Abstract
The simultaneous increase of atmospheric CO2 and nitrogen (N) deposition to terrestrial ecosystems is predicted to alter plant productivity and, consequently, to change the amount and quality of above- and belowground carbon entering forest soils. It is not known how such changes will impact the composition and function of soil fungal communities that play a key role in degrading complex carbon. We sequenced the fungal cellobiohydrolase I gene (cbhI) from soil DNA and cDNA to compare the richness and composition of resident and expressed cbhI genes at a U.S. Department of Energy free air-carbon dioxide enrichment (FACE) site (NC), which had been exposed to elevated atmospheric CO2 and/or N fertilization treatment for several years. Our results provide evidence that the richness and composition of the cellulolytic fungi surveyed in this study were distinct in the DNA- and cDNA-based gene surveys and were dominated by Basidiomycota that have low or no representation in public databases. The surveys did not detect differences in richness or phylum-level composition of cbhI-containing, cellulolytic fungi that correlated with elevated CO2 or N fertilization at the time of sampling.
INTRODUCTION
Increasing levels of atmospheric CO2 and nitrogen (N) deposition alter plant productivity and the amount and quality of above- and belowground carbon entering forest soils (e.g., see references 1, 8, 11, 14, 21, 25, 34, and 47). Carbon inputs to soil include deposition via plant litter (e.g., see reference 16), root exudates (e.g., see references 33 and 35), and root growth and turnover (e.g., see reference 31). Such altered carbon regimens are predicted to trigger a cascade of responses by soil microorganisms that are involved in carbon cycling and thus alter the magnitude and rate of carbon fluxes between land and atmosphere. This cascade of carbon cycling responses will be further altered by increased N deposition as a direct result of anthropogenic activities (e.g., land clearing and fertilizer runoff) and as an indirect result of altered chemistry of plant materials (11).
The composition and activity of soil fungi that decompose complex carbon will likely be impacted by altered soil carbon and N regimens. This may, in turn, alter net carbon fluxes, but the exact nature of soil fungal responses and their consequences remain poorly understood. Mechanisms underlying altered soil carbon fluxes may include competitive interactions that result in altered community richness, composition, and decomposition activity. Understanding these mechanisms is critical to determine whether increased carbon inputs will ultimately be sequestered or released to the atmosphere, further augmenting already rising atmospheric CO2 levels.
Many previous studies examining either the impact of elevated atmospheric CO2 or N deposition on microbial community composition, biomass, and function have produced mixed results. For instance, in response to elevated CO2, studies have noted an increase in fungal biomass, number of sporocarps, root tips (e.g., see reference 42), CFU (32), arbuscular mycorrhizal hyphae (41), and increased relative abundance, as determined by quantitative PCR (qPCR) and Sanger sequencing efforts (6, 23). However, contrasting results have been found in surveys of fungal saprotrophs using exoenzyme activities and rRNA genes amplified from soil DNA (e.g., see references 5, 7, 18, 20, 22, 23, and 28). Fungal ribosomal gene surveys that examined responses to N fertilization have documented increased abundances of Ascomycota in fertilized soils (e.g., see references 2 and 29). However, the feedback that this phylum-level shift has on carbon cycling remains poorly understood. The role of nitrogen in altering net primary productivity can vary, depending on the amount of CO2 that is available to drive photosynthesis (27), which may differentially impact the function of soil fungal communities. As a consequence of these mixed results, the impacts of simultaneous increases in N deposition and atmospheric CO2 levels on carbon sequestration and microbial feedbacks remain difficult to predict.
The studies described above were DNA-based surveys of rRNA gene fragments. Because all organisms contain ribosomal genes and cellulose-degrading microorganisms are not phylogenetically cohesive, ribosomal gene surveys are unable to detect specific responses of cellulolytic microorganisms that may be central in altered soil carbon cycling patterns. Furthermore, DNA-based studies probe all members of the community, whether or not they are actively participating at the time of sampling. The recent design of PCR primers that target the gene coding for the catalytic subunit of fungal glycosyl hydrolase family 7 cellobiohydrolase I (cbhI), a key enzyme involved in cellulose degradation, has enabled monitoring efforts of a subset of cellulolytic soil fungi using DNA- and RNA-based approaches (10). In a comparison of DNA- and RNA-based compositional surveys of cbhI in litter and soil, Baldrian et al. (3) demonstrated that some of the most abundant taxa in the DNA-based surveys did not dominate gene expression at the same time. This brings into question the functional importance of the most abundant taxa and provides a rationale for using both DNA- and RNA-based, functional gene approaches to examine functional group and individual taxon responses to environmental perturbations.
We generated DNA- and RNA-based profiles of the cbhI gene from soils collected from a U.S. Department of Energy free air-carbon dioxide enrichment (FACE) site in a loblolly pine plantation in North Carolina. This site contained replicate FACE plots that had been exposed to elevated atmospheric CO2 or ambient CO2 (control plots) for more than a decade (http://face.env.duke.edu/). Half of each CO2 treatment or control plot was fertilized with ammonium nitrate (http://face.env.duke.edu/fertilization.cfm), which allowed the combined effects of altered carbon and nitrogen regimens on soil fungal communities to be examined. The primary goals of the study were (i) to examine if the richness and composition of resident and expressed cbhI genes in soil have been impacted by long-term elevated atmospheric CO2 and/or N fertilization, (ii) to compare the DNA-based and -expressed cbhI gene profiles to determine the degree of overlap between the resident and active cbhI genes, and (iii) to expand the current database of cbhI sequences from named fungi in order to improve the capability to taxonomically classify environmental cbhI gene fragments and transcripts.
MATERIALS AND METHODS
Field site description, experimental design, and soil sample collection.
