the aligned SILVA 18S data set (version 10.4) (36) as the reference for calculating the expected percent differences. The deviation from expectation (DE) statistic was manually examined in each of the samples to determine the appropriate cutoff value for each sample. Of the initial 1,152 clones sequenced, 209 (18%) were suspected to be chimeras and were excluded from subsequent analyses.
(ii) OTU calling and taxonomic assignment.
Sequences from the six clone libraries were combined and classified into operational taxonomic units (OTUs) using the Microbial Eukaryote Species Assignment (MESA) program (5) at a sequence similarity cutoff of 95%. Using BLAST, the sequences in each OTU were searched against the curated SILVA eukaryotic small subunit database (36) and assigned to the highest common taxonomic level of the component sequences.
(iii) Diversity analyses.
The Species Prediction and Diversity Estimation (SPADE) program (8) was used to estimate commonly used nonparametric alpha diversity statistics (the inverse of the Simpson index [Ds−1], bias-corrected Chao1, and ACE1) for the total sample and for individual libraries.
(iv) Phylogenetic trees.
Maximum-likelihood trees of groups of interest were constructed using the PhyML plug-in (21) in Geneious. The Hasegawa-Kishino-Yano substitution model was used (24). One hundred bootstraps were generated with the following parameters: proportion of invariable sites, 0; number of substitution rate categories, 1.
RESULTS
Six libraries were constructed, and a total of 1,152 sequences were obtained. After removing low-quality and potentially chimeric sequences as well as 7 metazoan clones, 936 clones with an average length of 643 bp were retained for subsequent analysis. When all sequences were pooled, the 936 clones were grouped into 126 microbial eukaryotic OTUs using the MESA OTU-calling algorithm at a 95% sequence similarity cutoff (5). Diversity (richness) estimates (Ds−1, Chao1, and ACE1) for total, individual, and grouped libraries were estimated (9, 27) (Table 1). The inverse of the Simpson index (Ds−1) is a common diversity statistic that can range from 1 to the maximum number of OTUs in each sample; the higher the value, the greater the diversity. This simple biodiversity statistic accounts for taxonomic richness (number of OTUs), evenness (relative abundance of sequences within each OTU), and the sequencing effort of the clone library (10). The Chao1-bc species richness estimate (also called the rarefaction estimator) uses a bias-corrected method to estimate missing OTUs calculated from the number of singletons and doubletons to estimate total OTUs in a sample. ACE1 is a nonparametric abundance-based coverage estimator for highly heterogeneous communities that uses rare OTUs to estimate the number of missing OTUs. This estimator corrects from the observed number of OTUs but is not independent of sample size.
Table 1.
Protistan diversity estimates for each clone library, sample, and combined clone librariesa (n = 936 quality-passed sequences; 126 total OTUs at 95% identity)
| Library analysis type | n | No. of OTUs | Ds−1 | Richness (95% CI) |
|
|---|---|---|---|---|---|
| Chao1-bc | ACE1 | ||||
| Individual clone libraries | |||||
| 1 | 116 | 41 | 17.4 | 58.3 (46.9, 91.6) | 92.4 (59.4, 184.9) |
| 2 | 159 | 46 | 11.9 | 108.1 (69.6, 209.7) | 254.7 (118.2, 649.9) |
| 3 | 178 | 42 | 10.4 | 126.3 (68.5, 309.9) | 116.2 (67.4, 259.2) |
| 4 | 161 | 37 | 8.7 | 47.5 (40.1, 72.5) | 58.2 (43.6, 105.7) |
| 5 | 146 | 45 | 7.2 | 107.1 (68.6, 208.7) | 225.8 (109.6, 550.8) |
| 6 | 176 | 44 | 12.6 | 94.6 (61.0, 194.3) | 105.8 (65.5, 221.7) |
| Total | 936 | 126 | 22.4 | 265.2 (197.3, 398.8) | 380.7 (257.7, 619.4) |
| Combined clone libraries | |||||
| 1 + 4 | 277 | 65 | 15.5 | 101.25 (79.6, 155.3) | 135.8 (94.8, 232.9) |
| 2 + 5 | 305 | 74 | 14.9 | 177.5 (119.8, 307.7) | 335.5 (186.1, 684.0) |
| 3 + 6 | 354 | 67 | 19.1 | 144.5 (96.0, 273.9) | 135.2 (95.0, 233.0) |
Analyses included the following: 1, 0.8-μm filter, mechanical lysis, cDNA; 2, 0.8-μm filter, mechanical lysis, DNA; 3, 0.8-μm filter, chemical lysis, DNA; 4, 3.0-μm filter, mechanical lysis, cDNA; 5, 3.0-μm filter, mechanical lysis, DNA; 6, 3.0-μm filter, chemical lysis, DNA. Combined libraries are indicated for ML cDNA, ML DNA, and CL DNA comparisons. Ds−1 is the inverse of the Simpson's index. Chao1 and ACE1 are nonparametric richness estimators. Rare OTUs were defined as those with just singletons and doubletons (bias-corrected Chao1) or those with ≤10 members (ACE1) (11).
