Abstract
Salmonella enterica is one of the major food-borne pathogens associated with ready-to-eat fresh foods. Although polluted water might be a significant source of contamination in the field, factors that influence the transfer of Salmonella from water to the crops are not well understood, especially under conditions of low pathogen levels in water. The aim of this study was to investigate the short- and long-term (1 h to 28 days) persistence of Salmonella enterica serotype Typhimurium in the phyllosphere and the rhizosphere of parsley following spray irrigation with contaminated water. Plate counting and quantitative real-time PCR (qRT-PCR)-based methods were implemented for the quantification. By applying qRT-PCR with enrichment, we were able to show that even irrigation with water containing as little as ∼300 CFU/ml resulted in the persistence of S. Typhimurium on the plants for 48 h. Irrigation with water containing 8.5 log CFU/ml resulted in persistence of the bacteria in the phyllosphere and the rhizosphere for at least 4 weeks, but the population steadily declined with a major reduction in bacterial counts, of ∼2 log CFU/g, during the first 2 days. Higher levels of Salmonella were detected in the phyllosphere when plants were irrigated during the night compared to irrigation during the morning and during winter compared to the other seasons. Further elucidation of the mechanisms underlying the transfer of Salmonella from contaminated water to crops, as well as its persistence over time, will enable the implementation of effective irrigation and control strategies.
INTRODUCTION
Over the past few decades, fresh fruits and vegetables, and in particular leafy greens, sprouts, and herbs, have been increasingly recognized as significant reservoirs of food-borne pathogens (10, 11, 22). Such ready-to-eat foods are consumed raw with minimal or no processing aimed to destroy human pathogens. Hence, the presence of food-borne pathogens at the time of consumption hides potential health risks (10, 43). In the United States, for instance, the proportion of outbreaks associated with fresh produce, out of all reported food-borne outbreaks with an identified food source, has increased from 0.7% in the 1970s to 6% in the 1990s (51), to 13% in the 2000s (22), and to 33% in 2011 (15). Salmonella enterica is one of the most commonly identified food-borne pathogens associated with fresh produce. Between 2006 and 2008 S. enterica was the confirmed etiologic agent in 31 of 170 produce-related outbreaks reported to the Centers for Disease Control and Prevention (CDC) (16). It was also reported that ca. 16.5% of total Salmonella cases of illness and death in the United States in 2009 were associated with fresh produce (8). Recent surveys performed in different countries to investigate the occurrence of Salmonella spp. in fresh herbs and leafy vegetables identified Salmonella in up to 28% of the samples, prior to consumption (reviewed in reference 63). Since herbs and leafy vegetables are frequently traded internationally and are used as ingredients in many ways year-round, contaminated products can cause outbreaks with a wide geographic distribution (53).
The increase in reported food-borne illness cases linked to fresh produce can be attributed to increased consumption of ready-to-eat foods, the appearance of new minimally processed ready to eat products, globalization of the produce industry, and more effective surveillance (11, 43). In addition, recent evidence obtained by food microbiologists and plant pathologists supports the hypothesis that enteric pathogens have adapted to persist on or in plants as part of their natural life cycle between infecting hosts (57). It has been indicated that the connections between enteric pathogens such as Salmonella and plants may be more complicated than simple passive transfer, because the bacteria can attach, survive, and even colonize and grow on and in plants by affecting their immune defense response (5, 6, 18, 28, 32, 37, 39, 50).
Contamination of raw fruits and vegetables by human pathogens takes place along the “farm-to-fork” food production chain (13). For many years, water has been a Salmonella carrier (34) and, in fact, introducing Salmonella to leafy vegetables through irrigation water can be a major way of contamination (39, 50). However, little is known about environmental parameters that influence the rates of such transfer, especially under low levels of pathogens in irrigation water. Furthermore, there have been relatively few contradictory studies with regard to the survival of Salmonella and other enteric pathogens in the phyllosphere and the rhizosphere over extended periods (61). Therefore, the aim of the present study was to investigate the short-term (up to 2 days) and long-term (up to 4 weeks) persistence of S. enterica serotype Typhimurium in the phyllosphere and the rhizosphere of parsley plants following spray irrigation with contaminated water during the day and night and during different seasons.