Soils were collected from a U.S. Department of Energy FACE site located in the Blackwood Division of the Duke Forest (Chapel Hill, NC). The site is a Pinus taeda L. (loblolly pine) plantation that was established in 1983 (26). The soil in this plantation is an acidic clay loam of moderately low fertility in the Enon series (26). The six 30-m-diameter FACE plots sampled in this study were established in late August 1996; three plots (no. 2, 3, and 4) were fumigated with 571 ppm CO2 (ca. 200 ppm above ambient), and three plots (no. 1, 5, and 6) were fumigated with ambient CO2 (26) until October 2010. In 2005, two randomly selected quadrants of each of the six plots began receiving annual inputs of pellet ammonium nitrate at a rate of 11.2 g N m−2 (http://face.env.duke.edu/fertilization.cfm). Throughout the article, sequence libraries from the plots and treatments will be designated with a plot number (1, 2, 3, 4, 5, or 6) and field treatment (A, ambient CO2; E, elevated CO2; AF, ambient CO2 and nitrogen fertilization; and EF, elevated CO2 and N fertilization).
On 28 April 2010, triplicate soil cores (15-cm depth) were randomly collected from each of the fertilized and the nonfertilized quadrants in all six plots within a 4-h period of time. Triplicate cores from each quadrant were placed into a single plastic Ziploc bag and homogenized. A representative sample of homogenized soil was placed into a 50-ml Falcon tube and immediately flash frozen in liquid nitrogen. Frozen samples were transported back to the laboratory on dry ice, where they were stored at −70 to −80°C until nucleic acids were extracted (6 plots × 4 quadrants = 24 DNA extractions).
DNA and RNA extraction.
Prior to DNA and RNA extraction, each of the samples was crushed in a mortar and pestle under liquid nitrogen. From each of the 24 soil samples, DNA was extracted from triplicate 0.5-g subsamples using the FastDNA spin kit (MP Biomedicals, Solon, OH) according to the manufacturer's protocol. RNA was extracted from triplicate 2.0-g subsamples using the MoBio RNA Powersoil total RNA isolation kit (MoBio Laboratories, Carlsbad, CA) following the manufacturer's protocol. RNA extracts were treated with Turbo DNase (Ambion, Austin, TX) at 37°C for 1 h. DNase was inactivated using 0.2 volume of DNase inactivation reagent (Ambion, Austin, TX) and incubated for 2 min with occasional vortexing. The inactivation reagent was pelleted by centrifugation at 8,000 × g for 1.5 min, and RNA was transferred to sterile, nuclease-free, microcentrifuge tubes. RNA concentrations and 260/280 ratios were determined using the Nanodrop 2000c (Thermo Scientific, Wilmington, DE). All RNA and DNA extracts were diluted to a concentration of 50 ng μl−1 prior to pooling. Equimolar quantities of DNA or RNA were pooled from fertilized or unfertilized quarters in a given plot, which resulted in 12 pooled DNA extracts and 12 pooled RNA extracts across the six FACE plots sampled. All 12 RNA extracts were reverse transcribed to single-stranded cDNA as described below. Each DNA extract and synthesized cDNA was used for PCR amplification, cloning, and sequencing of the glycosyl hydrolase family 7 cellobiohydrolase I gene (cbhI) as described previously (44).
Reverse transcription of RNA.
Immediately following RNA extraction, single-stranded cDNA was synthesized. In preparation for reverse transcription, 3.5 μl of RNA (ca. 175 ng) and 1 μl of oligo(dT)18 primer (20 μM stock) were placed into PCR tubes and incubated in an Eppendorf Master Cycler Pro (Eppendorf North America, Hauppauge, NY) at 72°C for 3 min, followed by 2 min at 42°C. Immediately after, the following reagents were added (final concentrations listed), bringing the final reaction volume up to 10 μl: 1 μl diethyl pyrocarbonate (DEPC)-treated water, 1× First-Strand buffer, 2.5 mM dithiothreitol (DTT), 1 mM deoxynucleoside triphosphates (dNTP), 0.25 μl RNase inhibitor, and 1 μl SMARTscribe reverse transcriptase (Clontech, Mountain View, CA). The reaction mixture was incubated at 42°C for 90 min and terminated at 70°C for 10 min. The synthesized cDNA was diluted in 40 μl of 1× Tris-EDTA (TE) buffer (pH 8.0) (Roche, Indianapolis, IN) and used as a template for PCR amplification of cbhI as described below.
PCR amplification and cloning of cbhI from DNA and single-stranded cDNA.
From each DNA and cDNA template, a fragment of the cbhI gene encoding 166 to 173 amino acids was PCR amplified in triplicate 25-μl reaction mixtures and visualized as previously described (44). Triplicate PCR products amplified from single-stranded cDNA were pooled and purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA). All purified products were cloned using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA). From each cloning reaction, 96 to 192 clones were selected and bidirectionally sequenced using Sanger technology. Libraries generated from cDNA will be referred to as “cDNA-based libraries” throughout the remainder of the article.
Sequence assembly and analysis.
Fincon software (unpublished software, courtesy of Cliff Han, Los Alamos National Laboratory, NM) was used to assemble bidirectional reads. Sequences of less than 470 bp and/or containing ambiguous bases were eliminated from the libraries. Introns were located using Genewise 2.2.0 (4) on the basis of a hidden Markov model for glycosyl hydrolase family 7 (http://pfam.sanger.ac.uk/family?PF00840#tabview=tab5) and were excised from cbhI sequences in the DNA-based libraries. Inferred amino acid sequences were obtained using the batch translator on the Baylor College of Medicine Search Launcher (http://searchlauncher.bcm.tmc.edu/seq-util/seq-util.html). Inferred amino acid sequences in both DNA- and cDNA-based libraries were aligned together using Muscle 3.6 (9) and imported into ARB (37). After manually editing the alignment as necessary, a distance matrix was generated in ARB. This distance matrix was used as input to mothur software (39), for operational taxonomic unit (OTU)-based analyses as described below. A neighbor-joining tree was generated in ARB for the 15 most abundant OTUs in the cDNA- and the DNA-based libraries. Each of the OTUs in the phylogenetic tree made up ≥1.5% of the collective number of cDNA-based or DNA-based sequences sampled in this study and had an average of no less than one representative sequence in each of the 12 cDNA- or DNA-based libraries.