Total protistan diversity as measured by both parametric and nonparametric indexes showed higher diversity (Ds−1 = 22.4, Chao1-bc = 265.2, and ACE1 = 380.7) than any individual or paired sample (Table 1). Rarefaction plots of Chao1 and the number of OTUs indicated that additional protistan diversity remained undetected (data not shown). However, using the inverse Simpson's index, our range of values for individual samples processed from one water sample was 7.2 to 17.4, which is comparable to other estimates of Ds−1 taken at the same site and depth as part of another 18S rRNA gene protistan diversity study during a 4-month period in 2001 (6.0 to 28.8) (10). Combined DNA and cDNA samples that were extracted using an ML method had lower values than the combined samples using a CL extraction method (Ds−1 = 14.9 and 15.5 versus 19.1) (Table 1).
Higher-taxonomic (OTU) classifications of the clone libraries from each of the 6 samples revealed differences in the relative abundance of the protistan assemblages (Table 2; also see Fig. S1 in the supplemental material). When all groups were evaluated together, the alveolates made up the largest group, comprising 36.3% of the total clones (22.9% syndiniales, 12.7% ciliates, and 0.8% dinoflagellates). The stramenopile group had the second highest number of sequences at 26.1%; it consisted of diatoms (15.1%), MAST (8.3%), and other groups (2.7%). The contribution of the subsequent groups to the combined sample dropped precipitously, with the groups telonema, chlorophytes, haptophytes, and chrysophytes accounting for 8.5, 7.8, 5.2, and 4.1%, respectively, of the total sample. These were followed by the picobiliphytes, cercozoa, and choanoflagellates, which contribute 3.8, 3.2, and 2.3%, respectively. Finally, the cryptophytes and other protists (centroheliozoa, cryomonadida, katablepharidophyta, and rhodophyta) made up the final 2.6% of the sample (Table 2).
Table 2.