MATERIALS AND METHODS
Bacterial strains and preparation of bacterial suspension.
S. enterica serotype Typhimurium ATCC 14028 (S. Typhimurium) was used for contamination. S. Typhimurium is one of the most predominant Salmonella serotypes found in developed countries. It was the causative agent in food-borne outbreaks associated with different fruit and vegetables worldwide (17, 20) and has been detected in herbs and dried spices (48). Salmonella cells were transformed with pGFP plasmid (Clontech) by electroporation using a MicroPulser electroporator (Bio-Rad Laboratories), to obtain green fluorescent protein (GFP)-labeled cells. Transformants were selected by plating onto Luria-Bertani (LB) agar plates supplemented with ampicillin (100 μg/ml), and stored at −80°C in LB medium supplemented with 20% glycerol. The stability of GFP expression by S. Typhimurium ATCC 14028 on parsley leaves has been previously investigated and described (39). For contamination experiments, S. Typhimurium was cultured overnight in LB broth supplemented with ampicillin (100 μg/ml) at 37°C with aeration. Cells were harvested by centrifugation (4,000 × g for 20 min at 4°C), washed, and resuspended in sterile saline (0.85% NaCl). The culture was diluted to prepare 150-ml bacterial suspensions at concentrations ranging from ∼101 to ∼108 CFU/ml.
Growth of parsley plants.
Parsley plants (Petroselinum crispum var. neapolitanum) were grown in a glass greenhouse (3 by 6 by 2.5 m). Portions (2 g) of parsley seeds (∼800 seeds) were disseminated in each planter containing ∼12 liters of commercial nonsterile potting soil (Avital 11; Tuff Marom Golan, Marom Golan, Israel). Planters were automatically watered with tap water using drip irrigation. The amount of irrigation water was designed following a pilot soil capacity experiment aimed to keep the soil in the planter wet with minimal excess of water (39). The greenhouse has hatches to adapt the indoor conditions (temperature and humidity) to the seasonal weather changes in Haifa. Temperature and relative humidity in the greenhouse were automatically logged by a data logger (D-logMateTHD; MRC). At 10 to 12 weeks after dissemination, which is the common period of time for the seed to develop to a harvestable plant (3), parsley plants were used for irrigation assays. Preliminary experiments showed that plants of this age (and up to 4 weeks afterward) provide enough mass for sampling and processing.
Irrigation procedure with contaminated water.
Each planter was manually spray irrigated by spraying onto the phyllosphere three times during a 9-day period (3-day intervals) with 150 ml of freshly prepared contaminated water. Irrigation was performed by a hand sprayer from a distance of ∼20 cm from the phyllosphere. Each planter was spray irrigated by water harboring GFP-expressing S. Typhimurium at different bacterial concentrations, ranging from ∼101 to ∼108 CFU/ml. Day irrigations were conducted during late-morning hours, 10 to 11 a.m. Night irrigations were conducted between 10 to 11 p.m. To study the impact of seasonality on contamination, day irrigations were conducted in different seasons of the year. For safety reasons, the irrigation of each planter was conducted inside a glove box, from which the plant was removed 1 h after irrigation. In each experiment the plants in the control planters were grown and treated in a similar way using Salmonella-free water.
Sample collection and recovery of GFP-expressing S. Typhimurium for plate counting.
After the irrigation challenge, samples of leaves (20 g), stalks (20 g), and roots (10 g) were aseptically collected from each planter. Soil samples (20 g) were collected from 1 to 5 cm below the surface with a sterile spoon. Sampling was conducted in triplicates 48 h after the last irrigation step. For the study of persistence of S. Typhimurium in the plant environment, samples were collected at different time points (1 h, 10 h, 1 day, 2 days, 7 days, 2 weeks, and 4 weeks) after the last irrigation step. All samples were placed in sterile stomacher bags, brought immediately to the laboratory, and stored at 4°C until processing. For the recovery of plant- and soil-associated GFP-expressing S. Typhimurium, 180 ml of sterile saline was added to each leaf and stalk sample, and the samples were pummeled in a stomacher for 3 min at normal speed (620 paddle strokes per min). Root samples were washed three times by adding 100 ml of sterile saline and agitation for 15 s in order to wash off soil particles from the roots, after which 90 ml of saline was added, and the samples were pummeled in a stomacher. Soil samples were added with 180 ml of sterile saline and vigorously agitated by hand for 30 s. Serial dilutions (1:10 in saline) were prepared and plated for the enumeration of fluorescent colonies of S. Typhimurium on LB agar supplemented with ampicillin. The limit of detection for plating was 2 log CFU/g. Dilutions and plating for each sample were conducted in duplicates.
