Abstract
The genomes of human immunodeficiency virus type 1 (HIV-1) and hepatitis C virus (HCV) consist of single-stranded RNA encoding polyproteins, which are processed to individual functional proteins by virus-encoded specific proteases. These proteases have been used as targets for drug development. Here, instead of targeting these proteases to inhibit viral infection, we utilized the protease activity to activate a toxic protein to prevent viral infection. We engineered the MazE-MazF antitoxin-toxin system of Escherichia coli to fuse a C-terminal 41-residue fragment of antitoxin MazE to the N-terminal end of toxin MazF with a linker having a specific protease cleavage site for either HIV PR (HIV-1 protease), NS3 protease (HCV protease), or factor Xa. These fusion proteins formed a stable dimer (instead of the MazF2-MazE2-MazF2 heterohexamer in nature) to inactivate the ACA (sequence)-specific mRNA interferase activity of MazF. When the fusion proteins were incubated with the corresponding proteases, the MazE fragment was cleaved from the fusion proteins, releasing active MazF, which then acted as an ACA-specific mRNA interferase cleaving single-stranded MS2 phage RNA. The intramolecular regulation of MazF toxicity by proteases as demonstrated may provide a novel approach for preventive and therapeutic treatments of infection by HIV-1, HCV, and other single-stranded RNA viruses.
INTRODUCTION
The proteases encoded by a number of RNA viruses, such as human immunodeficiency virus (HIV-1) and hepatitis C virus (HCV), play an essential role in viral infection, as they are required for the processing of virus-encoded polyproteins (3, 9). Thus, HIV-1 and HCV proteases have been considered ideal drug targets (7, 10, 16). However, the major problem in using these proteases as drug targets is that viruses readily develop resistance to newly developed drugs, resulting in vicious cycles for drug development (2, 20). In the present report, to circumvent the problem of using the protease as a drug target, we attempted to positively use the activity of the viral proteases to activate a latent toxin (MazF) of Escherichia coli from a toxin-antitoxin (MazF-MazE) fusion protein by cleaving off antitoxin MazE fragment. MazF thus released functions as an ACA-specific mRNA interferase (25) to eliminate almost all cellular mRNA as well as single-stranded viral RNA in the virus-infected cells.
The HIV-1 genome, consisting of a single-stranded RNA of 9,749 bases, encodes two polyproteins (Pr55gag and Pr160gag-pol) which have to be processed by the HIV-1 protease, HIV PR (8). One of the proteins derived from Pr160gag-pol polyprotein is HIV PR, a small 99-residue aspartyl protease, that is essential for HIV-1 infection. Another RNA virus, HCV, causing chronic hepatitis and serious liver diseases, also consists of a single-stranded RNA of 9,600 nucleotides (12, 19). It encodes a polyprotein of about 3,000 amino acid residues, which is processed to smaller functional proteins by host and virus proteases such as NS3 protease (1).
Since these viral proteases cleave polyproteins at highly specific amino acid sequences, these specific protease cleavage sites for individual RNA viruses may be incorporated into the linker between MazE antitoxin and MazF toxin of the MazE-MazF fusion proteins so that viral proteases induced upon infection cleave the linker to activate MazF as an ACA-specific mRNA interferase. MazF has been shown to be a potent toxin, effectively causing apoptotic cell death in mammalian cells (22). In E. coli cells, two dimers of MazF form a stable heterohexamer complex with one MazE dimer in the center (Fig. 1C) (11). In this report, we fused a short 41-residue C-terminal fragment of MazE to the N-terminal end of MazF with a polypeptide linker which contains a specific cleavage site for HIV PR, HCV NS3 protease, or factor Xa (Fig. 1 and Table 1). We demonstrate that all of these fusion proteins can be activated to exert the MazF mRNA interferase activity only when treated with the corresponding specific proteases. We also demonstrate that the MazE-MazF fusion proteins form a stable dimer (Fig. 2B), in contrast to hexamer MazF2-MazE2-MazF2, which is formed in the cells. The present results suggest a novel preventive and therapeutic strategy against RNA viruses, such as HIV-1, HCV, and other single-stranded RNA viruses.