Environmental sequences were clustered into operational taxonomic units (OTUs) using the average neighbor algorithm on the basis of inferred amino acid sequences having ≥90% similarity (OTU90). In previous analyses, this OTU definition generally clustered cbhI sequences at the family level (44). Average richness recovered in each of the field treatments was determined based on normalized rarefaction analyses. Tentative taxonomic assignments of OTUs were made at the phylum level based on a BLAST analysis against the cbhI database compiled in this study, which is described below. Analysis of variance (ANOVA), Tukey's honestly significant difference (HSD) mean separation, and/or Student's t tests were conducted on richness and composition measures using JMP Statistical Discovery software version 5.1 (SAS, Cary, NC). Nonmetric multidimensional scaling analyses of DNA- and cDNA-based libraries based on Jaccard distances (binary) among OTU profiles were performed using the R software package (http://www.r-project.org).
Database of cbhI reference sequences from cellulolytic fungi.
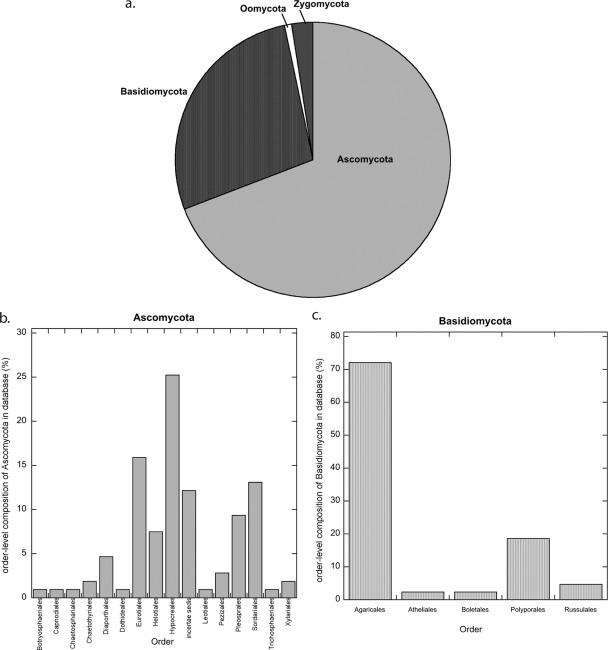
We compiled cbhI sequences available in GenBank and sequenced the cbhI gene from 38 fungal cultures from public and private collections. In addition, 19 cellulolytic cultures were isolated from plant tissue and soil samples collected in New Mexico and from the loblolly pine plantation FACE site using previously published procedures (36). This compilation resulted in a 234-sequence cbhI database. DNA extraction from fungal cultures, PCR amplification, and sequencing of the cbhI and internal transcribed spacer (ITS) gene fragments were performed as described by Weber et al. (44). Fungi isolated from soil and isolates from private collections were identified to the genus level, where possible, by BLAST analysis of the ITS sequence. The complete composition of the database and the sources of 38 cultures surveyed in this study are shown in Fig. 1 and in Table S1 in the supplemental material.
Fig 1.
(a) Percentage of phylum-level composition of the 155 fungi represented in the cbhI database. (b and c) Family-level composition (%) of the 107 Ascomycota (b) and 43 Basidiomycota (c) represented in the database.
Nucleotide sequence accession numbers.
Sequences representative of each of the OTUs recovered in DNA and cDNA-based libraries from soil samples have been deposited and made publically available in MG-RAST. The ITS sequences from the named fungal isolates have been deposited in GenBank under accession no. JQ775550 to JQ775581. The 234-cbhI sequence database has been deposited in the Ribosomal Database Project FunGene Repository (http://fungene.cme.msu.edu).
RESULTS
Characteristics, richness, and phylum-level composition of cbhI libraries.
An average of 82 cbhI clones (range, 25 to 152) (see Table SI2 in the supplemental material) and and average of 70 cbhI clones (range, 55 to 90) (see Table SI2) were sequenced in the cDNA- and DNA-based libraries, respectively. Richness was normalized to 25 sequences, the greatest common number of sequences present in all 24 libraries, for comparison between the cDNA- and DNA-based cbhI clone libraries. The normalized richness values (mean ± standard error [SE]) of the cDNA-based libraries (A, 10.1 ± 3.4; AF, 10.3 ± 2.8; E, 11.1 ± 0.8; and EF, 8.4 ± 2.2) were slightly lower (pairwise t across treatments, P < 0.079) than those of the respective DNA-based libraries (A, 12.8 ± 3.5; AF, 12.6 ± 3.6; E, 11.8 ± 1.8; and EF, 14.8 ± 0.3). The normalized richness values were not statistically different among the field treatments for either the cDNA or DNA data sets. When DNA-based libraries were normalized to 55 sequences, the greatest common numbers of sequences among them, mean richness values (±SE), were as follows: A, 20.5 ± 9.3; AF, 18.3 ± 9.0; E, 17.5 ± 5.9; and EF, 22.5 ± 1.4. These richness estimates were not statistically different. Collectively, these results provide evidence that cbhI richness in DNA-based surveys (resident) and cDNA-based surveys (active) may not be impacted by elevated CO2 or N fertilization.
