Abstract
Arsenic contamination of groundwater sources is a major issue worldwide, since exposure to high levels of arsenic has been linked to a variety of health problems. Effective methods of detection are thus greatly needed as preventive measures. In an effort to develop a fungal biosensor for arsenic, we first identified seven putative arsenic metabolism and transport genes in Aspergillus niger, a widely used industrial organism that is generally regarded as safe (GRAS). Among the genes tested for RNA expression in response to arsenate, acrA, encoding a putative plasma membrane arsenite efflux pump, displayed an over 200-fold increase in gene expression in response to arsenate. We characterized the function of this A. niger protein in arsenic efflux by gene knockout and confirmed that AcrA was located at the cell membrane using an enhanced green fluorescent protein (eGFP) fusion construct. Based on our observations, we developed a putative biosensor strain containing a construct of the native promoter of acrA fused with egfp. We analyzed the fluorescence of this biosensor strain in the presence of arsenic using confocal microscopy and spectrofluorimetry. The biosensor strain reliably detected both arsenite and arsenate in the range of 1.8 to 180 μg/liter, which encompasses the threshold concentrations for drinking water set by the World Health Organization (10 and 50 μg/liter).
INTRODUCTION
Arsenic is a ubiquitous toxic metalloid that contaminates both groundwater sources (38, 43, 45) and soils (14) worldwide. Strikingly, more than 100 million people in the world are at risk from consuming water contaminated with arsenic (20), and strategies to detect and prevent this global problem are urgently required.
Arsenic has numerous valence states, but its two predominant forms in nature are arsenate(V), present in more oxidized conditions, and arsenite(III), found in reducing environments (40, 44, 46). Arsenate mimics phosphate and inhibits the production of ATP (25). Moreover, arsenite binds to cellular enzymes containing −SH groups and thus disrupts their function (25). The long-term exposure to unsafe levels of arsenic leads to arsenicosis, which is characterized by skin lesions, such as melanosis, leukomelanosis, and hyperkeratosis, and ultimately death (32, 60). Arsenic was also one of the first elements to be recognized as a carcinogen (40, 52), and chronic exposure has been associated with bladder, lung, and skin cancer (World Health Organization [WHO], http://www.who.int/topics/arsenic/en/).
The WHO has proposed guidelines for acceptable concentrations in drinking water. Although these guidelines suggest a maximum exposure concentration of arsenic of 10 μg/liter for drinking water, numerous countries continue to use a cutoff of 50 μg/liter (20). In many parts of the world, arsenic concentrations far exceed both of these limits, particularly in West Bengal and Bangladesh (5). There are currently two widely used methods of arsenic detection in drinking water: laboratory-based analytical methods and field-based testing methods (5). The laboratory-based analytical methods require highly trained personnel and expensive analytical machinery, such as inductively coupled plasma mass spectroscopy (ICPMS) and atomic absorption spectroscopy. Further, the delay in turnaround time between specimen collection and result availability limits their day-to-day use. Field-based testing methods are largely chemical colorimetric assays, such as the Gutzeit method, which generate toxic arsine gas by reducing arsenic with a strong acid (20). These tests require the use of hazardous chemicals and can generate toxic by-products. Biosensors and bioreporters are beginning to emerge as safe, alternate methods to detect environmental pollutants such as arsenic (22, 29, 50). Although several arsenic biosensors have been reported, these traditionally rely on bacterial reporter systems (7), which have a relatively narrow tolerance to variation in culture systems, transportation, and storage. No commercial biosensors are currently in use in affected countries. One approach to developing a field-use-friendly biosensor is the use of a more robust host strain, such as fungi.
Filamentous fungi are highly adaptable organisms that can grow in extreme environmental conditions, such as in contaminated areas with potentially toxic chemical compounds and elements (10). This intrinsic resistance, combined with the long-term stability of resting fungal spores, suggests that they may serve as useful organisms for the development of biodetection or bioremediation strategies. Arsenic-hyperresistant filamentous fungi, such as Aspergillus sp. P37, have been isolated from nature, and some biochemical pathways of arsenic detoxification have been characterized in these strains (14–17, 54, 58). However, the genetics of arsenic metabolism remain unknown in filamentous fungi (14, 59).
Aspergillus niger is a filamentous fungus commonly associated with wood and is routinely used for the industrial production of enzymes and organic acids (6, 19). This organism has been observed to survive and propagate under concentrations of arsenic as high as 300 mg/liter (18). A. niger is efficient in bioleaching of several heavy metals (19), possibly due to its ability to produce large amounts of oxalic acids in culture. This organism also produces catalase, which allows the fungus to protect itself against environmental stress, including arsenic (11). The intrinsic arsenic resistance of A. niger coupled with the availability of genomic sequence data led us to select this organism as a model system to examine arsenic resistance and develop an arsenic biosensor system in filamentous fungi.
Although the mechanisms of arsenic detoxification remain unstudied in filamentous fungi, several pathways mediating detoxification of arsenic have been elucidated in the yeast Saccharomyces cerevisiae. One mechanism of resistance is mediated by the products of three contiguous genes, ACR1, ACR2, and ACR3 (9, 15, 37). ACR1 encodes a transcriptional regulator, ACR2 encodes an arsenate reductase that converts arsenate into arsenite, and ACR3 encodes a plasma membrane arsenite efflux transporter that pumps arsenite out of the cell, allowing resistance (9, 15, 26, 37). In addition, conjugation of arsenite with glutathione (GSH) to form As(GS)3 and subsequent transport into vacuoles by Ycf1p for detoxification purposes have been proposed to increase arsenic resistance in S. cerevisiae (14, 26).
We therefore used bioinformatics to identify putative genes involved in the arsenic homeostasis of A. niger and developed a hypothetical model of A. niger arsenic metabolism and transport based on S. cerevisiae. To begin to validate this model, we characterized the function of one of these genes, acrA, in detail. This gene is the putative orthologue of the S. cerevisiae ACR3 gene, which encodes an arsenite efflux pump (66). Consistent with its role as an efflux pump, an AcrA-enhanced green fluorescent protein (eGFP) fusion localized to the plasma membrane in A. niger, and disruption of A. niger acrA was associated with increased accumulation of and sensitivity to arsenic species. Moreover, we discovered that the expression of the acrA gene was rapidly and highly induced in the presence of very low concentrations of arsenate. Based on this observation, we tested a strain containing the construct of the native promoter of acrA fused with egfp as a potential biosensor for arsenic and confirmed the ability of this strain to detect levels of arsenic at and below the WHO thresholds for arsenic levels in drinking water in the developed and developing worlds (24).
MATERIALS AND METHODS
Fungal strain culture conditions.
Aspergillus niger strains were propagated on potato dextrose agar plates (PDA; 0.4% potato starch, 2% dextrose, 1.5% agar) and incubated at 37°C. Other media used were yeast peptone dextrose (YPD; 1% yeast extract, 2% peptone, 2% glucose) and Sabouraud (1% enzymatic digest of casein, 2% dextrose). All media were purchased from GIBCO (Invitrogen).
Nucleic and proteic sequence comparison.