Number of microbial eukaryote sequences by group for each sample typea
| Taxonomic group | ML cDNA |
ML DNA |
CL DNA |
Total |
||||
|---|---|---|---|---|---|---|---|---|
| No. of sequences | Relative distribution | No. of sequences | Relative distribution | No. of sequences | Relative distribution | No. of sequences | Relative distribution | |
| Alveolates | ||||||||
| Ciliates | 58 (3) | 20.9 | 55 (4) | 18.1 | 6 (2) | 1.7 | 119 (6) | 12.7 |
| Dinoflagellates | 0 (0) | 0.0 | 2 (2) | 0.7 | 5 (3) | 1.4 | 7 (4) | 0.8 |
| Syndiniales group I | 6 (3) | 2.2 | 65 (7) | 21.4 | 60 (5) | 16.9 | 131 (11) | 14.0 |
| Syndiniales group II | 1 (1) | 0.4 | 29 (7) | 9.5 | 52 (11) | 14.7 | 82 (12) | 8.8 |
| Syndiniales group V | 0 (0) | 0.0 | 1 (1) | 0.3 | 0 (0) | 0.0 | 1 (1) | 0.1 |
| Cercozoa | 19 (8) | 6.9 | 4 (3) | 1.3 | 7 (5) | 2.0 | 30 (12) | 3.2 |
| Chlorophytes | 17 (6) | 6.1 | 29 (7) | 9.5 | 27 (5) | 7.6 | 73 (9) | 7.8 |
| Choanoflagellates | 11 (5) | 4.0 | 8 (5) | 2.6 | 3 (2) | 0.8 | 22 (6) | 2.4 |
| Chrysophytes | 17 (6) | 6.1 | 7 (4) | 2.3 | 14 (4) | 4.0 | 38 (8) | 4.1 |
| Cryptophytes | 3 (2) | 1.1 | 2 (2) | 0.7 | 3 (1) | 0.8 | 8 (3) | 0.9 |
| Haptophytes | 20 (4) | 7.2 | 22 (6) | 7.2 | 7 (2) | 2.0 | 49 (7) | 5.2 |
| Stramenopiles | ||||||||
| Diatoms | 30 (6) | 10.8 | 25 (8) | 8.2 | 86 (9) | 24.3 | 141 (12) | 15.1 |
| MAST | 29 (7) | 10.5 | 10 (4) | 3.3 | 39 (10) | 11.0 | 78 (11) | 8.3 |
| Other | 9 (8) | 3.2 | 11 (8) | 3.6 | 5 (5) | 1.4 | 25 (17) | 2.7 |
| Picobiliphytes | 24 (2) | 8.7 | 8 (2) | 2.6 | 4 (1) | 1.1 | 36 (2) | 3.9 |
| Telonema | 25 (2) | 9.0 | 24 (2) | 7.9 | 30 (2) | 8.5 | 79 (2) | 8.5 |
| Other protists | 8 (2) | 2.9 | 2 (1) | 0.7 | 6 (3) | 1.7 | 16 (3) | 1.7 |
| Total | 277 (65) | 304 (73) | 354 (70) | 935 (126) | ||||
Relative distributions are provided as percent composition. ML, mechanical lysis extraction; CL, chemical lysis extraction. Numbers in parentheses indicate the number of OTUs at 95% similarity.
Comparison of taxonomic composition of DNA and cDNA libraries.
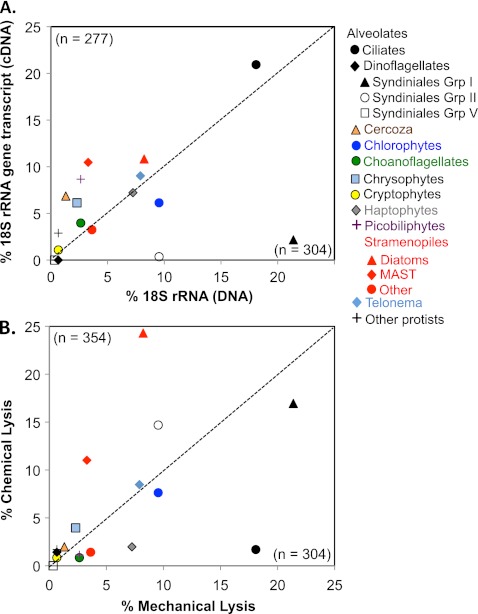
DNA samples extracted by ML had 304 clones (73 OTUs). RNA was reverse transcribed to cDNA and yielded 277 clones and 65 OTUs (Table 2). Forty-one OTUs were shared between the cDNA and DNA samples, while the rest were unique to each sample. Biases that could be attributed to nucleic acid (DNA versus RNA) are shown in Fig. 1A. The biggest difference between the two data sets was for a dominant novel alveolate, group I Syndiniales. This group was highly represented in the DNA library (21.4%) but not in the cDNA sample (0.4%). Syndiniales group II also showed higher representation in DNA than cDNA libraries (9.5 and 0.4%, respectively) (Table 2 and Fig. 1A). The ciliates were the next most dominant group and were almost equally represented in both DNA and cDNA libraries (18.1 and 20.9%, respectively). The MAST were slightly favored in cDNA libraries versus DNA libraries (10.5 and 3.3%, respectively), but other stramenopiles (including diatoms) were generally equally represented. Other groups that demonstrated increased representation in the cDNA sample compared to that in the DNA sample were the cercozoa (6.9 and 1.3%, respectively) and the picobiliphytes (8.7 and 2.6%, respectively). The contribution of the next most numerous groups dropped to less than 10% of the samples, making comparisons less clear. Chlorophytes, haptophytes, and telonema were all fairly equally represented. The clonal representation of the remaining groups was too low (<5%) to compare (Table 2 and Fig. 1A).