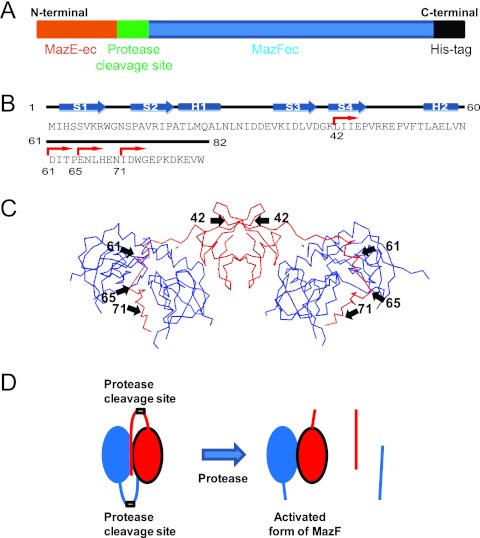
Fig 1.
Schematic presentation of the MazE-MazF fusion protein with a linker containing a specific cleavage site for factor Xa, HIV PR, or HCV NS3 protease. (A) Schematic presentation of the MazE-MazF fusion protein with a linker (in green) containing a protease cleavage site and a His6 tag at the C-terminal end. (B) The amino acid sequence of MazE. The secondary structures, determined by X-ray crystallography (11), are also indicated above the sequence. Note that it contains a 22-residue-long unstructured C-terminal extension. β-Strands and α-helices are indicated with blue arrows and rectangular shapes, respectively. Red arrows indicate the starting residues for the C-terminal segments used in the present study. The residue number of the first residue of each segment is also indicated. (C) Crystal structure of the MazE-MazF complex (Protein Data Bank entry 1UB4). Black arrows and numbers indicate the sites and the residue numbers of truncated MazE fragments. (D) Schematic presentation of the activation of the MazE-MazF fusion protein having a protease cleavage site in the linker. Blue and red molecules of the MazE-MazF fusion protein form a dimer as shown, and the yellow box indicates the protease cleavage site between the MazE fragment and MazF.
Table 1.
Activation of various MazE-MazF fusion proteins by factor Xa, HIV-1 PR, and HCV NS3 protease
| Name | MazE regiona (aa) | Protease cleavage site | Expressionb | Toxicityc |
|---|---|---|---|---|
| MazEF-FXa | 42 to 82 | Factor Xa | +++ | − |
| MazE(61)F-FXa | 61 to 82 | Factor Xa | + | + |
| MazE(65)F-FXa | 65 to 82 | Factor Xa | − | +++ |
| MazE(71)F-FXa | 71 to 82 | Factor Xa | − | +++ |
| MazEF-HIV | 42 to 82 | HIV-1 PR | +++ | − |
| MazEF-HCV | 42 to 82 | HCV NS3 protease | +++ | − |
The residue numbers from the N-terminal end of MazE are shown (11).
A plus sign indicates expression, and three plus signs indicate very high expression of the fusion proteins. A minus indicates no expression.
A minus indicates that the fusion protein is not toxic even in the presence of IPTG. A plus sign indicates that it is toxic only in the presence of IPTG. Three plus signs indicate that it is toxic even in the absence of IPTG.
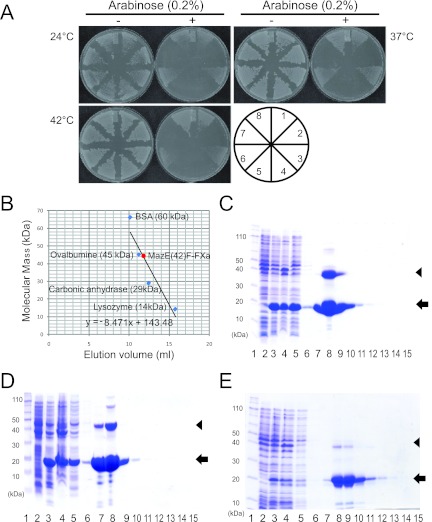
Fig 2.