The A. niger genomic sequences were obtained from the Central Aspergillus Data Repository (CADRE) (www.cadre-genomes.org.uk/Aspergillus_niger/). The putative arsenic homeostasis genes in Aspergillus niger were found by comparing gene and protein sequences with those of S. cerevisiae (12, 35, 39, 49, 61, 62, 66) using nucleotide-BLAST and protein-BLAST from the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Identification and phylogenetic analysis of A. niger arsenic transporter.
Select arsenic transporter amino acid sequences found in literature (1, 13) and highly homologous sequences to A. niger AcrA identified through BLAST searches (3) were aligned with A. niger AcrA using the ClustalW multiple sequence alignment (MSA) tool (34). The Gonnet substitution matrix and neighbor-joining clustering model were used in ClustalW MSA, and results were exported and formatted to analyze phylogenetic relationships and build a cladogram using TreeVector (42). To ensure that only sequences within the functional domain related to arsenic transport were analyzed, each candidate sequence was queried to identify conserved domain regions using the NCBI Conserved Domain Database (CDD) query protocol (36). The CDD domain cluster, TIGRFAMs, and InterPro matches were further drilled and cross-referenced to assess and verify classification of A. niger AcrA (4, 28).
Isolation of DNA and RNA.
Conidia of A. niger were inoculated into 20 ml of YPD broth and incubated with shaking (200 rpm) at 37°C for 16 to 18 h. Subsequently, mycelia were harvested by filtration through a P5 Whatman paper (GE Healthcare) and then ground to fine powder under liquid nitrogen in order to break the fungal cell wall. Samples were then processed for the extraction of RNA or DNA. DNA extraction was conducted by resuspending the ground hyphae in 500 μl of DNA extraction buffer [0.7 M NaCl, 0.1 M Na2(SO3), 0.05 M EDTA, 1% SDS, 0.1 M Tris-HCl (pH 7.5)]. Following resuspension, DNA was extracted with phenol-chloroform and precipitated with ethanol. RNA extraction was conducted according to the manufacturer's instructions of the Nucleospin RNA plant minikit (Macherey-Nagel, Germany).
Stimulation and quantification of expression of putative genes involved in arsenic homeostasis.
Conidia were inoculated into 20 ml of YPD broth and subsequently incubated (200 rpm, 37°C) for 16 h. The resulting mycelia were then stimulated by incubation in YPD containing different concentrations of arsenate (potassium arsenate, monobasic; KH2AsO4; Sigma-Aldrich). RNA was harvested and extracted at different time points of culture and then analyzed using real-time reverse transcription (RT)-PCR. The priming oligonucleotides for the genes of interest were designed using Primer-BLAST from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The primers used for each gene are shown in Table S1 in the supplemental material. The synthesis of cDNA from 1 μg of RNA was performed in 20 μl using Quantitect reverse transcriptase (Qiagen, Mississauga, Canada) with random primers according to the manufacturer's recommendations. Subsequently, 0.2 μl of the synthesized cDNA was analyzed in real-time RT-PCR with SYBR green Quantitect System (Fermentas, Canada), using an ABI 7000 thermocycler (Applied Biosystems, Streetsville, Canada). The fungal gene expression was normalized to Aspergillus niger TEF1 (transcription elongation factor 1) expression, which was used as the endogenous control.
Disruption of the acrA gene.
All PCRs were prepared using KAPA HiFi HotStart ReadyMix according to the manufacturer's instructions (KAPA Biosystems; Boston, MA). The Aspergillus fumigatus split marker transformation protocol previously developed in our laboratory for the generation of homologous integrants was adapted for use in A. niger (55). This technique relies on cotransformation of A. niger protoplasts with DNA fragments, each containing half of the marker (hph encoding hygromycin phosphotransferase) fused to one flanking sequence of the gene to delete. Successful homologous integration of these DNA fragments regenerates the hygromycin marker to permit selection. Fragments were generated using a modified GATEWAY system cloning approach. Briefly, the amplification of the upstream and downstream flanking sequences was performed using the priming oligonucleotides AcrA gates 1 and 2 and 3 and 4, respectively (see Table S1 in the supplemental material). These primers were created using Primer3 (53). Subsequently, the amplified flanking sequences were cloned into pENTR/D-TOPO vector (Invitrogen, Canada) and transformed into DH5α Escherichia coli cells. The plasmids were then isolated from putative transformants and analyzed using restriction enzymes to verify if the fragments were inserted in the correct orientation. The flanking sequence was then transferred from the pENTR/D-TOPO plasmid to the attR-ccdB-modified pAN7.1, which contains the hph resistance cassette (27), using LR clonase (Invitrogen, Canada). Consequently, E. coli DH5α cells were transformed and selected on ampicillin plates. The insertions in the plasmids containing the correct products allowed amplification by PCR with the long-range PCR enzyme (Fermentas, Canada) of one flanking sequence fused with half of the marker cassette. Aspergillus niger protoplasts were then produced from young hyphae after digesting the cell wall for 3 h at 30°C (60 rpm) with protoplasting solution containing Driselase from Basidiomycetes sp. and lysing enzymes from Trichoderma harzianum (Sigma-Aldrich, Canada). Subsequently, these protoplasts were cotransformed with 5 μg of each DNA fragment (63). Transformants were selected on PDA plates with 250 μg/ml of hygromycin. The deletion of the acrA open reading frame (ORF) was confirmed by long-range PCR with primers AcrA gate 1, AcrA gate 4, AcrA-RT sense, and AcrA-RT antisense, as well as by real-time RT-PCR using primers AcrA-RT sense and AcrA-RT antisense (see Table S1). The isolation of a pure clone was done by single-spore purification.
Construction of the acrA-complemented strain.
To verify that any phenotypes observed in the acrA-null mutant strain were due to specific deletion of acrA, we complemented the acrA-null mutant with a wild-type allele of acrA. We used the same GATEWAY cloning technique as the one used for the gene deletion in order to replace the hph cassette by the acrA ORF fused to the ble cassette (coding for resistance to phleomycin). Variations were as follows. Priming oligonucleotides AcrA gates 1 and 5 were used to amplify the upstream flanking sequence and the ORF, while the downstream flanking sequence was amplified by AcrA gate 3 and gate 4 (see Table S1 in the supplemental material). Cloned sequences were transferred from pENTR/D-TOPO to a modified p402 vector, which contains the ble resistance cassette (27). Transformants were selected on PDA plates with 150 μg/ml of phleomycin. The reinsertion of the acrA gene was confirmed by long-range PCR and real-time RT-PCR.
Construction of the AcrA-GFP overexpression strain.
The GFP-encoding gene (egfp) was amplified by PCR using the primers GFP-F and GFP-R (see Table S1 in the supplemental material) and using the plasmid p123 (57) as a template. After PCR amplification, the PCR product was cloned into the plasmid pEYFPC (31) using NcoI and NotI. The resulting plasmid was designated pGFP and contains egfp under the expression of the A. nidulans gpdA promoter. Afterwards, the 1,278-bp AcrA-encoding gene (acrA) was amplified by PCR using the primers acrA-F and acrA-R (see Table S1). The PCR product was cloned into pGFP using EcoRV and NotI. The resulting plasmid contains acrA tagged with egfp at the N terminus and under the expression of the A. nidulans gpdA promoter. Finally, the 1,423-bp phleomycin cassette, which contains the A. nidulans gpdA promoter, phleomycin resistance gene (ble), and the cyc1 terminator, was amplified by PCR using the primers Phleo-EcoRI and Phleo-SacI (see Table S1) and p402 as a template and cloned into the acrA-egfp overexpression plasmid using EcoRI and SacI. The resulting plasmid was designated pAcrA-GFP. This plasmid was then used for the transformation of the wild-type A. niger strain.