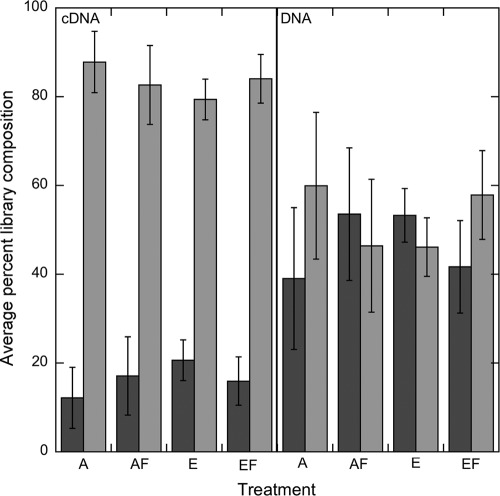
Sequences in the cDNA-based and DNA-based libraries were classified at the phylum level based on a protein BLAST analysis against the 234-sequence database compiled in this study (Fig. 1; see Table SI1 in the supplemental material). No significant differences in phylum composition were found among the four field treatments in either the cDNA- or DNA-based surveys (Fig. 2). The proportions of the two major phyla were significantly different between the cDNA-based surveys and the DNA-based surveys (e.g., pairwise t test of Basidiomycota proportion, P < 0.0001). The cDNA-based survey was highly enriched in Basidiomycota sequences (83.4%) relative to Ascomycota sequences (16.5%) (pairwise t test, P < 0.0001), while the proportions of Basidiomycota (52.5%) to Ascomycota (46.9%) sequences in the DNA-based surveys were very similar (Fig. 2). Less than 1% of all cDNA or DNA sequences were classified as Oomycota or Zygomycota.
Fig 2.
Phylum-level composition of cbhI sequences in DNA- and cDNA-based libraries from each of the field treatments. Each bar represents an average percent ± 1 SE (n = 3). Ascomycota are in dark gray, and Basidiomycota are in light gray. A, ambient CO2; AF, ambient CO2, N fertilization; E, elevated CO2; EF, elevated CO2, N fertilization.
OTU-based compositional analysis.
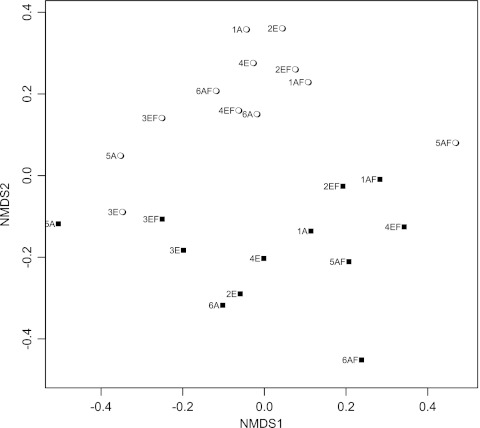
The cDNA- and DNA-based libraries have different compositions based on nonmetric multidimensional scaling analysis of the Jaccard distances between the 24 cbhI libraries (Fig. 3). This indicated that the composition of the community actively expressing cbhI was different from that of the most abundant residents possessing the gene. However, there was no distinct clustering pattern among the cDNA-based or DNA-based libraries by field treatment. This indicated that the sampling, sequencing, and analysis approach used in this study could not detect a compositional shift of resident and active taxa possessing cbhI in response to elevated CO2 and/or N fertilization at the time of sampling.
Fig 3.
Nonmetric multidimensional scaling (NMDS) plot based on the Jaccard distance metric among OTU profiles (OTUs defined at ≥90% similarity) for each of the 12 DNA-based (○) and 12 cDNA-based (■) libraries. Numbers indicate field plot numbers, and letters represent the field treatment as follows: A, ambient CO2; E, elevated CO2; AF, ambient CO2 and N fertilization; and EF, elevated CO2 and N fertilization.
Lack of distinct compositional clustering patterns can be attributed to the relatively low number of OTUs that were shared among all four treatments or replicate libraries within a given treatment. From 12 to 21.8% of the 133 OTUs identified across all of the cDNA-based libraries were unique to a given treatment. These unique OTUs were among some of the most abundant within these libraries. For instance, OTUs 329, 59, 194, and 327 in the E libraries contained 52, 27, 22, and 13 sequences, respectively, and comprised 4 to 16% of the sequences in the libraries. OTUs 84 and 83 were unique to the AF cDNA-based libraries and contained 19 and 10 sequences, or 9.6 and 5.1% of the collective AF library sequences, respectively. OTUs 61 and 344 were unique to the A cDNA-based libraries, containing 32 and 26 sequences (17.1 and 14%), respectively. OTU 365 was unique to the EF cDNA-based library and contained 10 sequences (3.7%). All other OTUs that were unique to a given treatment contained less than 5% of the sequences for a given treatment. Because these unique OTUs had low relative abundance and were variably present among the libraries, it cannot be determined whether or not they represent a subset of the population that is responsive to field treatment.
All sequences from the DNA-based libraries clustered into 189 OTUs, and 14.8 to 20.1% of the OTUs were unique to a given treatment. With the exception of one OTU that was unique to the A DNA-based libraries and contained 56 sequences (26% of the A libraries, OTU 344), each of these unique OTUs contained eight or fewer sequences per OTU (<5% of all sequences), indicating that the majority of the sequences are within OTUs that are shared among two or three treatments.
Overall, only five OTUs were recovered in cDNA-based libraries that were present in all four of the field treatments (OTUs 69, 41, 135, 213, and 153), while 11 OTUs were recovered in DNA-based libraries from all four field treatments (OTUs 475, 272, 342, 90, 443, 431, 162, 434, 69, 93, and 423). This indicates that resident taxa are heterogeneously distributed across the site. Expression patterns were even more variable, suggesting that local environmental parameters exerted a stronger influence over expression patterns than any of the field treatments.