Characterization of MazE-MazF fusion protein. (A) Toxicity of MazE(42)F-FXa in E. coli. The transformants harboring pBAD24 vector (1 and 8), mazF-ec (2 and 7), mazF-bs (4 and 5), and mazE(42)F-FXa (3 and 6) were streaked on M9 plates with or without 0.2% arabinose and incubated at three different temperatures, 24, 37, and 42°C. (B) Gel filtration of MazE(42)F-FXa. The linear trend line is used for calculating the molecular mass for the MazE-MazF fusion protein. The purification of MazE(42)F-FXa (C), MazEF-HIV (D), and MazEF-HCV (E) are also shown. Lane 1, molecular mass markers; lane 2, the whole-cell lysate without induction; lane 3, the whole-cell lysate after incubation for 4 h at 37°C; lane 4, the cell pellet; lane 5, flowthrough fraction; lane 6, wash fraction; and lanes 7 to 15, elution fractions. Arrows indicate the positions of monomers of the fusion proteins, and arrowheads indicate the positions of their dimers.
MATERIALS AND METHODS
Construction of MazE-MazF fusion proteins.
Four kinds of lengths of C-terminal fragments of Escherichia coli MazE (Fig. 1B and C) were fused to E. coli MazF with protease cleavage sites and four extra residues (Gly-Gly and Gly-Ser at the N-terminal and the C-terminal ends, respectively) (Fig. 1A). For amplifying mazE fragments, the following primers were used: MazE(42) (forward, TATACATATGTTAATTATTGAGCCA), MazE(61) (forward, TATACATATGGACATCACGCCGGAA), MazE(65) (forward, TATACATATGGAAAACCTCCACGAG), MazE(71) (forward, TATACATATGATCGACTGGGGAGAGCCG), MazE(FXa) (reverse, TATAGGATCCACGACCTTCAATACCTCCCCAGACTTCCTTATC), MazE(HCV) (reverse, TATAGGATCCCACCCAGGTGCTGGTCACCACTTCCAGATCACCTCCCCAGACTTCCTTATC), and MazE(HIV) (reverse, TATAGGATCCCGCTTCCGCCAGCACACGCGCACCTCCCCAGACTTCCTTATC).
Each PCR fragment was cloned into pET21c (Novagen) with a C-terminal His6 tag (24). pET21c-mazE(42)F-Fxa, pET21c-mazE(61)F-Fxa, pET21c-mazE(65)F-Fxa, and pET21c-mazE(71)F-Fxa have a sequence corresponding to a factor Xa cleavage site between mazE and mazF genes. pET21c-mazEF-HIV and pET21c-mazEF-HCV include HIV PR and HCV NS3 serine protease cleavage sites, respectively.
Toxicity of MazE(42)F-FXa in vivo.
mazE(42)F-Fxa was cloned in pBAD24 (23). pBAD24-mazF-ec (mazF from E. coli) and pBAD24-mazF-bs (mazF from Bacillus subtilis [18]) were used for positive controls. E. coli BW25113 cells were used for transformation, and the transformants harboring mazF-ec, mazF-bs, and mazE(42)F-Fxa were streaked on M9 plates in the presence or absence of 0.2% arabinose.
Expression and purification of MazE-MazF with protease cleavage sites.
To purify MazE-MazF fusion proteins containing protease cleavage sites, pET21c-mazE(42)F-Fxa, pET21c-mazE(61)F-Fxa, pET21c-mazE(65)F-Fxa, pET21c-mazE(71)F-Fxa, pET21c-mazEF-HIV, and pET21c-mazEF-HCV were introduced into E. coli BL21(DE3). The fusion proteins were induced by the addition of 0.5 mM isopropyl-β-d-1-thiogalactoside (IPTG) for 4 h at 37°C and then purified with Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) according to the manufacturer's protocol. The HIV PR and HCV NS3 proteases were kind gifts from Edward Arnold (Rutgers University) and Smita Patel (University of Medicine and Dentistry of New Jersey), respectively.
Dimer formation of the MazE-MazF fusion proteins.