GFP localization of AcrA.
The strain containing the AcrA-eGFP fusion construct was examined under confocal microscopy to identify the subcellular localization and trafficking of AcrA. A total of 105 conidia/ml was inoculated in 1 ml of YPD broth and incubated at 37°C (200 rpm). After 6 h of incubation, samples were taken for observation of the germlings under an Olympus Fluoview confocal microscope at 100× oil immersion using a fluorescein isothiocyanate (FITC) filter with excitation at 488 nm and emission at 507 nm.
We also observed protoplasts at 100× oil immersion under confocal microscopy after the cell wall was digested for 3 h at 30°C (60 rpm) with protoplasting solution used in the fungal transformation protocol.
Growth inhibition assay.
Arsenic sensitivity of the different strains was determined by growth of conidia in Sabouraud broth supplemented with arsenate in a gradient of increasing concentration (0 to 2,048 mg/liter arsenate). In a 96-well plate containing Sabouraud liquid medium, a total of 2 × 105 conidia/ml was incubated in a final volume of 200 μl at 37°C. For a total volume of 200 μl per well, 100 μl of conidia suspended in liquid medium was added to each well containing 100 μl of arsenate dilution. After 24 h of incubation, fungal growth was determined by the absorbance taken at 630 nm using a spectrophotometer. The optical density at 630 nm (OD630) in each well was compared to the same strain growth control in the absence of arsenate and expressed as percent normal growth.
Arsenic content.
To determine the intracellular concentration of individual strains, A. niger strains were grown in YPD liquid broth overnight at 37°C (200 rpm) at 5 × 105 conidia/ml. The culture medium was removed by filtration through P5 Whatman paper (GE Healthcare). The mycelial biomass was cut into 1-cm2 squares and weighed. Half of the cut squares were exposed to chloroform vapor for 20 min to kill them. Subsequently, all the squares were inoculated into dishes containing YPD liquid broth with 180 mg/liter of arsenate. The plates were incubated for 24 h at 37°C. The cultures were then harvested, and the mycelia were washed with 5 ml of 0.1 N HCl for 30 min (23, 47). The mycelia were then dried by vacuum and inactivated in liquid nitrogen. Samples were dried at room temperature and weighed. The dry samples were subsequently digested using 1 ml of trace metal-grade Aqua Regia (3 parts HCl:1 part HNO3) in a closed Teflon container at 95°C for 30 min. The HCl wash was also kept for arsenic analysis. The quantification of total arsenic species for both killed and live biomass was determined by using a Perkin Elmer AAnalyst800 atomic absorption spectrometer with a graphite furnace at the Trace Element Analysis Lab, Department of Earth & Planetary Sciences of McGill University (Montréal, Québec, Canada).
Construction of the ΔacrA::PacrA-GFP strain.
The 1-kb upstream sequence of acrA was amplified by PCR with priming oligonucleotides AcrA-Bios sense and AcrA-Bios antisense (see Table S1 in the supplemental material). Using the Infusion HD cloning kit (Clontech, USA), the PCR fragment was cloned into the pGFP plasmid previously digested with AgeI and EcoRV. The resulting plasmid was designated pPacrA-GFP. Subsequently, the acrA deletion strain was transformed ectopically with the pPacrA-GFP plasmid.
Fluorescence detection.
Fluorescence was determined by growth of conidia in YPD broth or RPMI 1640 with MOPS (morpholinepropanesulfonic acid; Sigma-Aldrich, Canada), supplemented with arsenate in a gradient of increasing concentration (0 to 180 μg/liter). Other metals and metalloids, including sodium arsenite, cadmium, antimony(III) chloride, iron(III) phosphate, cobalt(II) sulfate, and copper(II) sulfate, were used to test the specificity of the biosensor (Fisher Scientific, Canada). In a 96-well black plate with a clear bottom (Corning, USA), a total of 2 × 105 conidia/ml was incubated in each well at 37°C. At different time points, fungal growth was determined by the absorbance taken at 630 nm, and the GFP signal was measured at excitation of 485 nm and emission of 535 nm using the SpectraMax microplate reader. For visualization, conidia were incubated in YPD broth supplemented with arsenate (0, 18, 90 μg/liter) on glass coverslips at 37°C for 24 h. Subsequently, coverslips were placed on microscope slides and pictures were taken using confocal microscopy at ×40 magnification using a FITC filter. The ΔacrA strain was used as a control for autofluorescence. Hyphae of the biosensor strain and the ΔacrA strain were also pregrown for 24 h at 37°C in a 96-well black plate with a clear bottom and then exposed to a gradient of increasing concentrations of arsenate (0 to 180 μg/liter). At the indicated time points, GFP signal was measured using a SpectraMax microplate reader as described above.
RESULTS
Putative arsenic transport and metabolism genes are present in A. niger.
Using the recently completed whole-genome sequence of A. niger, we performed a homology sequence alignment of putative arsenic transport and metabolism genes in A. niger by comparing these sequences with elements of known arsenic resistance pathways in well-characterized organisms, including E. coli and S. cerevisiae, using BLAST. Using this approach, we identified seven candidate A. niger genes predicted to encode proteins involved in mediating arsenic metabolism and transport (Table 1).
Table 1.
Putative arsenic metabolism and transport genes in Aspergillus niger and Saccharomyces cerevisiae identified by bioinformatics analysis
| Putative function | A. niger gene | S. cerevisiae gene | E value | Reference |
|---|---|---|---|---|
| Arsenate reductase (Arc2) | ANI_1_452044 | ACR2 | 3e−13 | 39 |
| Arsenic methyltransferase (Cyt19) | ANI_1_448044 | MTQ2 | 7e−41 | 49 |
| Arsenite efflux transporter (AcrA) | ANI_1_1278164 | ACR3 | 5e−79 | 66 |
| Plasma membrane phosphate transporter (Pho87) | ANI_1_2404014 | PHO87 | 3e−168 | 12 |
| Aquaglyceroporin (GlpF), trivalent metalloid transporter | ANI_1_1314144 | FPS1 | 1e−36 | 35 |
| Glutathione S-conjugate transporter (GstA) | ANI_1_1332014 | GTT1 | 3e−43 | 62 |
| ABC metal ion transporter | ANI_1_480034 | YCF1 | 0.0 | 61 |
Expression levels of the putative arsenic transport and metabolism genes are affected by arsenate exposure.
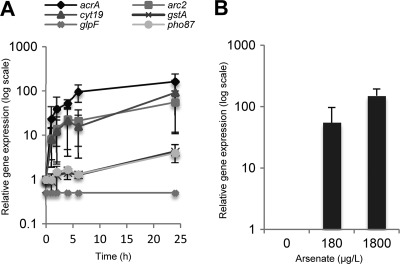
To examine the roles of these candidate genes in arsenic homeostasis, we determined the effects of arsenate on the expression levels of a subset of these genes in wild-type Aspergillus niger as measured by real-time RT-PCR (Table 1). Exposure to high levels of arsenate (180 mg/liter) for 1 h stimulated a significant increase in the expression of several of these genes, in particular gene locus ANI_1_1278164, identified in our search as an orthologue of the S. cerevisiae transporter ACR3 (Fig. 1A). Exposure of hyphae of A. niger to this level of arsenate for 24 h resulted in an over 200-fold increase in the mRNA levels of the ANI_1_1278164 gene locus. Although other putative resistance genes also showed increased expression in the presence of arsenate, the fold increase was highest for this gene. Similar levels of ANI_1_1278164 expression were also seen in response to markedly lower concentrations of arsenate (180 μg/liter; Fig. 1B), suggesting that expression of this gene is highly sensitive to arsenate exposure.