Rank abundance comparison.
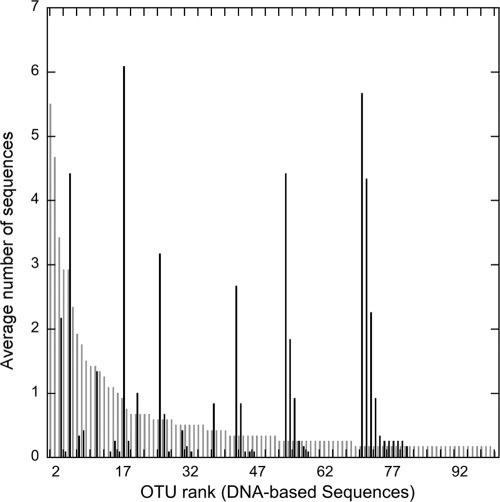
Rank abundance comparison of the collective cDNA and DNA libraries further identified the compositional and structural disparities between active and resident taxa. Overall, only 59 of the total of 133 cbhI OTUs (44%) that were recovered in the cDNA-based libraries were also found in the DNA-based libraries. For the OTUs recovered in both library types, the ranks in the DNA- and cDNA-based libraries were not the same (Fig. 4). For instance, the OTU rank number 17 in the DNA-based libraries, which contained a total of 12 sequences from DNA-based libraries (1.4% of the 835 DNA-based sequences), was the most abundant transcript (73 total sequences; 7% of the 980 cDNA-based sequences) detected in the cDNA-based libraries (Fig. 4). In addition, some of the “rare” types or singletons in the DNA-based library comprise as much as 3% of the 980 total cbhI transcripts sequenced (Fig. 4). The rank order of OTUs in the DNA-based libraries did not parallel the rank order of cbhI transcripts at the time we sampled. This suggests that taxa producing the majority of the transcripts were not necessarily the most abundant cbhI gene copies at the time of sampling.
Fig 4.
Rank abundance plot of the 100 most abundant OTUs (based on average number of sequences in each OTU across all 12 DNA-based libraries) recovered in the collective DNA-based libraries (gray bars) and the average abundance of the sequences recovered from the same OTUs in the cDNA libraries (black bars). The OTUs displayed represent 89% of the DNA-based sequences (746) and 60.4% of the cDNA-based sequences (592).
There were 74 OTUs recovered in the cDNA-based libraries that were not present in any of the DNA-based libraries; 30 of these OTUs were singletons, 37 OTUs contained 2 to 9 transcripts each, and 7 OTUs contained between 10 and 26 sequences each. In total, these 74 OTUs represented 302 sequences, or 31% of the collective 980 sequences sampled across all 12 of the cDNA-based libraries. This indicates that almost one-third of the sequences expressed at the time of the soil sampling were not captured by the DNA-based libraries.
Rank abundance analysis for cDNA- and DNA-based libraries within each of the treatments revealed similar disparity between the two library types for each of the treatments (see Fig. SI1 in the supplemental material). Only 25% of the OTUs in the DNA-based A libraries were also recovered in the cDNA-based A libraries from April 2010. Recoveries of similar magnitude were also observed for the AF (23.6%; 13 out of 55 OTUs), E (23.7%; 14 out of 59 OTUs), and EF (36%; 13 out of 36 OTUs) libraries, indicating that the degree of compositional overlap between DNA- and cDNA-based libraries may not have been impacted by field treatment (see Fig. SI1).
Phylogenetic analysis of the most abundant DNA- and cDNA-based sequences.
Phylogenetic analysis was performed on representative sequences from the 15 most abundant OTUs in the DNA- and cDNA-based libraries along with the 234 cbhI sequences in the database compiled in this study. The 15 most abundant OTUs in the cDNA-based libraries contained almost 55% of the 980 total sequences; 14 of these OTUs were classified as Basidiomycota, and 1 was classified as Ascomycota (Table 1). The 15 most abundant OTUs in the DNA-based libraries contained almost 50% of the 839 total sequences: 7 of these OTUs were classified as Basidiomycota, and 8 were classified as Ascomycota (Table 1). Three of the 15 most abundant OTUs in the DNA-based libraries were also among the 15 most abundant OTUs in the cDNA-based libraries (OTUs 136, 342, and 344) (Table 1).
Table 1.
Abundance of the 15 most abundant OTUs in the DNA-based and cDNA-based libraries and their phylum-level classificationa
| Phylum-level classification and OTU ID no. | % of sequence inb: |
|
|---|---|---|
| cDNA-based library | DNA-based library | |
| Ascomycota | ||
| 401 | 0 | 7.9 |
| 475 | 0.1 | 4.91 |
| 272 | 0 | 4.19 |
| 383 | 0.51 | 2.75 |
| 443 | 0 | 2.51 |
| 431 | 0 | 2.16 |
| 136 | 1.63 | 2.04 |
| 434 | 0.1 | 1.8 |
| Basidiomycota | ||
| 344 | 2.65 | 6.71 |
| 342 | 5.41 | 4.19 |
| 90 | 0.41 | 3.35 |
| 75 | 0 | 2.04 |
| 162 | 0 | 1.92 |
| 74 | 0.1 | 1.56 |
| 87 | 0.31 | 1.56 |
| 69 | 7.45 | 1.44 |
| 135 | 3.88 | 0.84 |
| 61 | 3.27 | 0.48 |
| 41 | 5.41 | 0.36 |
| 194 | 2.24 | 0.36 |
| 43 | 6.94 | 0.24 |
| 59 | 2.76 | 0.24 |
| 329 | 5.31 | 0.24 |
| 37 | 1.73 | 0 |
| 42 | 1.53 | 0 |
| 84 | 1.94 | 0 |
| 284 | 2.65 | 0 |
Each OTU comprises ≥1.5% of the total number of cDNA or DNA sequences sampled. Collectively, the 15 most abundant OTUs in the cDNA-based library and the DNA-based library make up 54% and 50% of the respective total libraries.