Gel filtration chromatography was performed on an ÄKTA purifier system (GE Healthcare) with a Superdex 75 column (GE Healthcare). Five hundred micrograms of MazE(42)F-Fxa protein was applied to the column and equilibrated with a buffer containing 10 mM Tris-HCl (pH 7.0) and 100 mM NaCl. Protein markers were separated under the same conditions and detected by the UV light detector (280 nm) of the ÄKTA purifier.
Activation of the MazE-MazF fusion proteins.
To measure the activity of MazF from the MazE-MazF fusion proteins, purified fusion proteins were incubated with protease in a reaction buffer (10 mM Tris-HCl, pH 7.0, and 1 mM dithiothreitol [DTT]) at 37°C for 1 h. As a control, only protease or only MazE-MazF fusion proteins were incubated under the same conditions. The reaction mixture containing 0.1 μg MazE-MazF fusion proteins was mixed with MS2 phage RNA (Roche) for 1, 2, 5, and 10 min in the same buffer at 37°C. The reaction was stopped by the addition of 2× RNA dye containing 6 M urea, 2× TBE (178 mM Tris, 178 mM boric acid, and 4 mM EDTA), bromophenol blue, and xylene cyanol. The samples were incubated at 65°C for 2 min and then analyzed in 1.2% agarose gel in 1× TBE buffer.
RESULTS
In the 82-residue MazE, the N-terminal 47 residues are involved in dimer formation with an intertwined β-barrel, from which two 35-residue C-terminal segments are extended like wings covering one of the interfaces of a MazF dimer (11). In this MazE C-terminal segment there is one helical structure (residues 55 to 59; α-helix H2), and this helix, together with a short segment of residues 51 to 54, is considered to play a crucial role in the trajectory of MazE interacting with MazF (in Fig. 1B and C, α-helix H2 locates on top of a MazF dimer, from which the unstructured C-terminal segment is extended downward). The C-terminal fragment of MazE interacts with a MazF dimer through many van der Waals interactions, polar interaction, and salt bridges. The C-terminal (residues 55 to 77) domain of MazE contains a total of six negatively charged residues that directly interact with the MazF homodimer. The side chain of Trp73 in MazE composes the major interaction with the loop region (S1-S2) of MazF (11). Besides these interactions, Ile71, Glu69, and His68 play important roles in interacting with the MazF dimer, adopting an extended strand-like structure.
On the basis of these specific interactions, we used fused the 41-resdue C-terminal fragment of MazE to the N-terminal end of MazF (Fig. 1). Gly-Gly and Gly-Ser were added at the N- and C-terminal ends of the factor Xa cleavage site, respectively, for the flexibility of the molecule. The mazE(42)F-Fxa fusion construct was cloned in plasmid pBAD24 and transformed in E. coli BW25113. All constructs, including the mazE(42)F-Fxa fusion protein, were able to form colonies in the presence of 0.2% arabinose (Fig. 2A), indicating that the MazE-MazF fusion protein is not toxic to the cells. Notably, MazF from the pBAD24-mazF plasmid showed high toxicity, as colonies were not formed even in the presence of 0.025% arabinose on plates (data not shown). We also constructed other MazE-MazF fusion proteins containing 22-, 18-, and 12-residue C-terminal fragments of MazE, termed mazE(61)F-Fxa, mazE(65)F-Fxa, and mazE(71)F-Fxa, respectively (Fig. 1B and C). Among the MazE-MazF fusion proteins, MazE(42)F-FXa was expressed at a very high yield when induced (Fig. 2C, lane 8), while the others were found to be poorly expressed. Although MazE(61)F-FXa containing the 18-residue MazE C-terminal fragment was expressed, this fusion protein showed toxicity in the toxicity assay without treatment with factor Xa. On the other hand, when the MazE C-terminal fragment was extended to 41 residues, the resulting MazE-MazF fusion protein, MazE(42)F-FXa, was no longer toxic without treatment with factor Xa (Table 1). Thus, MazE(42)F-FXa was subsequently purified with the use of a Ni-NTA agarose column. The apparent molecular mass of MazE(42)F-FXa calculated from the gel filtration was 45 kDa (Fig. 2B), which agrees well with the size of a dimer of MazE(42)F-FXa, indicating that MazE(42)F-FXa also forms a dimer like MazF-ec (11).