Fig 1.
Effect of arsenate on Aspergillus niger. (A) Wild-type A. niger was incubated in YPD broth containing 180 mg/liter of arsenate or in YPD broth alone for 24 h. RNA extraction was performed at different time points, and the expression levels of the indicated genes were analyzed through real-time RT-PCR. Six putative arsenic homeostasis genes, acrA, arc2, cyt19, gstA, glpF, and pho87, were tested for expression levels in response to arsenate. (B) acrA mRNA expression in wild-type A. niger exposed to 180 or 1,800 μg/liter of arsenate for 24 h. Error bars correspond to the standard deviations of the means for three biological replicates, each performed in triplicate. Gene expression was normalized to the endogenous gene TEF1.
The A. niger gene locus ANI_1_1278164 encodes a putative arsenic transporter of the Acr3 family of arsenite transporter proteins.
The A. niger gene locus ANI_1_1278164 has been annotated as arsB in the National Center for Biotechnology Information (NCBI) database; however, it was identified in our search as bearing significant homology with the S. cerevisiae arsenite transporter gene ACR3. In order to classify the A. niger arsenic transporter and determine its relationship with other known arsenic transporters, a phylogenetic tree was built (see Fig. S1 in the supplemental material), and conserved domain sequences were examined. This analysis indicates that the putative A. niger arsenic transporter encoded by locus ANI_1_1278164 is a member of the Acr3 subfamily (TIGR00832 and COG0798). We therefore designated this gene as acrA, in accordance with Aspergillus gene nomenclature.
Disruption of acrA induces hypersensitivity to arsenic.
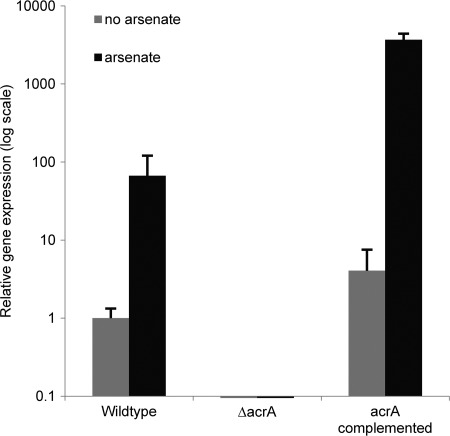
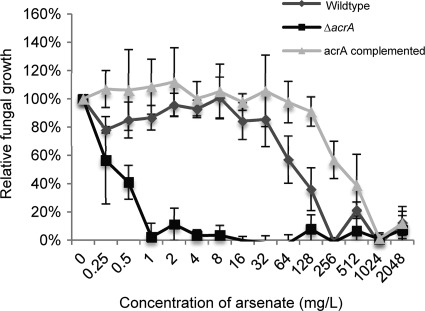
In light of the strong upregulation of acrA in response to arsenate, we focused our initial studies on this gene. To determine the role of acrA in mediating arsenic resistance, we constructed a ΔacrA mutant strain in which the entire coding region of the acrA gene was replaced with the drug resistance marker hygromycin phosphotransferase. To confirm the specificity of this mutation, we complemented this mutant strain with a wild-type allele of acrA at the original locus and confirmed that complementation restored acrA expression (Fig. 2). To determine the consequences of acrA deletion on arsenic sensitivity, we performed a microtiter MIC assay to compare the sensitivity of the wild-type, ΔacrA, and acrA complemented strains to arsenate. Growth of the ΔacrA mutant strain was markedly inhibited by arsenate concentrations as low as 0.5 mg/liter (Fig. 3). In contrast, the wild-type strain produced significant growth at concentrations as high as 64 mg/liter. Complementation of the ΔacrA mutant strain with a wild-type allele of acrA restored arsenic resistance to wild-type levels, confirming that this phenotype was due to the absence of acrA and not an undetected secondary mutation. Collectively, these data strongly suggest that the disruption of acrA results in hypersensitivity to arsenic and that the AcrA protein is important in arsenic detoxification of Aspergillus niger.
Fig 2.
Effects of deletion and reinsertion of acrA on expression levels. Wild-type A. niger, the ΔacrA mutant, and acrA complemented strains were incubated in YPD broth with or without 180 mg/liter of arsenate for 1 h. RNA extraction was performed, and acrA expression levels were analyzed using real-time RT-PCR. Error bars correspond to the standard deviations of the means from three biological replicates. Gene expression was normalized to the endogenous gene TEF1. Results are presented as mean gene expression, normalized to mRNA levels in arsenate-free YPD.
Fig 3.
Hypersensitivity of the ΔacrA mutant strain to arsenate. Serial dilutions of arsenate in liquid Sabouraud medium were prepared in a 96-well plate. All strains were incubated for 24 h with the indicated concentrations of arsenate. Fungal growth was quantified by measuring the OD630, and the absorbance in each well was compared to the same strain growth control in the absence of arsenate and expressed as percent normal growth. Error bars correspond to the standard deviations of the means from three independent experiments.
AcrA is localized to the fungal plasma membrane.
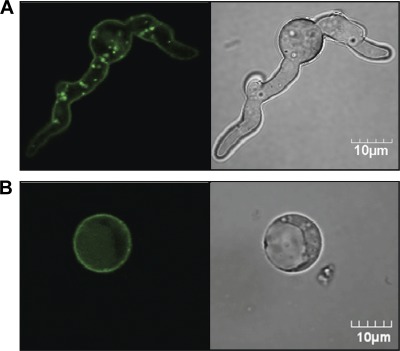
The acrA gene was identified and annotated based on homology to the S. cerevisiae arsenite efflux transporter ACR3. If the A. niger AcrA also functions as an efflux pump, it would be predicted to be located at the plasma membrane. To test this hypothesis, we constructed an acrA-egfp fusion cassette and expressed this construct in wild-type A. niger. Confocal microscopy demonstrated peripheral fungal cell fluorescence, consistent with localization of the AcrA-eGFP fusion protein to the fungal plasma membrane (Fig. 4A). In addition, accumulation of GFP within intracellular vacuoles was observed, likely due to accumulation of degraded fusion protein within the vacuole (41). To confirm that the peripheral fluorescence was localized to the plasma membrane and not the cell wall of A. niger, protoplasts in which the fungal cell wall had been removed by enzymatic digestion were examined. Removal of the fungal cell wall had no effect on the fluorescence of cells, confirming that AcrA-eGFP localized to the plasma membrane and not the cell wall of A. niger (Fig. 4B). Collectively, these findings are consistent with a role for AcrA as a plasma membrane efflux transporter.
Fig 4.