Surface disinfection of leaves.
Samples of parsley leaves were dipped in 80% ethanol for 10 s, followed by immersion in 0.1% HgCl2 (wt/vol) for 10 min (55). Leaves were washed in sterile saline twice and processed for the recovery of internal S. Typhimurium by viable counting on LB plates, as described above.
Sample collection and enumeration by qRT-PCR without enrichment.
Five leaves (each about 0.1 g) were aseptically collected from each planter and processed for the quantification of leaf-associated S. Typhimurium by quantitative real-time PCR (qRT-PCR) as described recently (36) with some modifications. In the laboratory setting of our previous study, we utilized primers targeting the rrn gene for the detection of S. Typhimurium inoculated on leaves. However, the use of these primers in a greenhouse study occasionally resulted in false-positive detection, possibly due to the presence of indigenous bacteria or soil bacteria with homologous rrn sequences. Thus, we decided to modify the protocol and to target the Salmonella-specific gene sirA (2). Briefly, each individual leaf was immersed in liquid nitrogen for 10 min in an Eppendorf tube. Samples were ground to a fine powder (for 2 min) on liquid nitrogen. Total bacterial DNA was extracted using ZR soil microbe DNA kit (Zymo Research) according to the manufacturer's instructions in a final volume of 50 μl. qRT-PCR analysis was performed using 2 μl of each isolated DNA sample, 175- and 250-nmol/liter concentrations of forward (ACTCGCGTTCAGACAAACTG) and reverse (CGCTATTCGTTCGGTGTA) primers, respectively, targeting the sirA gene, and 5 μl of ABsolute QPCR SYBR green mix (ABgene) in a 10-μl total reaction volume. A three-step protocol was used in a Rotor-Gene 3000 (Corbett Research): (i) denaturation (15 min at 95°C), (ii) an amplification and extension program repeated 50 times (1 s at 95°C, 15 s at 57°C, and 20 s at 72°C), and (iii) a melting-curve program of heating from 72 to 99°C, at a heating rate of 1°C per 5 s. The concentration of experimental samples was calculated from the linear regression of a standard curve, obtained by purified DNA (isolated by the same procedure) from bacterial suspensions at known concentrations (5 × 101 to 5 × 105 CFU/ml).
Detection of S. Typhimurium by RT-PCR after enrichment.
Samples of leaves (10 g) were aseptically collected from each planter and placed in sterile stomacher bags as described above. Portions (50 ml) of buffered peptone water were added, and the samples were incubated at 37°C. After 5 and 20 h of incubation, 1 ml of the enrichment medium was collected. The enriched cultures were pelleted by centrifugation, washed in sterile saline, and repelleted. Total bacterial DNA was extracted using a ZR soil microbe DNA kit, and qRT-PCR analysis was performed as described above.
Statistics.
Unless mentioned specifically, all experiments were conducted at least twice. Sampling was conducted in triplicates. The data were analyzed using InStat 3.10 (GraphPad Software, Inc.) by one-way analysis of variance and Tukey-Kramer test. A P value of < 0.05 was accepted as indicating significance.
RESULTS
Recovery of S. Typhimurium from parsley and soil following spray irrigation with water containing different concentrations of the pathogen.