Fig 1.
Taxonomic ratio differences in DNA versus cDNA (A) and mechanical versus chemical lysis extraction methods (B) for specified clone libraries. The dotted line represents a 1:1 line. The numbers of sequences are included in the corner corresponding to the library type.
Comparison of taxonomic compositions of mechanical and chemical lysis libraries.
Taxonomic affiliations of clone libraries generated from DNA using an ML lysis method versus a CL extraction method from replicate filters were compared. There were 304 and 354 clones and 73 and 70 OTUs in the ML and CL samples, respectively. Among OTUs, 41 were common to both samples. Although the ML and CL clone libraries were constructed using the same seawater sample, each method produced a strikingly different picture of natural protistan diversity (Fig. 1B and Table 2), especially for ecologically important groups. In the largest groups, ciliates were overrepresented in ML samples (18.1% for ML and 1.7% for CL samples). Of the abundant stramenopiles, both diatoms and MAST groups were overrepresented in CL samples compared to ML samples (diatoms; 24.3 and 8.2%, respectively) and MAST (11.0 and 3.3%, respectively). The dominant group in the ML sample, syndiniales group I (21.4%), had a slightly decreased representation in the CL sample (16.9%). Other groups were represented at similar levels in both ML and CL groups (Fig. 1B and Table 2).
Phylogenetic affiliations of OTUs.
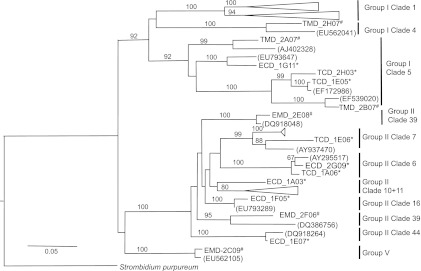
Maximum-likelihood trees were constructed for the major taxonomic groups to confirm the taxonomic affiliations of sequences obtained in this study and to investigate their phylogenetic diversity. Two are included here. Alveolate sequences belonged to five different groups: syndiniales groups I, II, and V, ciliates, and dinoflagellates. Although more sequences were affiliated with syndiniales group I (n = 131), they comprised only three previously defined (20) independent clades: clades 1, 4, and 5 (Fig. 2). Within the more diverse Syndiniales group II, 82 clones from this study belonged to seven independent clades. Only one sequence was retrieved that belonged to syndiniales group V, which is a less diverse group with fewer sequences retrieved thus far relative to groups I and II (20) (see Table S2 in the supplemental material). While the ML method was more successful at retrieving a diversity of clones than the CL method, there were a few OTUs that were only retrieved by the CL method.
Fig 2.
Maximum-likelihood tree of Syndiniales groups I, II, and V based on the analysis of partial 18S rRNA gene sequences. Representative sequences from this data set obtained by chemical lysis (*) or mechanical lysis (#) are shown. The ciliate Strombidium purpureum was used as an outgroup.
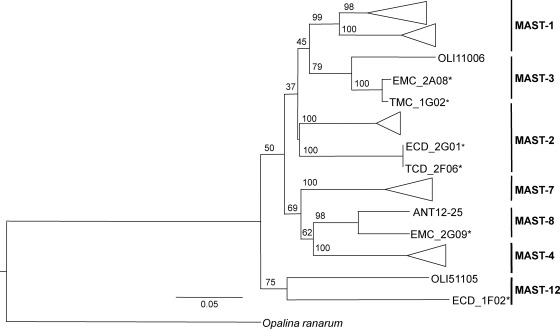
The MAST groups were also well represented in our samples. Sequences from 7 out of the 12 previously defined clusters (29) were retrieved. MAST-1 comprised 2 distinct clades (Fig. 3), with each clade being formed by a different OTU. Each of the other clades were comprised of 1 OTU. The CL method was successful at retrieving a few more OTUs than the ML method, which did not produce any unique OTUs (see Table S2 in the supplemental material).