GFP localization of AcrA in A. niger. (A) Germlings of the AcrA-GFP overexpression strain grown for 6 h in YPD broth. Pictures were taken with confocal microscopy at 100× oil immersion. (B) Protoplasts of the AcrA-GFP overexpression strain after enzymatic digestion of the cell wall, observed under confocal microscopy at 100× oil immersion. GFP images are shown on the left, and differential interference contrast (DIC) pictures are shown on the right.
AcrA plays a role in arsenic transport in Aspergillus niger.
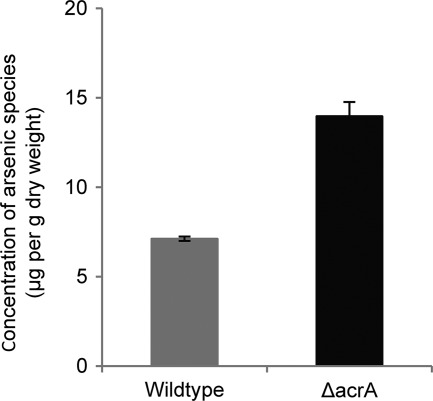
The results of our MIC and localization studies supported the hypothesis that AcrA functions as an arsenite efflux transporter. To test this hypothesis specifically, we compared the accumulation of total arsenic species within mycelia of the wild-type and ΔacrA mutant strain using a Perkin Elmer AAnalyst800 atomic absorption spectrometer with a graphite furnace (Fig. 5). To ensure that only intracellular and not adsorbed arsenic species were measured, the fungal biomass was washed with HCl before the cellular arsenic levels were measured (23). Very low levels of arsenic (0.6 to 0.9 μg per gram [dry weight]) were found in dead hyphae of both the wild-type and ΔacrA mutant strains incubated with 180 mg/liter of arsenate, and there was no significant difference between the strains (data not shown). In contrast, live hyphae incubated under these conditions contained significant levels of arsenic, with a significantly higher level of arsenic detected in the ΔacrA mutant compared with wild-type A. niger (Fig. 5). These results are consistent with the hypothesis that AcrA functions in the efflux of arsenic in A. niger.
Fig 5.
Quantification of arsenic uptake by live A. niger strains. A. niger strains were grown in YPD liquid broth overnight. The mycelial biomass was then cut into 1-cm2 squares, inoculated in YPD liquid broth with 180 mg/liter of arsenate, and incubated for 24 h. The cultures were then harvested, and the mycelia were washed with 0.1 N HCl to remove adsorbed arsenate. The mycelial biomass samples were dried and digested, and the quantification of total arsenic species was determined by Perkin Elmer AAnalyst800 atomic absorption spectrometer with graphite furnace. Graphs indicate means ± standard errors. Data comprise three independent experiments, each performed in duplicate. P < 0.05, by factor analysis of variance (ANOVA).
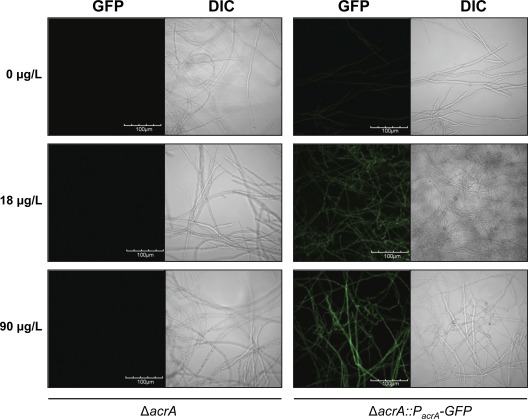
Use of a ΔacrA::PacrA-GFP strain as a potential biosensor of arsenic.
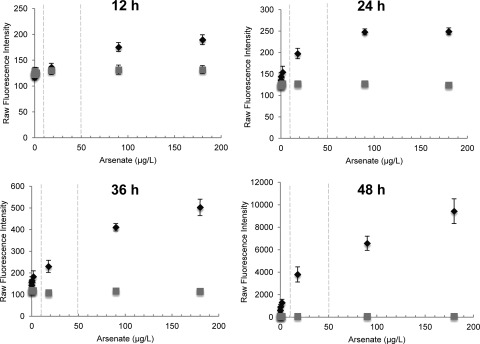
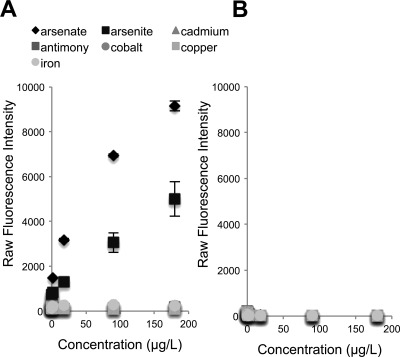
The dramatic upregulation of acrA expression in response to low levels of arsenate suggested that this gene might be useful as a potential biosensor for arsenic. To test this hypothesis, we created a fusion construct in which egfp was fused to the acrA promoter. This plasmid was then expressed in the ΔacrA strain. Through confocal microscopy, we observed that the strain fluoresced even at concentrations of arsenate as low as 18 μg/liter (Fig. 6). To determine the sensitivity and kinetics of the biosensor strain in response to arsenate, we tested the ability of conidia of the biosensor strain to germinate and fluoresce in the presence of arsenate. Exposure of conidia of this strain to a gradient of arsenate produced a dose-dependent fluorescence that was detectable as early as 12 h after incubation and increased dramatically by 48 h of arsenate exposure (Fig. 7). At this time point, reproducible dose-dependent fluorescence was observed throughout the range of the maximum safe limit of arsenic detection (10 to 50 μg/liter) suggested by the World Health Organization (48, 56). In parallel, we also tested the effects of exposing pregrown hyphae of the biosensor strain to the same range of arsenate concentrations. Although greater overall fluorescence was seen with pregrown hyphae, the background autofluorescence of hyphae alone increased to the same degree, and no significant difference in the fluorescent signal was found compared with exposure of conidia (data not shown). In addition, we tested the specificity of the biosensor strain for arsenate by testing several other metals and metalloids, including arsenite, antimony(III), copper(II), cobalt(II), iron(III), and cadmium (Fig. 8). Interestingly, the biosensor strain was specific to both arsenate and arsenite species and did not produce significant fluorescence with any other metals tested. Collectively, these data suggest that a biosensor strategy based on the A. niger acrA promoter sequence may be a useful approach for the detection of low-level arsenic contamination of water, although further studies to examine the reproducibility and specificity of this biosensor in field specimens should be performed.
Fig 6.
Fluorescence of the biosensor strain in response to arsenate. Conidia of the ΔacrA and ΔacrA::PacrA-GFP strains were incubated in YPD broth with arsenate at the indicated concentrations on glass coverslips at 37°C. After 24 h of incubation, confocal images of fungi on coverslips were obtained using confocal microscopy. The ΔacrA strain was used as a control for autofluorescence.
Fig 7.
Arsenate detection using conidia of the A. niger biosensor strain. Conidia of the ΔacrA and ΔacrA::PacrA-GFP strains were grown in YPD broth supplemented with arsenate at the indicated concentrations. At 12 h, 24 h, 36 h, and 48 h, GFP expression was measured by determining the raw fluorescence intensity using a Spectramax fluorometer. Dashed lines indicate the arsenic threshold concentrations set by the WHO for drinking water in developed and developing countries (10 μg/liter and 50 μg/liter, respectively). Results are shown as means ± standard errors. Data comprise two independent experiments, each performed in triplicate. ♦ indicates ΔacrA::PacrA-GFP, and  indicates ΔacrA. Note that the scale of the y axis increases over time.
indicates ΔacrA. Note that the scale of the y axis increases over time.