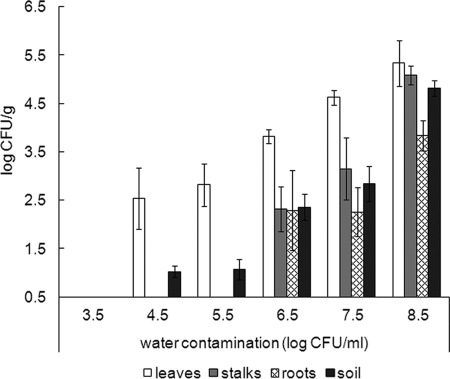
Day irrigation of parsley plants grown in a greenhouse with water carrying the human pathogen S. Typhimurium resulted in the contamination of leaves, stalks, and roots, as well as the soil in which they were grown. S. Typhimurium was not detected by plate counting in any of the samples after irrigations with water harboring <4.5 log CFU of the pathogen/ml, but above this concentration the level of water contamination was correlated with the levels of the pathogen's abundance detected in the phyllosphere, the rhizosphere, and the soil (Fig. 1). The contaminated water was sprayed directly toward the plant phyllosphere, and consequently the leaves were the most susceptible spheres to contamination. The pathogen was readily detected by plate counting following irrigation with heavily contaminated water (i.e., 6.5 to 8.5 log CFU/ml), while irrigation with water harboring intermediate levels of S. Typhimurium (i.e., 4.5 to 5.5 log CFU/ml) resulted in detection of the pathogen only in parsley leaves and in some but not all of the soil samples. At 48 h after irrigation, the levels of the culturable bacteria on the leaves were ca. 2 to 3 log lower than the respective concentration of the pathogen in the irrigation water (Fig. 1). S. Typhimurium was not detected in the control planters that were grown and treated in a similar way using Salmonella-free water.
Fig 1.
S. Typhimurium recovered from parsley phyllosphere, rhizosphere, and soil 48 h after spray irrigations during morning hours (10 to 11 a.m.) with water harboring S. Typhimurium at levels of 3.5 to 8.5 log CFU/ml. After recovery of the bacteria, GFP-tagged S. Typhimurium was enumerated by plating. The results are the averages of at least two independent repeats analyzed in triplicates. The experiments were conducted in June and July.
Internalization of the pathogen into the leaves.
S. Typhimurium was also recovered from surface-sterilized leaves, indicating endophytic properties of leaf-associated S. Typhimurium. After day irrigation with heavily contaminated water (i.e., water harboring 8.5 log CFU/ml), for example, surface-sterilized leaves harbored 4.5 log CFU/g, which represent ca. 1.5% of the total leaf-associated S. Typhimurium (6.3 log CFU/g) recovered from leaves that were not surface sterilized.
Time of irrigation affects the levels of contamination.
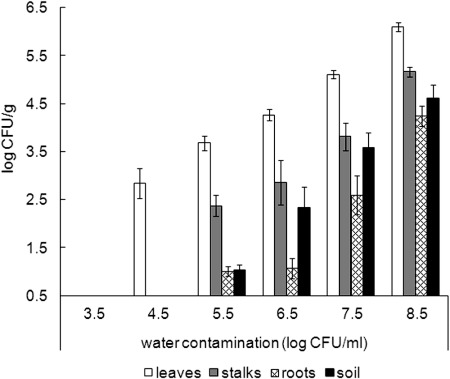
When irrigation was applied during the night, the contamination of the phyllosphere and rhizosphere exhibited a similar pattern as that found following day irrigation application, with the phyllosphere being the most susceptible sphere to contamination (Fig. 2). Leaves harbored significantly (P < 0.05) higher levels of S. Typhimurium in comparison to stalks, root, and soil. Interestingly, the leaves harbored significantly (P < 0.05) higher levels of the pathogen, 0.5 to 0.9 log CFU/g higher, after irrigations conducted during the night compared to the levels of contamination observed after irrigations during the morning, when irrigation water carried 5.5 to 8.5 log CFU/ml. Similarly to irrigations performed during morning hours, the presence of culturable S. Typhimurium was not detected in any of the samples after irrigations conducted during the night with water harboring <4.5 log CFU/ml.
Fig 2.
S. Typhimurium recovered from parsley phyllosphere, rhizosphere, and soil 48 h following spray irrigation during night hours (10 to 11 PM) with water harboring S. Typhimurium at levels of 3.5 to 8.5 log CFU/ml. After recovery of the bacteria, GFP-tagged S. Typhimurium was enumerated by plating. The results are the averages of at least two independent repeats analyzed in triplicates. The experiments were conducted in June and July.