As shown in Table 1, the two other fusion proteins were constructed by the same strategy as that for MazE(42)F-FXa, in which the linkers contained a cleavage site for either HIV PR or HCV NS3 protease. For the HIV PR cleavage site, RVL▾AEA (p24/p2) was used (6, 13, 14). For the HCV NS3 protease cleavage site, the minimal substrate length has to be 10 residues (15), so EDVVCC▾SMSY (5A/5B) was used. These fusion proteins were designated MazEF-HIV and MazEF-HCV, respectively. As shown in Table 1, these fusion proteins also were not toxic to E. coli cells, even if their expression was induced, indicating that these MazE-MazF fusion proteins are resistant to proteases from E. coli. MazEF-HIV and MazEF-HCV were also well expressed without showing toxicity (Fig. 2D and E, respectively).
To evaluate the activation of MazEF-HIV by HIV PR, the various molar ratios of HIV PR to MazEF-HIV were examined (Fig. 3A). MazEF-HIV was indeed cleaved by HIV PR (Fig. 3A, lanes 10 to 15) and induced its endoribonuclease activity, even at a ratio of 100 to 1 (data not shown). The incubation of only MazEF-HIV without HIV PR did not generate active MazF (Fig. 3A, lane 4). The bands of MazEF-HIV and HIV PR are seen at 19 and 10.1 kDa, respectively (Fig. 3A, first and third arrows, respectively). After MazEF-HIV was treated with HIV PR, the size of MazF was reduced to 13.6 kDa (Fig. 3A, second arrow). As shown in Fig. 3B, when the MazE(42)F-FXa fusion protein was treated with factor Xa, its endoribonuclease activity was effectively activated to cleave MS2 phage RNA (3.5 kb). This factor Xa activation was highly efficient, as the full-size MS2 phage RNA completely disappeared within 1 min and the RNA digestion appeared to be almost completed in 5 min, indicating that the linker is fully susceptible to protease digestion.
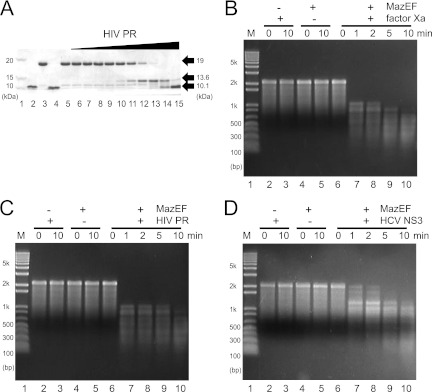
Fig 3.
Activation of MazF from MazE(42)F-FXa, MazEF-HIV, and MazEF-HCV proteins. (A) Cleavage of MazEF-HIV. Black arrows indicate, from top to bottom, MazE-MazF fusion protein, MazF, and HIV PR. Lane 1, molecular mass markers; lane 2, HIV PR; lane 3, MazE-MazF fusion protein; lane 4, HIV PR by itself incubated for 30 min; lane 5, MazE-MazF fusion protein by itself incubated for 30 min. For lanes 6 to 15, the concentration of HIV PR was increased five times for each lane with a constant amount of the MazE-MazF fusion protein (280 μM). Thus, the molar ratios between HIV PR and MazE-MazF fusion protein changed from 3 × 10−6:1 (lane 6) to 1.6:1 (lane 15). The activation of MazF from the MazE-MazF fusion protein by factor Xa, HIV PR, and HCV NS3 protease is shown in panels B, C, and D, respectively. The MazF activity was measured with MS2 phage RNA as described previously (17).
To examine the MazF activation of both MazEF-HIV and MazEF-HCV by HIV PR and NS3 protease, respectively, mRNA interferase activity was analyzed with the use of MS2 phage RNA. The full-size 3.5-kb MS2 phage RNA was digested only when MazEF-HIV and MazEF-HCV were treated with individual proteases (Fig. 3C and D, lanes 7). Notably, these proteases did not show endoribonuclease activity. This result clearly demonstrates that MazE-MazF fusion proteins are specifically cleaved and activated by viral proteases only when their specific cleavage sites exist in the linker between MazE and MazF.