Fig 8.
Specificity of the A. niger biosensor strain for arsenic species. (A) Conidia of the ΔacrA::PacrA-GFP strain was grown in YPD broth supplemented with different metals and metalloids, including arsenate, arsenite, cadmium, antimony(III), cobalt(II), copper(II), and iron(III) at the indicated concentrations. At 48 h, GFP expression was measured by determining the raw fluorescence intensity using a Spectramax fluorometer. (B) Conidia of the ΔacrA strain were used as a control for autofluorescence. Results are shown as means ± standard errors. Data comprise two independent experiments, each performed in triplicate.
DISCUSSION
Homology searches based on known bacterial and fungal arsenic resistance pathways identified seven putative arsenic metabolism and transport genes in Aspergillus niger (Table 1). Interestingly, although an A. niger orthologue of S. cerevisiae ACR3 was found, it was not located in a gene cluster as is the case in S. cerevisiae. Indeed, no orthologue for the S. cerevisiae ACR1 regulatory gene could be found in A. niger, and an orthologue of ACR2 encoding a putative arsenate reductase was found clustered with genes encoding a putative arsenic methyltransferase and putative arsenic resistance protein gene, arsH. Interestingly, the arsH gene is not found in S. cerevisiae, although it is found in different bacterial species, most notably in Shigella flexneri (65). This cluster found in A. niger contrasts with Aspergillus fumigatus, in which the putative arsenate reductase, arsenite efflux transporter, and arsenic methyltransferase genes are found together in a duplicated cluster on chromosomes 1 and 5, highlighting the genomic diversity of this genus.
By extrapolating the function of the products of these A. niger genes from the role of their orthologues in yeast arsenic metabolism and transport, we propose a hypothetical model of arsenic homeostasis in A. niger (8). In this predicted model based on homology of A. niger genes with their yeast orthologues, arsenic from the environment is transported into the cell either as arsenite through GlpF or as arsenate through Pho87. Intracellular arsenate is reduced to arsenite by Arc2 and then detoxified by one of the three methods: glutathionation by GstA and sequestration into vacuoles by an ABC metal ion transporter, biovolatilisation via methylation by Cyt19, or direct export from the cell by the AcrA efflux transporter. To begin to test this model, we selected acrA, the A. niger homologue of S. cerevisiae ACR3, for further study (9).
In S. cerevisiae, the ACR3 gene encodes an arsenite transporter whose expression is strongly induced by the presence of arsenite and arsenate (51). Multiple lines of evidence suggest that this function is preserved in A. niger: acrA mRNA expression was markedly upregulated in the presence of arsenate, an AcrA-eGFP fusion construct localized to the fungal plasma membrane, and deletion of acrA resulted in arsenate hypersensitivity and accumulation of intracellular arsenic species. Although the loss of acrA resulted in marked hypersensitivity to arsenate, this phenotype was associated with only a modest increase in intracellular arsenic levels. These data suggest a low tolerance for increased intracellular arsenic levels in the A. niger ΔacrA strain, beyond which the toxicity of arsenic causes cell death and therefore prevents further cellular accumulation of arsenic. Collectively, these data provide support to our model in which AcrA-mediated efflux plays a key role in arsenic homeostasis in A. niger, although experimental confirmation of the other elements of this system is still required. Further, as our approach was based on the detection of known pathways of arsenic homeostasis in other organisms, it is possible that other mechanisms of arsenic detoxification, unique to filamentous fungi, contribute to arsenic resistance in this strain.
The marked upregulation of acrA expression in the face of low concentrations of arsenate suggests that the acrA promoter is quite sensitive to arsenic (Fig. 1B). Based on this observation, we performed proof-of-principle studies to determine if an A. niger acrA promoter fusion could function as a biosensor for arsenic. Using an egfp fusion construct, we developed a strain that produced dose-dependent fluorescence in response to arsenic concentrations in the range of the recommended WHO limit of 10 to 50 μg/liter of arsenic for drinking water (Fig. 7) and did not react to a range of other metals and metalloids (Fig. 8). This approach therefore holds promise for use in testing water samples from regions, such as Bangladesh, where 43% of more than 50,000 hand tube well water samples analyzed were found to have arsenic concentrations above 10 μg/liter and 27% had arsenic concentrations above 50 μg/liter (48).
Currently, numerous arsenic detection methods exist for the detection of arsenic contamination in drinking water. The laboratory-based analytical methods are extremely quantitative and accurate but require extensive training and are costly. In addition, transport times limit their use in field situations (20). Other field use strategies are therefore preferable for the testing of arsenic-contaminated water samples from countries like Bangladesh and India (5). The field-based methods in current use are cost-effective but produce toxic by-products and use dangerous reagents like mercury (5, 21) and hydrochloric acid, which are not ideal to transport and handle in the field (5). Most whole-cell biosensors reported to date are based on bacterial systems (7, 22, 29). The ΔacrA::PacrA-GFP biosensor strain is the first fungal biosensor for arsenic detection. There are numerous advantages of using fungi as whole-cell biosensors (7). Bacterial cells have short shelf and application lives since they are relatively fragile under arsenic exposure. They are also limited in terms of tolerance to different pH, temperature, and osmotic environments (7). On the other hand, fungi are more tolerant to and robust in diverse environmental conditions, a significant advantage for a biosensor-based assay for use in the field. Further, fungal spores are extremely robust and can be stored in desiccated form for years without loss of viability. Moreover, A. niger is a nonpathogenic strain widely used in industrial applications, is considered a “generally regarded as safe” organism, and can be further modified to produce auxotrophic strains that are nonviable outside the laboratory.
The choice of a target gene for construction of a biosensor is critical, as the degree of target upregulation governs the reporter output, and specificity of the target gene for arsenic exposure will determine the specificity of the assay. The majority of arsenic-responsive biosensor bacterial strains use reporter constructs based on the transcriptional repressor arsR (22, 64). Cross-reactions with antimony and other metalloids have been described using this gene (50). Our choice of the acrA system for the reporter fusion was highly specific for arsenite and arsenate and did not cross-react with antimony or other metalloids. Further, the very high levels of upregulation of acrA expression seen in response to arsenic exposure translated to a highly sensitive arsenic detection assay in vitro.
Despite these advantages, the A. niger biosensor strain has several limitations. First, in this proof-of-principle study, we used eGFP and fluorimetry as a detection system. For use in the field, a colorimetric detector such as glucuronide or X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (2, 20, 30, 33) would be preferable, as this system would obviate detection equipment and allow for a point-of-use testing. Second, signal detection using fluorescence required a minimum of 12 h of incubation, and this might be prolonged further using a colorimetric detector. Finally, before actual field testing, a thorough investigation of the effects of other parameters will be required, including specificity of the biosensor for arsenic in field water samples and the effects of growth media, storage conditions, and incubation temperatures. Although these experiments are beyond the scope of the current study, this proof-of-principle study suggests the possibility that an A. niger acrA reporter-based bioassay could provide the basis for a simple and economical test kit for measuring and monitoring arsenic levels in contaminated groundwater of developing nations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Glenna Keating (Trace Element Analysis Lab, Department of Earth & Planetary Sciences at McGill University) for arsenic quantification and Axel Brakhage (Leibniz Institute for Natural Product Research and Infection Biology-HKI, Germany) for providing the pEYFPC and p123 plasmids.