Implementation of qRT-PCR with or without enrichment for detection of Salmonella in plants irrigated with low levels of bacteria.
In light of the limited ability to detect Salmonella by recovery and plating following irrigation with water carrying low levels of the pathogen, we decided to extract DNA from leaves and to implement qRT-PCR for the quantification of leaf-associated S. Typhimurium. qRT-PCR proved to be a more sensitive method for bacterial detection (Table 1). S. Typhimurium DNA was detected in all of the sampled leaves irrigated with water carrying 4.5 to 5.5 log CFU/ml. Furthermore, the detection of S. Typhimurium was feasible even in 60% of leaves irrigated with water carrying low level of 3.5 log CFU/ml. Still, leaf-associated S. Typhimurium was undetectable by this method following irrigation with water carrying very low levels of 1.5 to 2.5 log CFU/ml.
Table 1.
Quantification of S. Typhimurium on parsley leaves by qRT-PCR
| Level of S. Typhimurium in water (log CFU/ml) | Frequencya | Amt of S. Typhimurium on leaves (mean log CFU/g ± SD)b |
|---|---|---|
| 5.5 | 5/5 | 5.3 ± 0.3 |
| 4.5 | 5/5 | 4.3 ± 0.4 |
| 3.5 | 3/5 | 3.2 ± 0.8c |
| 2.5 | 0/5 | NDd |
| 1.5 | 0/5 | ND |
Calculated as the average number of leaf samples that tested positive for S. Typhimurium of the total sampled leaves.
Calculated as the concentration of S. Typhimurium on leaves; the mean log CFU/g was quantified by qRT-PCR. Melting-curve analysis of the amplified PCR products confirmed positive amplification of the target gene only in the contaminated leaves and not in the Salmonella-free control leaves. The results are the averages of at least two independent determinations. The experiments were conducted in June and July. ND, not detected.
This calculation is based on the positive samples.
Positive after enrichment for 20 h.
In order to further extend our ability to detect Salmonella on plants irrigated with water carrying very low levels of the pathogen, we decided to add an enrichment step before the PCR for the detection of leaf-associated S. Typhimurium. Parsley plants were spray irrigated as described with water carrying 1.5 and 2.5 log CFU/ml and processed by enrichment of leaf samples. After a 5-h enrichment step, all samples tested negative for Salmonella. However, after 20 h of enrichment, all leaf samples from plants irrigated with water carrying 2.5 log CFU/ml were found to be positive for Salmonella. Melting-curve analysis of the amplified PCR products confirmed the positive amplification of the target gene only in the contaminated leaves and not in the Salmonella-free control leaves.
Thus, leaves irrigated with water containing at least 4.5 log CFU/ml were positive for S. Typhimurium by both quantification methods: viable counts and qRT-PCR. After irrigation with water containing 3.5 log CFU/g, S. Typhimurium was detected on leaves only by qRT-PCR, and after irrigation with water containing 2.5 log CFU/g, S. Typhimurium was detected only by RT-PCR following enrichment for 20 h. Leaf-associated S. Typhimurium was proven to be undetectable by all three tested methods after irrigation with water carrying 1.5 log CFU/ml.
Short- and long-term persistence of S. Typhimurium.