Fig 3.
Maximum-likelihood tree of marine stramenopile (MAST) groups based on the analysis of partial 18S rRNA sequences. Only representative sequences from this study (indicated by asterisks) are shown on this condensed tree. Clones beginning with EMC and TMC were extracted by mechanical lysis. Clones beginning with ECD and TCD were extracted by chemical lysis. Opalina ranarun, an opalinid, was used as an outgroup.
DISCUSSION
Microbial eukaryotes are ubiquitous in marine systems and crucial to the structure and function of ecosystems (7). They have important roles as part of the microbial loop as photosynthesizers, grazers, and remineralizers of nutrients (39). They also may lend resiliency to whole ecosystems in the face of changing conditions (6). The growing awareness of their importance has resulted in increased focus on studying community composition and distribution, and 18S rRNA gene surveys have provided an attractive alternative (or complement) to traditional microscopic assessments of eukaryotic communities. These surveys have revealed the presence of high-level taxonomic groups that are comprised mostly or entirely of uncultured environmental clones and a patchy distribution of communities.
Different PCR primers can bias results (30); however, less is known about the biases resulting from extraction methods and nucleic acid extraction types. Our results will be useful for researchers who are trying to compare past studies that have used different methods or want to avoid methodological biases that could mask diverse or transient assemblages. While many eukaryotic groups appeared to have similar representation in libraries, we found that a few ecologically significant group-specific biases can be introduced depending on the extraction method chosen.
The release of nucleic acids during extraction is dependent on the structure of the membranes. Rigorous lysis procedures may be required for some groups but detrimental to the target genes of other groups. For example, in our sample, ciliates were more represented in libraries generated using an ML method. Conversely, relatively few ciliate clones were observed in filters that were extracted with a chemical lysis method, while the stramenopiles (e.g., diatoms and MAST groups) are much more represented in libraries that have been extracted using chemical lysis. Diatoms are common members of the phytoplankton assemblages in southern California waters near the SPOTs sampling station, but their relative abundance in clone libraries often does not reflect typical numerical dominance over dinoflagellates during periods of the spring bloom (10), and it has been suggested that this is due to an extraction, amplification, or cloning bias. Our study results show a clear bias for extraction methods (CL versus ML), with chemical lysis yielding a higher proportion of (especially diatom) clones. To date, organisms within the MAST groups have not been cultured, and little is known about their morphology. However, a pelagic protist known as Solenicola setigera was observed to form colonies on the diatom Leptocylindrus mediterranus (24a). This Solenicola-Leptocylindrus consortium has been found to be ubiquitous in marine environments ranging from the tropical to Artic and Antarctic waters (18). Recently, the phylogenetic position of Leptocylindrus has been clarified and shown to branch within the MAST-3 group (19). If a similar lifestyle holds true for the MAST-3 and other MAST organisms at the study site, as diatoms are preferentially lysed by the CL method, any organism living in consortium with the diatoms may also show up at greater frequency. We also observed the cooccurrence of potentially different but closely related Solenicola strains in their sample of colonized diatoms; therefore, it is possible that ML will miss strains of commensals or parasites that are more closely attached to the host.