DCS was supported by a Canadian Institutes of Health Research (CIHR) Clinician Scientist Award, a Burroughs Welcome Fund Career Award in the Biomedical Sciences, and a Chercheur-Boursier Clinicien Award from the Fonds de la Recherche en Santé du Québec. This work was supported by an operating grant from CIHR.
Footnotes
Published ahead of print 30 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Achour AR, Bauda P, Billard P. 2007. Diversity of arsenite transporter genes from arsenic-resistant soil bacteria. Res. Microbiol. 158:128–137 [DOI] [PubMed] [Google Scholar]
- 2. Adams MR, Grubb SM, Hamer A, Clifford MN. 1990. Colorimetric enumeration of Escherichia coli based on beta-glucuronidase activity. Appl. Environ. Microbiol. 56:2021–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 4. Apweiler R, et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29:37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arora M, Megharaj M, Naidu R. 2009. Arsenic testing field kits: some considerations and recommendations. Environ. Geochem. Health 31(Suppl 1):45–48 [DOI] [PubMed] [Google Scholar]
- 6. Baker SE. 2006. Aspergillus niger genomics: past, present and into the future. Med. Mycol. 44(Suppl 1):S17–S21 [DOI] [PubMed] [Google Scholar]
- 7. Baronian KHR. 2004. The use of yeast and moulds as sensing elements in biosensors. Biosens. Bioelectron. 19:953–962 [DOI] [PubMed] [Google Scholar]
- 8. Bhattacharjee H, Rosen BP. 2007. Arsenic metabolism in prokaryotic and eukaryotic microbes, p 371–406 In Nies D, Silver S. (ed), Molecular microbiology of heavy metals, vol 6 Springer Berlin, Heidelberg, Germany [Google Scholar]
- 9. Bobrowicz P, Wysocki R, Owsianik G, Goffeau A, Ulaszewski S. 1997. Isolation of three contiguous genes, ACR1, ACR2 and ACR3, involved in resistance to arsenic compounds in the yeast Saccharomyces cerevisiae. Yeast 13:819–828 [DOI] [PubMed] [Google Scholar]
- 10. Bučková M, Godočíková J, Polek B. 2007. Responses in the mycelial growth of Aspergillus niger isolates to arsenic contaminated environments and their resistance to exogenic metal stress. J. Basic Microbiol. 47:295–300 [DOI] [PubMed] [Google Scholar]
- 11. Bučková M, Godočíková J, Simonovičová A, Polek B. 2005. Production of catalases by Aspergillus niger isolates as a response to pollutant stress by heavy metals. Curr. Microbiol. 50:175–179 [DOI] [PubMed] [Google Scholar]
- 12. Bun-ya M, et al. 1996. Two new genes, PHO86 and PHO87, involved in inorganic phosphate uptake in Saccharomyces cerevisiae. Curr. Genet. 29:344–351 [PubMed] [Google Scholar]
- 13. Cai L, Liu G, Rensing C, Wang G. 2009. Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils. BMC Microbiol. 9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cánovas D, de Lorenzo V. 2007. Osmotic stress limits arsenic hypertolerance in Aspergillus sp. P37. FEMS Microbiol. Ecol. 61:258–263 [DOI] [PubMed] [Google Scholar]
- 15. Cánovas D, Durán C, Rodríguez N, Amils R, de Lorenzo V. 2003. Testing the limits of biological tolerance to arsenic in a fungus isolated from the River Tinto. Environ. Microbiol. 5:133–138 [DOI] [PubMed] [Google Scholar]
- 16. Cánovas D, Mukhopadhyay R, Rosen BP, de Lorenzo V. 2003. Arsenate transport and reduction in the hyper-tolerant fungus Aspergillus sp. P37. Environ. Microbiol. 5:1087–1093 [DOI] [PubMed] [Google Scholar]
- 17. Cánovas D, Vooijs R, Schat H, de Lorenzo V. 2004. The role of thiol species in the hypertolerance of Aspergillus sp. P37 to arsenic. J. Biol. Chem. 279:51234–51240 [DOI] [PubMed] [Google Scholar]
- 18. Cernanský S, Kolencík M, Sevc J, Urík M, Hiller E. 2009. Fungal volatilization of trivalent and pentavalent arsenic under laboratory conditions. Bioresour. Technol. 100:1037–1040 [DOI] [PubMed] [Google Scholar]
- 19. Clausen C. 2004. Improving the two-step remediation process for CCA-treated wood: part I. Evaluating oxalic acid extraction. Waste Manag. 24:401–405 [DOI] [PubMed] [Google Scholar]
- 20. de Mora K, et al. 2011. A pH-based biosensor for detection of arsenic in drinking water. Anal. Bioanal. Chem. 400:1031–1039 [DOI] [PubMed] [Google Scholar]
- 21. Dhar RK, Zheng Y, Rubenstone J, van Geen A. 2004. A rapid colorimetric method for measuring arsenic concentrations in groundwater. Anal. Chim. Acta 526:203–209 [Google Scholar]
- 22. Diesel E, Schreiber M, van der Meer JR. 2009. Development of bacteria-based bioassays for arsenic detection in natural waters. Anal. Bioanal. Chem. 394:687–693 [DOI] [PubMed] [Google Scholar]
- 23. Gadd GM. 1993. Interactions of fungi with toxic metals. New Phytol. 124:25–60 [Google Scholar]
- 24. Galal-Gorchev H, Ozolins G, Bonnefoy X. 1993. Revision of the WHO guidelines for drinking water quality. Ann. Ist. Super. Sanita 29:335–345 [PubMed] [Google Scholar]
- 25. Geng C, Zhu Y, Hu Y, Williams P, Meharg A. 2006. Arsenate causes differential acute toxicity to two P-deprived genotypes of rice seedlings (Oryza sativa L.). Plant Soil 279:297–306 [Google Scholar]
- 26. Ghosh M, Shen J, Rosen BP. 1999. Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 96:5001–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gravelat FN, et al. 2010. Aspergillus fumigatus MedA governs adherence, host cell interactions and virulence. Cell. Microbiol. 12:473–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haft DH, Selengut JD, White O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res. 31:371–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hansen LH, Sørensen SJ. 2001. The use of whole-cell biosensors to detect and quantify compounds or conditions affecting biological systems. Microb. Ecol. 42:483–494 [DOI] [PubMed] [Google Scholar]
- 30. Hirt H. 1991. A novel method for in situ screening of yeast colonies with the beta-glucuronidase reporter gene. Curr. Genet. 20:437–439 [DOI] [PubMed] [Google Scholar]
- 31. Hoff B, Kück U. 2005. Use of bimolecular fluorescence complementation to demonstrate transcription factor interaction in nuclei of living cells from the filamentous fungus Acremonium chrysogenum. Curr. Genet. 47:132–138 [DOI] [PubMed] [Google Scholar]
- 32. Jain CK, Ali I. 2000. Arsenic: occurrence, toxicity and speciation techniques. Water Res. 34:4304–4312 [Google Scholar]
- 33. Joshi N, Wang X, Montgomery L, Elfick A, French CE. 2009. Novel approaches to biosensors for detection of arsenic in drinking water. Desalination 248:517–523 [Google Scholar]