DISCUSSION
Here, we demonstrated that the fusion of a C-terminal 41-residue fragment of antitoxin MazE to the N-terminal end of toxin MazF, with a linker having a specific viral protease cleavage site, resulted in the formation of a stable dimer which could be activated only when the fusion protein was treated with the corresponding viral protease, such as HIV PR (HIV-1 protease) and NS3 protease (HCV protease). Factor Xa also could be used as a protease. When MazF and MazE are separately produced, they form a heterohexamer consisting of one MazE dimer and two MazF dimers in which a MazE dimer is sandwiched by two MazF dimers. Therefore, to release enzymatically active MazF dimers from the MazF2-MazE2-MazF2 hexamer, the central MazE dimer has to be destroyed. In E. coli, MazE dimers are much more unstable than MazF dimers when they exist by themselves, since MazE contains an unstructured long C-terminal segment, which is extended from a MazE dimer (11). In the MazE-MazF complex, this MazE C-terminal segment binds to the cleft formed between two MazF molecules in a MazF dimer, where the MazF active center also exists. In the present study, this C-terminal fragment is fused to the N-terminal end of MazF so that the MazF enzymatic activity is fully suppressed, which can, however, be activated by cleaving the MazE fragment off at the linker between MazE and MazF molecules. Importantly, this linker was found to be resistant to endogenous proteases in E. coli cells, while it is sensitive to specific proteases from RNA viruses if the linker contains a specific cleavage site for the proteases, which are thus able to activate the ACA-specific endoribonuclease activity of MazF.
Using the MazE-MazF fusion protein, we are now able to regulate MazF activity intramolecularly by a highly sequence-specific protease. This opens a few interesting approaches which were previously inaccessible. First, as we can completely suppress MazF toxicity in the cells, the MazE-MazF fusion protein can be produced in large amounts, and after the purification of the fusion protein active MazF can be generated by protease treatment. Notably, the yield of this processing is very high and the final MazF is highly purified, as His6 tag purification is carried out in two steps, the first step for the fusion protein and the second step for cleaved MazF (the His6 tag is added at the C-terminal end of the MazE-MazF fusion protein). In addition, the fusion protein may be used as a tool for the preventive as well as therapeutic treatment of RNA virus infection, as cells carrying the fusion protein with a linker cleavable with a specific RNA viral protease may attain resistance to RNA viral infection.
We have previously shown that CD4+ cells can be engineered to be resistant to HIV-1 infection by transfecting CD4+ cells with the mazF gene under the control of the HIV-1 long terminal repeat (4). More recently, the in vivo safety of this strategy and the persistence of the transfected CD4+ cells has been tested using monkeys (5). MazF induction in mammalian cells has been shown to lead to BAK-dependent apoptotic cell death (22). These results suggest that the strategy presented in this paper is applicable to many other single-stranded RNA viruses, such as poliovirus, influenza virus, and measles virus, for the prevention and treatment of their infection. The effect of the MazEF fusion protein with an HIV-1 PR cleavage site in eukaryotic cells is under investigation. After the submission of the present paper, Shapira et al. published a paper (21) on the MazE-MazF fusion protein in which the 35-residue C-terminal fragment of MazE was fused to the N-terminal end of MazF with the linker containing the HCV P10-P10′ NS3 cleavage sequence.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health grants 1RO1GM081567 and 3RO1GM081567-02S1.
We thank Sehrish Ajmal for the critical reading of the manuscript and Kuen-Phon Wu for advice on the gel filtration experiment.