The last group that has biased representation was the syndiniales, a novel alveolate group related to dinoflagellates (20, 23). The syndiniales can be quite diverse (Fig. 2) and can contribute up to 50% of sequences retrieved in clone libraries from coastal and oceanic regions around the world (20, 38). Many are thought to be parasitic on other protists, zooplankton, and fish. In surface samples from SPOTs collected during a 4-month period in 2001, syndiniales contributed between 3.5 and 8.5% of the community (10). Syndiniales groups in our combined samples made up 22.9% of the sample but were more represented in 18S rRNA gene libraries than 18S rRNA gene transcript (cDNA) libraries. Overrepresentation of the Syndniales (novel marine alveolates) in rRNA-gene-versus-gene-transcript libraries was also seen in a study of Arctic picoeukaryotes (40a). However, they appeared to be more equally represented in both chemical and mechanical lysis extraction methods. This result suggests that this group is abundant but in an inactive state. Alternatively, the overrepresentation of 18S DNA sequences compared to RNA sequences could be due to multiple copies of the gene in the genome (46). For this group the nucleic acid type, whether DNA or RNA, is important.
Many microbial eukaryotes can have high levels of gene duplication (41). While precise information on the gene copy number of the syndiniales is not known, the number of 18S gene copies in microbial eukaryotes can vary by 4 orders of magnitude and is generally correlated to genome size (35). The syndiniales groups are closely related to dinoflagellates, which are renowned for their large genomes (22). In addition, the low and sometimes nonexistent branch lengths of the syndiniales tree support the multiple gene copy explanation. On the other hand, although a trophic mode cannot be inferred based on phylogenetic affiliation, if the syndiniales are parasites like the Duboscquellidae, it makes sense for there to be a lot of dormant cells or cysts in the water column that are ready to take advantage of changing conditions to begin infecting other cells.
Picobiliphytes, cercozoa, and chrysophytes showed slight biases, having higher proportions in samples with cDNA than with DNA. Our phylogenetic trees showed a greater number and diversity of transcript clones, including some clades that exclusively contained clones only found in the transcript samples (trees not shown). This pattern is indicative of rare but highly active organisms. Alternatively, they may have fewer rRNA gene copies , which would result in underrepresentation alongside organisms with higher gene copy numbers in 18S rRNA gene clone libraries. The picobiliphytes are a recently discovered group of pigmented protists that are related to telonemids, cryptophytes, and katablepharids (11, 34, 45). A previous study found that the picobiliphytes may be underrepresented using conventional PCR-based clone libraries (26). Again, a lower gene copy number may have allowed their presence to be masked by the presence of eukaryotes with higher gene copy numbers. Fluorescence in situ hybridization (FISH) enumeration estimated this group to be present in low abundance (<1%) in the Arctic Ocean, Norwegian Sea, and other places off the coast of Europe (34). The current study indicates that while this group of organisms is present in low abundance, they are nonetheless active and could be significant in the environment, an observation that would have been lost by just looking at conventional 18S rRNA gene clone libraries.
Conclusions.
Our results indicate that certain types of 18S rRNA gene and gene transcript clone libraries do not always represent abundant organisms or groups that are ecologically important and active in the environment at sampling time. An 18S rRNA transcript clone library may be more likely to reflect the portion of the microbial community that was active at the time that the sample was taken. Therefore, organisms that are retrieved in low numbers in DNA clone libraries cannot be assumed to be minor components of the community, as they may not have been effectively sampled or may not contribute significantly to the activity of the community. For example, diatoms have historically been underrepresented in SPOTs libraries, especially during the spring bloom, when they may dominate the community in microscopic counts. That diatoms are not well lysed by the more commonly used mechanical extraction methods may explain some of this discrepancy. The abundances of siliceous organisms were greater in clone libraries created using the chemical lysis method. Recognizing that the uncultured group II novel alveolates (syndiniales) also tend to also be more abundant in chemically lysed samples provides more insight into their proposed parasitic/commensal lifestyle with diatoms.
This result further underscores the need for caution when using relative abundances alone in clone libraries to infer natural abundances. While no method is perfect or bias free, teasing out the groups that are preferentially amplified by the different methods can inform decisions on the best method for a target application. We recommend that combinations of methods and 18S rRNA gene and rRNA gene transcripts be used to optimize targeted sampling. While it is intuitive that a combination of ML and CL and DNA and cDNA methods will yield a higher number and diversity of clones, this research provides an important look into group-specific biases caused by different sampling techniques, which may help interpret past studies or optimize DNA yields for targeted future studies when costs and time are constrained.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Science Foundation (NSF) grant EF 0626526 to K.B.H. A.K. acknowledges funding from an NIH Cellular, Biochemical and Molecular Sciences Training Program grant (#T32GM67587) for salary.