- 34. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 35. Maciaszczyk-Dziubinska E, Migdal I, Migocka M, Bocer T, Wysocki R. 2010. The yeast aquaglyceroporin Fps1p is a bidirectional arsenite channel. FEBS Lett. 584:726–732 [DOI] [PubMed] [Google Scholar]
- 36. Marchler-Bauer A, et al. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Messens J, Silver S. 2006. Arsenate reduction: thiol cascade chemistry with convergent evolution. J. Mol. Biol. 362:1–17 [DOI] [PubMed] [Google Scholar]
- 38. Mukherjee A, et al. 2010. Tolerance of arsenate-induced stress in Aspergillus niger, a possible candidate for bioremediation. Ecotoxicol. Environ. Saf. 73:172–182 [DOI] [PubMed] [Google Scholar]
- 39. Mukhopadhyay R, Shi J, Rosen BP. 2000. Purification and characterization of ACR2p, the Saccharomyces cerevisiae arsenate reductase. J. Biol. Chem. 275:21149–21157 [DOI] [PubMed] [Google Scholar]
- 40. Páez-Espino D, Tamames J, de Lorenzo V, Cánovas D. 2009. Microbial responses to environmental arsenic. Biometals 22:117–130 [DOI] [PubMed] [Google Scholar]
- 41. Petersson J, Pattison J, Kruckeberg AL, Berden JA, Persson BL. 1999. Intracellular localization of an active green fluorescent protein-tagged Pho84 phosphate permease in Saccharomyces cerevisiae. FEBS Lett. 462:37–42 [DOI] [PubMed] [Google Scholar]
- 42. Pethica R, Barker G, Kovacs T, Gough J. 2010. TreeVector: scalable, interactive, phylogenetic trees for the web. PLoS One 5:e8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pokhrel D, Viraraghavan T. 2006. Arsenic removal from an aqueous solution by a modified fungal biomass. Water Res. 40:549–552 [DOI] [PubMed] [Google Scholar]
- 44. Pokhrel D, Viraraghavan T. 2008. Arsenic removal from an aqueous solution by modified A. niger biomass: batch kinetic and isotherm studies. J. Hazard. Mater. 150:818–825 [DOI] [PubMed] [Google Scholar]
- 45. Pokhrel D, Viraraghavan T. 2006. Arsenic removal from aqueous solution by iron oxide-coated fungal biomass: a factorial design analysis. Water Air Soil Pollut. 173:195–208 [Google Scholar]
- 46. Pokhrel D, Viraraghavan T. 2008. Arsenic removal in an iron oxide-coated fungal biomass column: analysis of breakthrough curves. Bioresour. Technol. 99:2067–2071 [DOI] [PubMed] [Google Scholar]
- 47. Price MS, Classen JJ, Payne GA. 2001. Aspergillus niger absorbs copper and zinc from swine wastewater. Bioresour. Technol. 77:41–49 [DOI] [PubMed] [Google Scholar]
- 48. Rahman MM, Naidu R, Bhattacharya P. 2009. Arsenic contamination in groundwater in the Southeast Asia region. Environ. Geochem. Health 31:9–21 [DOI] [PubMed] [Google Scholar]
- 49. Ren X, et al. 2011. Involvement of N-6 adenine-specific DNA methyltransferase 1 (N6AMT1) in arsenic biomethylation and its role in arsenic-induced toxicity. Environ. Health Perspect. 119:771–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roberto FF, Barnes JM, Bruhn DF. 2002. Evaluation of a GFP reporter gene construct for environmental arsenic detection. Talanta 58:181–188 [DOI] [PubMed] [Google Scholar]
- 51. Rosen BP, Tamás MJ. 2010. Arsenic transport in prokaryotes and eukaryotic microbes. Adv. Exp. Med. Biol. 679:47–55 [DOI] [PubMed] [Google Scholar]
- 52. Rosen P. 1971. Theoretical significance of arsenic as a carcinogen. J. Theor. Biol. 32:425–426 [DOI] [PubMed] [Google Scholar]
- 53. Rozen S, Skaletsky HJ. 2000. Primer3 on the WWW for general users and for biologist programmers, p 365–386 In Krawetz S, Misenser S. (ed), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 54. Sharples JM, Meharg AA, Chambers SM, Cairney JW. 2000. Mechanism of arsenate resistance in the ericoid mycorrhizal fungus Hymenoscyphus ericae. Plant Physiol. 124:1327–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sheppard DC, et al. 2005. The Aspergillus fumigatus StuA protein governs the up-regulation of a discrete transcriptional program during the acquisition of developmental competence. Mol. Biol. Cell 16:5866–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Smith AH, Lingas EO, Rahman M. 2000. Contamination of drinking-water by arsenic in Bangladesh: a public health emergency. Bull. World Health Organ. 78:1093–1103 [PMC free article] [PubMed] [Google Scholar]
- 57. Spellig T, Bottin A, Kahmann R. 1996. Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol. Gen. Genet. 252:503–509 [DOI] [PubMed] [Google Scholar]
- 58. Su S, Zeng X, Bai L, Jiang X, Li L. 2010. Bioaccumulation and biovolatilisation of pentavalent arsenic by Penicillin janthinellum, Fusarium oxysporum and Trichoderma asperellum under laboratory conditions. Curr. Microbiol. 61:261–266 [DOI] [PubMed] [Google Scholar]
- 59. Tamás MJ, Wysocki R. 2001. Mechanisms involved in metalloid transport and tolerance acquisition. Curr. Genet. 40:2–12 [DOI] [PubMed] [Google Scholar]
- 60. Thakur JK, Thakur RK, Ramanathan A, Kumar M, Singh SK. 2011. Arsenic contamination of groundwater in Nepal—an overview. Water 3:1–20 [Google Scholar]
- 61. Thorsen M, et al. 2009. Genetic basis of arsenite and cadmium tolerance in Saccharomyces cerevisiae. BMC Genomics 10:10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Todorova T, Vuilleumier S, Kujumdzieva A. 2007. Role of glutathione s-transferases and glutathione in arsenic and peroxide resistance in Saccharomyces cerevisiae: a reverse genetic analysis approach. Biotechnol. Biotechnol. Eq. 21:348–352 [Google Scholar]
- 63. Twumasi-Boateng K, et al. 2009. Transcriptional profiling identifies a role for BrlA in the response to nitrogen depletion and for StuA in the regulation of secondary metabolite clusters in Aspergillus fumigatus. Eukaryot. Cell 8:104–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. van der Meer JR, Belkin S. 2010. Where microbiology meets microengineering: design and applications of reporter bacteria. Nat. Rev. Microbiol. 8:511–522 [DOI] [PubMed] [Google Scholar]
- 65. Vorontsov II, et al. 2007. Crystal structure of an apo form of Shigella flexneri ArsH protein with an NADPH-dependent FMN reductase activity. Protein Sci. 16:2483–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wysocki R, Bobrowicz P, Ulaszewski S. 1997. The Saccharomyces cerevisiae ACR3 gene encodes a putative membrane protein involved in arsenite transport. J. Biol. Chem. 272:30061–30066 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.