Boldface indicates the presence of a particular OTU in the cDNA- or DNA-based library.
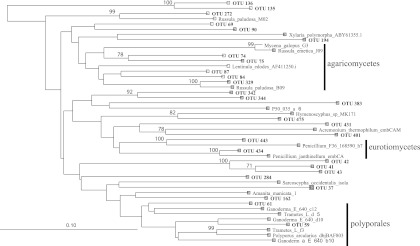
The majority of the OTUs, including the most abundant OTU in the cDNA library (OTU 69) (Fig. 5 and Table 1), did not cluster closely with any of the known reference sequences and often formed separate and distinct clades from the rest of the reference and environmental sequences (e.g., OTUs 135 and 136) (Fig. 5). The OTU that was most closely related to any of the reference sequences was OTU 329, the fifth most abundant OTU in the cDNA libraries, which shared 95.7% identity with Russula paludosa B09 (Fig. 5). OTU 329 and Russula paludosa, along with sequences representing OTUs 75, 74, 87, and 84, populated a clade containing sequences from Lentinula edodes, Mycena galopus, and Russula emetica; however, these OTUs were only between 78.6 and 84.7% similar to any of the reference sequences within this clade. Reference sequences from Russula spp. do not appear to be monophyletic, and OTU 272 (74.4% similar) clustered separately with Russula paludosa M02, away from the larger Russula-containing clade described above (Fig. 5). Members of the Polyporales (Trametes, Polyporus, and Ganoderma) formed a single clade containing sequences representing OTUs 59 and 61, which were 91.3 and 84% similar to sequences obtained from Ganoderma E. Sequences from Penicillium spp. formed a distinct clade containing representative sequences from OTUs 443 and 434, which were 84.8 and 95% similar to Penicillium F36 and Penicillium janthinellum, respectively.
Fig 5.
Neighbor-joining analysis (1,000 bootstrap replicates) of representative cbhI sequences from the 15 most abundant OTUs in the cDNA-based and DNA-based libraries. Only bootstrap values greater than 70 are displayed. An endoglucanse gene sequence in the glycosyl hydrolase 7 family from Aspergillus oryzae was used as the outgroup (BAE66197). Branches on the tree that contain sequences from a fungal order are highlighted but are not meant to imply a strictly cohesive phylogeny for these orders.
DISCUSSION
The impacts of elevated CO2, N fertilization, and other environmental perturbations on soil microbial communities have been frequently assessed by measuring richness and composition, using DNA-based techniques and rRNA gene sequencing (e.g., see references 6, 12, 23, and 38). To focus on a subset of cellulolytic fungi, we used DNA- and cDNA-based approaches to examine cbhI gene composition of resident and active fungal populations in a large-scale factorial field experiment. Our results provide evidence that the richness and composition of the cellulolytic fungi surveyed in this study were distinct in the DNA- and cDNA-based gene surveys and were dominated by Basidiomycota for which cbhI is not or is poorly represented in public sequence databases.
We did not detect a significant change in the richness and composition of DNA-based and cDNA-based cbhI libraries in response to elevated CO2 and/or N fertilization at the time we obtained the samples. This result corroborates a DNA-based cbhI survey conducted at the same field site in July 2007 (44). These results also parallel results of some previous studies in a variety of ecosystem types that have found cellobiohydrolase activity and gene expression to remain unchanged by elevated CO2 or N fertilization (11, 20). Like many of these previous studies, our study was conducted at a single time point. It is possible that responses to the treatments are manifest at other times of the year (20). Furthermore, it is possible that subtle compensatory dynamics of various lesser abundant taxa may have gone undetected in our relatively shallow sequencing effort.
Surveys using the cbhI gene detect a subset of fungi involved in cellulose degradation. Cellulose degradation requires a suite of exoenzymes (e.g., cellobiohydrolases, β-glucosidases, and endoglucanases), and the activities of other fungi such as brown-rot fungi can use nonenzymatic mechanisms to degrade cellulose (46). The apparent lack of response to field treatment by the cbhI-containing fungi surveyed here does not preclude that other subsets of the cellulolytic guild may be responding to field treatment.
Expressed gene (cDNA) profiles provide another layer of insight to identify taxa that may play active roles in biogeochemical processes in soil. Despite the relatively equal proportions of Ascomycota and Basidiomycota sequences recovered in the DNA-based libraries, Basidiomycota transcripts dominated the cDNA-based libraries (Fig. 2). In addition, some of the most abundant OTUs in the cDNA libraries were recovered only once in the DNA libraries or not at all (Fig. 4). Such disconnects between dominant members of DNA profiles and expression profiles have been noted in the past for bacteria (e.g., see references 17, 19, 30, and 40) and have recently been noted for fungal laccase genes (24), nitrate reductase genes (13), and cellobiohydrolase genes (3). Collectively, our results, combined with those of others, indicate that “rare” taxa in DNA-based libraries may have some of the most abundant gene transcripts and may be making substantial contributions to cellulose degradation, but the link between transcript numbers and relative contribution to activity remains to be explicitly demonstrated. This provides a rationale for sequencing more deeply to recover “rare” taxa and calibrating transcript numbers and contributions to net activity in situ (30, 43).
The incongruency between the DNA- and cDNA based surveys could be due to metabolic diversity or to an abundance of inactive fungal spores. The cbhI-containing fungi in soil are likely to be metabolically diverse, performing a wide array of functions in addition to cellulose degradation, and their abundance patterns are likely to be under selection by a multitude of intersecting factors (e.g., competition and resource availability). Future studies should aim at understanding the conditions under which particular taxa become active, particularly those fungi that form spores.