We thank David A. Caron, Peter D. Countway, Keith Stormo, and John F. Heidelberg for constructive discussions on the research presented in this paper.
Footnotes
Published ahead of print 23 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF. 2005. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl. Environ. Microbiol. 71:8966–8969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aguilera A, Gómez F, Lospitao E, Amils R. 2006. A molecular approach to the characterization of the eukaryotic communities of an extreme acidic environment: methods for DNA extraction and denaturing gradient gel electrophoresis analysis. Syst. Appl. Microbiol. 29:593–605 [DOI] [PubMed] [Google Scholar]
- 3. Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berney C, Fahrni J, Pawlowski J. 2004. How many novel eukaryotic “kingdoms? ” Pitfalls and limitations of environmental DNA surveys. BMC Biol. 2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caron DA, et al. 2009. Defining DNA-based operational taxonomic units for microbial-eukaryote ecology. Appl. Environ. Microbiol. 75:5797–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caron D, Countway P. 2009. Hypotheses on the role of the protistan rare biosphere in a changing world. Aquat. Microb. Ecol. 57:227–238 [Google Scholar]
- 7. Caron DA, Worden AZ, Countway PD, Demir E, Heidelberg KB. 2009. Protists are microbes too: a perspective. ISME J. 3:4–12 [DOI] [PubMed] [Google Scholar]
- 8. Chao A, Chazdon RL, Colwell RK, Shen TJ. 2005. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol. Lett. 8:148–159 [Google Scholar]
- 9. Colwell RK, Coddington JA. 1994. Estimating terrestrial biodiversity through extrapolation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 345:101–118 [DOI] [PubMed] [Google Scholar]
- 10. Countway P, Vigil P, Schnetzer A, Moorthi S, Caron D. 2010. Seasonal analysis of protistan community structure and diversity at the USC Microbial Observatory (San Pedro Channel, North Pacific Ocean). Limnol. Oceanogr. 55:2381–2396 [Google Scholar]
- 11. Reference deleted.
- 12. DeLong E. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U. S. A. 89:5685–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Díez B, Pedrós-Alió C, Massana R. 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 67:2932–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ewing B, Green P. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Gen. Res. 8:86–194 [PubMed] [Google Scholar]
- 15. Ewing B, Hillier L, Wendl M. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Gen. Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- 16. Fuhrman JA, McCallum K, Davis AA. 1993. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl. Environ. Microbiol. 59:1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giovannoni SJ, Britschgi TB, Moyer CL, Field KG. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60–63 [DOI] [PubMed] [Google Scholar]
- 18. Gómez F. 2007. The consortium of the protozoan Solenicola setigera and the diatom Leptocylindrus mediterraneus in the Pacific Ocean. Acta Protozool. 46:15–24 [Google Scholar]
- 19. Gómez F, Moreira D, Benzerara K, López-García P. 2011. Solenicola setigera is the first characterized member of the abundant and cosmopolitan uncultured marine stramenopile group MAST-3. Environ. Microbiol. 13:193–202 [DOI] [PubMed] [Google Scholar]
- 20. Guillou L, et al. 2008. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata). Environ. Microbiol. 10:3349–3365 [DOI] [PubMed] [Google Scholar]
- 21. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 22. Hackett J, Anderson D, Erdner D, Bhattacharya D. 2004. Dinoflagellates: a remarkable evolutionary experiment. Am. J. Bot. 91:1523–1534 [DOI] [PubMed] [Google Scholar]
- 23. Harada A, Ohtsuka S, Horiguchi T. 2007. Species of the parasitic genus Duboscquella are members of the enigmatic marine alveolate group I. Protist 158:337–347 [DOI] [PubMed] [Google Scholar]
- 24. Hasegawa M, Kishino H, Yano T-A. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160–174 [DOI] [PubMed] [Google Scholar]