Footnotes
Published ahead of print 23 March 2012
REFERENCES
- 1. Brass V, et al. 2008. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc. Natl. Acad. Sci. U. S. A. 105:14545–14550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brik A, Wong CH. 2003. HIV-1 protease: mechanism and drug discovery. Org. Biomol. Chem. 1:5–14 [DOI] [PubMed] [Google Scholar]
- 3. Carrere-Kremer S, et al. 2004. Regulation of hepatitis C virus polyprotein processing by signal peptidase involves structural determinants at the p7 sequence junctions. J. Biol. Chem. 279:41384–41392 [DOI] [PubMed] [Google Scholar]
- 4. Chono H, et al. 2011. Acquisition of HIV-1 resistance in T lymphocytes using an ACA-specific E. coli mRNA interferase. Hum. Gene Ther. 22:35–43 [DOI] [PubMed] [Google Scholar]
- 5. Chono H, et al. 2011. In vivo safety and persistence of endoribonuclease gene-transduced CD4+ T cells in cynomolgus macaques for HIV-1 gene therapy model. PLoS One 6:e23585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Oliveira T, et al. 2003. Variability at human immunodeficiency virus type 1 subtype C protease cleavage sites: an indication of viral fitness? J. Virol. 77:9422–9430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Foote BS, Spooner LM, Belliveau PP. 2011. Boceprevir: a protease inhibitor for the treatment of chronic hepatitis C. Ann. Pharmacother. 45:1085–1093 [DOI] [PubMed] [Google Scholar]
- 8. Frankel AD, Young JA. 1998. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 67:1–25 [DOI] [PubMed] [Google Scholar]
- 9. Freed EO. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1–15 [DOI] [PubMed] [Google Scholar]
- 10. Ghosh AK, et al. 2011. Design of HIV-1 protease inhibitors with C3-substituted hexahydrocyclopentafuranyl urethanes as P2-ligands: synthesis, biological evaluation, and protein-ligand X-ray crystal structure. J. Med. Chem. 54:5890–5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kamada K, Hanaoka F, Burley SK. 2003. Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol. Cell 11:875–884 [DOI] [PubMed] [Google Scholar]
- 12. Kato N. 2000. Genome of human hepatitis C virus (HCV): gene organization, sequence diversity, and variation. Microb. Comp. Genomics 5:129–151 [DOI] [PubMed] [Google Scholar]
- 13. Katoh E, et al. 2003. A solution NMR study of the binding kinetics and the internal dynamics of an HIV-1 protease-substrate complex. Protein Sci. 12:1376–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kupiec JJ, Hazebrouck S, Leste-Lasserre T, Sonigo P. 1996. Conversion of thymidylate synthase into an HIV protease substrate. J. Biol. Chem. 271:18465–18470 [DOI] [PubMed] [Google Scholar]
- 15. Lin C. 2006. HCV NS3-4A serine protease. In Tan S-L. (ed), Hepatitis C viruses: genomes and molecular biology. Lilly Research Laboratories, Norfolk, United Kingdom [Google Scholar]
- 16. McHutchison JG, Patel K. 2002. Future therapy of hepatitis C. Hepatology 36:S245–S252 [DOI] [PubMed] [Google Scholar]
- 17. Nariya H, Inouye M. 2008. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 132:55–66 [DOI] [PubMed] [Google Scholar]
- 18. Park JH, Yamaguchi Y, Inouye M. 2011. Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett. 585:2526–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seeff LB. 2002. Natural history of chronic hepatitis C. Hepatology 36:S35–S46 [DOI] [PubMed] [Google Scholar]
- 20. Shafer RW, et al. 2007. HIV-1 protease and reverse transcriptase mutations for drug resistance surveillance. AIDS 21:215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shapira A, et al. 2012. Removal of hepatitis C virus-infected cells by a zymogenized bacterial toxin. PLoS One 7:e32320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shimazu T, et al. 2007. NBK/BIK antagonizes MCL-1 and BCL-XL and activates BAK-mediated apoptosis in response to protein synthesis inhibition. Genes Dev. 21:929–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yamaguchi Y, Park JH, Inouye M. 2009. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J. Biol. Chem. 284:28746–28753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang J, Zhang Y, Inouye M. 2003. Characterization of the interactions within the mazEF addiction module of Escherichia coli. J. Biol. Chem. 278:32300–32306 [DOI] [PubMed] [Google Scholar]
- 25. Zhang Y, et al. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12:913–923 [DOI] [PubMed] [Google Scholar]