Most of the dominant OTUs in the cDNA-based libraries are not closely related to any cbhI gene sequences in current public databases (Fig. 5). The closest BLAST hit of the top 15 OTUs in the cDNA- and DNA-based libraries to a sequence in our compiled database was Russula paludosa. This genus contains many mycorrhizal taxa that have not been generally perceived as key participants in cellulose degradation. However, recent studies have begun to reveal otherwise (15). Our soil cbhI sequences are very distant from reference sequences in our database (Fig. 5), which contains many of the prototypical cellulose degraders that are being used for bioenergy applications (e.g., Trichoderma, Chaetomium, and Phanerochaete) (45). This indicates that much remains to be learned about the taxa that actively degrade cellulose in soils. Expanded cbhI databases that facilitate improved classification at subphylum levels will be necessary to gain ecological insights regarding this process.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rytas Vilgalys, Robert B. Jackson, Charles “Will” Cook, Terri M. Porter, Greg Bonito, and several dedicated undergraduate students and postdocs for access to the FACE site and assistance with sample collection, storage, and shipping. We thank Brian Nowak-Thompson (Cornell College, Mount Vernon, IA) for providing fungal cultures from his private collection.
This study was funded by a United States Department of Energy-Science Focus Area grant (C.R.K.), a Los Alamos National Laboratory Director's Postdoctoral fellowship (C.F.W.), and a Western Illinois University Undergraduate Research and Scholarly Activity grant (Z.G.).
Footnotes
Published ahead of print 30 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ainsworth EA, Long SP. 2004. What have we learned from 15 years of free-air CO2 enrichment (FACE)? A meta-analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2. New Phytol. 165:351–372 [DOI] [PubMed] [Google Scholar]
- 2. Allison SD, Gartner TB, Mack MC, McGuire K, Treseder K. 2010. Nitrogen alters carbon dynamics during early succession in boreal forest. Soil Biol. Biochem. 42:1157–1164 [Google Scholar]
- 3. Baldrian P, et al. 2011. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 6:248–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Birney E, Clamp M, Durbin R. 2004. GeneWise and Genomewise. Genome Res. 14:988–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carney KM, Hungate BA, Drake BG, Megonigal JP. 2007. Altered soil microbial community at elevated CO2 leads to loss of soil carbon. Proc. Natl. Acad. Sci. U. S. A. 104:4990–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castro HF, Classen AT, Austin EE, Norby RJ, Schadt CW. 2010. Soil microbial community responses to multiple experimental climate change drivers. Appl. Environ. Microbiol. 76:999–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chung H, Zak DR, Lileskov EA. 2006. Fungal community composition and metabolism under elevated CO2 and O3. Oecologia 147:143–154 [DOI] [PubMed] [Google Scholar]
- 8. Couteaux M, Bottner P, Berg B. 1995. Litter decomposition, climate and litter quality. Trends Ecol. Evol. 10:62–66 [DOI] [PubMed] [Google Scholar]
- 9. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edwards IP, Upchurch RA, Zak DR. 2008. Isolation of fungal cellobiohydrolase I genes from sporocarps and forest soils by PCR. Appl. Environ. Microbiol. 74:3481–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Edwards IP, Zak DR, Kellner H, Eisenlord SD, Pregitzer KS. 2011. Simulated atmospheric N deposition alters fungal community composition and suppresses ligninolytic gene expression in a northern hardwood forest. PLoS One 6:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Enwall K, et al. 2007. Long-term impact of fertilization on activity and composition of bacterial communities and metabolic guilds in agricultural soil. Soil Biol. Biochem. 39:106–115 [Google Scholar]
- 13. Gorfer M, et al. 2011. Community profiling and gene expression of fungal assimilatory nitrate reductases in agricultural soil. ISME J. 5:1771–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haase S, et al. 2007. Elevation of atmospheric CO2 and N-nutritional status modify nodulation, nodule carbon supply, and root exudation of Phaseolus vulgaris L. Soil Biol. Biochem. 39:2208–2221 [Google Scholar]
- 15. Hibbett DS, Gilbert L-B, Donoghue MJ. 2000. Evolutionary instability of ectomycorrhizal symbioses in basidiomycetes. Lett. Nat. 407:506–508 [DOI] [PubMed] [Google Scholar]
- 16. Hoosbeek MR, Scarascia-Mugnozza GE. 2009. Increased litter build up and soil organic matter stabilization in a polar plantation after 6 years of atmospheric CO2 enrichment (FACE): final results of POP_EuroFACE compared to other forest FACE experiments. Ecosystems 12:220–238 [Google Scholar]
- 17. Izumi H, Anderson AC, Alexander IJ, Kilham K, Moore ERB. 2006. Diversity and expression of nitrogenase genes (nifH) from ectomycorrhizas of Corsican pine (Pinus nigra). Environ. Microbiol. 8:2224–2230 [DOI] [PubMed] [Google Scholar]
- 18. Janus LR, et al. 2005. Elevated atmospheric CO2 alters soil microbial communities associated with trembling aspen (Populus tremuloides) roots. Microb. Ecol. 50:102–109 [DOI] [PubMed] [Google Scholar]
- 19. Jia Z, Conrad R. 2009. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11:1658–1671 [DOI] [PubMed] [Google Scholar]