- 24a. Hasle GR, Syvertsen EE. 1997. Marine diatoms, p 5–385 In Tomas CR. (ed), Identifying marine phytoplankton. Academic Press, San Diego, CA [Google Scholar]
- 25. Heidelberg KB, Gilbert JA, Joint I. 2010. Marine genomics: at the interface of marine microbial ecology and biodiscovery. Microb. Biotechnol. 3:531–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heywood J, Sieracki M, Bellows W, Poulton NJ, Stepanauskas R. 2011. Capturing diversity of marine heterotrophic protists: one cell at a time. ISME J. 5:674–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJ. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. López-García P, Rodríguez-Valera F, Pedrós-Alió C, Moreira D. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603–607 [DOI] [PubMed] [Google Scholar]
- 29. Massana R, et al. 2004. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl. Environ. Microbiol. 70:3528–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Massana R, Terrado R, Forn I, Lovejoy C, Pedrós-Alió C. 2006. Distribution and abundance of uncultured heterotrophic flagellates in the world oceans. Environ. Microbiol. 8:1515–1522 [DOI] [PubMed] [Google Scholar]
- 31. Medlin L, Elwood HJ, Stickel S, Sogin ML. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491–499 [DOI] [PubMed] [Google Scholar]
- 32. Miller DN, Bryant JE, Madsen EL, Ghiorse WC. 1999. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 65:4715–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moon-van der Staay SY, De Wachter R, Vaulot D. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607–610 [DOI] [PubMed] [Google Scholar]
- 33a. Narasingarao P, et al. 2012. De novo metagenomic assembly reveals abundant novel major lineage of archaea in hypersaline microbial communities. ISME J. 6:81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Not F, et al. 2007. Picobiliphytes: a marine picoplanktonic algal group with unknown affinities to other eukaryotes. Science 315:253–255 [DOI] [PubMed] [Google Scholar]
- 35. Prokopowich CD, Gregory TR, Crease TJ. 2003. The correlation between rDNA copy number and genome size in eukaryotes. Genome 46:48–50 [DOI] [PubMed] [Google Scholar]
- 36. Pruesse E, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Richards TA, Bass D. 2005. Molecular screening of free-living microbial eukaryotes: diversity and distribution using a meta-analysis. Curr. Opin. Microbiol. 8:240–252 [DOI] [PubMed] [Google Scholar]
- 38. Romari K, Valout D. 2004. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol. Oceanogr. 49:784–798 [Google Scholar]
- 39. Sherr B, Sherr E, Caron D, Vaulot D, Worden A. 2007. Oceanic protists. Oceanography 20:130–134 [Google Scholar]
- 40. Stoeck T, Hayward B, Taylor GT, Varela R, Epstein SS. 2006. A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist 157:31–43 [DOI] [PubMed] [Google Scholar]
- 40a. Terrado R, et al. 2011. Protist community composition during spring in an Arctic flaw lead polynya. Polar Biol. 34:1901–1914 [Google Scholar]
- 41. van Dolah FM, et al. 2009. The Florida red tide dinoflagellate Karenia brevis: new insights into cellular and molecular processes underlying bloom dynamics. Harmful Algae 8:562–572 [Google Scholar]
- 42. von Wintzingerode F, Göbel U. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213–229 [DOI] [PubMed] [Google Scholar]
- 43. Weekers P, Gast R, Fuerst P, Byers T. 1994. Sequence variations in small-subunit ribosomal RNAs of Hartmannella vermiformis and their phylogenetic implications. Mol. Biol. Evol. 11:684–690 [DOI] [PubMed] [Google Scholar]
- 44. Worden A. 2006. Picoeukaryote diversity in coastal waters of the Pacific Ocean. Aquat. Microb. Ecol. 43:165–175 [Google Scholar]
- 45. Yoon HS, et al. 2011. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science 332:714–717 [DOI] [PubMed] [Google Scholar]
- 46. Zhu F, Massana R, Not F, Marie D, Vaulot D. 2005. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol. Ecol. 52:79–92 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.