- 20. Kelley AM, Fay PA, Polley HW, Gill RA, Jackson RB. 2011. Atmospheric CO2 and soil extracellular enzyme activity: a meta-analysis and CO2 gradient experiment. Ecosphere 2:1–20 [Google Scholar]
- 21. King JS, et al. 2001. Fine-root biomass and fluxes of soil carbon in young stands of paper birch and trembling aspen as affected by elevated atmospheric CO2 and tropospheric O3. Oecologia 128:237–250 [DOI] [PubMed] [Google Scholar]
- 22. Larson JL, Zak DR, Sinsabaugh RL. 2002. Extracellular enzyme activity beneath temperate trees growing under elevated carbon dioxide and ozone. J. Soil Sci. Soc. Am. 66:1848–1856 [Google Scholar]
- 23. Lesaulnier S, et al. 2008. Elevated atmospheric CO2 affects soil microbial diversity associated with trembling aspen. Environ. Microbiol. 10:926–941 [DOI] [PubMed] [Google Scholar]
- 24. Luis P, Kellner H, Martin F, Buscot F. 2005. A molecular method to evaluate basidiomycete laccase gene expression in forest soils. Geoderma 128:18–27 [Google Scholar]
- 25. Lukac M, et al. 2009. Forest soil carbon cycle under elevated CO2—a case of increased throughput. Forestry 82:75–86 [Google Scholar]
- 26. McCarthy HR, et al. 2010. Re-assessment of plant carbon dynamics at the Duke free-air CO2 enrichment site: interactions of atmospheric [CO2] with nitrogen and water availability over stand development. New Phytol. 185:514–528 [DOI] [PubMed] [Google Scholar]
- 27. McGuire AD, Milillo JM, Joyce LA. 1995. The role of nitrogen in the response of forest net primary production to elevated atmospheric carbon dioxide. Annu. Rev. Ecol. Syst. 26:43–503 [Google Scholar]
- 28. Moorhead DL, Linkins AE. 1997. Elevated CO2 alters belowground exoenzyme activities in tussock tundra. Plant Soil 189:321–329 [Google Scholar]
- 29. Nemergut DR, et al. 2008. The effects of chronic nitrogen fertilization on alpine soil microbial communities: implications for carbon and nitrogen cycling. Environ. Microbiol. 10:3093–3105 [DOI] [PubMed] [Google Scholar]
- 30. Nicol GW, Leininger S, Schleper C, Prosser JI. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10:2966–2978 [DOI] [PubMed] [Google Scholar]
- 31. Norby RJ, Ledford J, Reilly CD, Miller NE, O'Neill EG. 2004. Fine-root production dominates response of a deciduous forest to atmospheric CO2 enrichment. Proc. Natl. Acad. Sci. U. S. A. 101:9689–9693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Olszyk DM, et al. 2001. Interactive effects of CO2 and O3 on ponderosa pine plant/litter/soil mesocosm. Environ. Pollut. 115:447–462 [DOI] [PubMed] [Google Scholar]
- 33. Paterson E, et al. 2008. Labile and recalcitrant plant fractions are utilized by distinct microbial communities in soil: independent of the presence of root and mycorrhizal fungi. Soil Biol. Biochem. 40:103–113 [Google Scholar]
- 34. Phillips RL, Zak DR, Homes WE, White DC. 2002. Microbial community composition and function beneath temperate trees exposed to elevated atmospheric carbon dioxide and ozone. Oecologia 131:236–244 [DOI] [PubMed] [Google Scholar]
- 35. Phillips RP, Bernhardt ES, Schlesinger WH. 2009. Elevated CO2 increases root exudation from loblolly pine (Pinus taeda) seedlings as an N-mediated response. Tree Physiol. 29:1513–1523 [DOI] [PubMed] [Google Scholar]
- 36. Porras-Alfaro A, Bayman P. 2007. Mycorrhizal fungi of Vanilla: specificity, phylogeny and effects on seed germination and plant growth. Mycologia 99:510–525 [DOI] [PubMed] [Google Scholar]
- 37. Pruesse E, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramirez KS, Lauber CL, Knight R, Bradford MA, Fierer N. 2010. Consistent effects of nitrogen fertilization on soil bacterial communities in contrasting systems. Ecology 91:3463–3470 [DOI] [PubMed] [Google Scholar]
- 39. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sharma S, Szele Z, Schilling R, Munch JC, Schloter M. 2006. Influence of freeze-thaw stress on the structure and function of microbial communities and denitrifying populations in soil. Appl. Environ. Microbiol. 72:2148–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Treseder KK, Egerton-Warburton LM, Allen MF, Cheng Y, Oechel WC. 2003. Alteration of soil carbon pools and communities of mycorrhizal fungi in chaparral exposed to elevated carbon dioxide. Ecosystems 6:786–796 [Google Scholar]
- 42. Treseder KK. 2004. Nutrient acquisition strategies of fungi and their relation to elevated atmospheric CO2, p 713–731 In Dighton J, White JF, Oudemans P. (ed), The fungal community: its organization and role in the ecosystem, 3rd ed CRC Press, Boca Raton, FL [Google Scholar]
- 43. Turk KA, et al. 2011. Nitrogen fixation and nitrogenase (nifH) expression in tropical waters of the eastern North Atlantic. ISME J. 5:1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weber CF, et al. 2011. Responses of cellulolytic fungal communities to elevated atmospheric CO2 are complex and variable across five ecosystems. Environ. Microbiol. 13:2778–2793 [DOI] [PubMed] [Google Scholar]
- 45. Webster J, Weber RWS. 2007. Introduction to fungi, 3rd ed, p 514–576 Cambridge University Press, New York, NY [Google Scholar]
- 46. Xu G, Goodell B. 2001. Mechanisms of wood degradation by brown-rot fungi: chelator-mediated cellulose degradation and binding of iron by cellulose. J. Biotechnol. 87:43–57 [DOI] [PubMed] [Google Scholar]
- 47. Zak DR, et al. 1993. Elevated atmospheric CO2 and feedback between carbon and nitrogen cycles. Plant Soil 151:105–117 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.