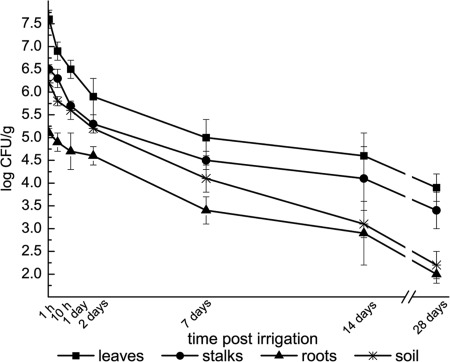
Fig. 3 presents the short- and long-term persistence of culturable S. Typhimurium (1 h to 28 days) in plant and soil samples after irrigation with contaminated water. The pathogen proved to have the ability to survive in the phyllosphere, in the rhizosphere, and in the soil for at least 28 days after the irrigation challenge. Parsley phyllosphere harbored the highest initial levels of S. Typhimurium 1 h after the irrigation challenge, followed by the soil and the rhizosphere. A significant decline of 0.7 log CFU/g of leaves was observed within the first 10 h. The rapid decline continued and, after 24 h, the pathogen counts were 1.1 log CFU/g lower. The levels of S. Typhimurium in the rhizosphere and in the soil also declined rapidly, and an ∼0.5-log CFU/g reduction was observed within 24 h. Within 48 h, levels of S. Typhimurium on leaves declined by ∼2 log CFU/g. Pathogen abundance continued to decrease both in parsley and in the soil for the entire period of sampling, but at a lower rate. At 1 and 2 weeks after plant contamination, the phyllosphere still harbored S. Typhimurium at a level of 4 to 5 log CFU/g, and 4 weeks into the experiment the parsley leaves and stalks still harbored high counts of 3.9 and 3.4 log CFU/g, respectively (0.02 and 0.08% of the counts at the first hour, respectively). S. Typhimurium counts in the rhizosphere and in soil reached levels of 2.0 and 2.2 log CFU/g (0.08 and 0.01%), respectively, 4 weeks after the irrigation challenge.
Fig 3.
Persistence of S. Typhimurium in parsley. Plants were spray irrigated with water harboring 8.5 log CFU/ml. Bacterial abundance was determined at different time points (1 h, 10 h, 1day, 2 days, 7 days, 14 days, and 28 days) postinoculation. After recovery of the bacteria, GFP-tagged S. Typhimurium was enumerated by plating. The results are the averages of at least two independent repeats analyzed in triplicates. The experiments were conducted in October and November.
Impact of seasonality on contamination.
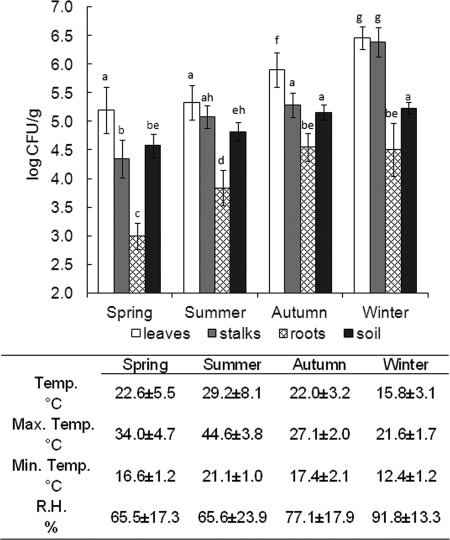
Application of contaminated irrigation water in different seasons affected the level of S. Typhimurium counts in the phyllosphere and the rhizosphere (Fig. 4). S. Typhimurium counts on parsley leaves and stalks were 1.3 and 2.1 log CFU/g higher during the winter compared to irrigation conducted during the spring, the seasons with the highest and the lowest recorded contamination levels, respectively (P < 0.05). Contamination of the rhizosphere varied significantly with up to 1.6 log CFU/g difference between the seasons. Similarly to the phyllosphere, the lowest level of S. Typhimurium in the rhizosphere was recorded during the spring.
Fig 4.
Impact of irrigation in different seasons on S. Typhimurium contamination levels in parsley in the phyllosphere, rhizosphere, and soil. Plants were spray irrigated with water harboring 8.5 log CFU/ml. After recovery of the bacteria, GFP-tagged S. Typhimurium was enumerated by plating. Air temperature (Temp. °C) and relative humidity (R.H. %) were automatically logged, and the averaged values are shown, as well as the average minimum night temperature and maximum day temperature. The results are the average of at least two independent repeats analyzed in triplicates. Columns assigned different letters indicate the statistical significance between bacterial counts (P < 0.05).
DISCUSSION
Sprinkler/spray irrigation is the main irrigation regime of fruits and vegetables in countries such as the United States and the United Kingdom, in which over half of total irrigated acreage is subjected to overhead irrigation (56, 58). It is anticipated that spray irrigation results in a great frequency of contamination of greens due to direct contact of the leaves with the pathogen source (10, 22, 56). On the other hand, the ability of enteric bacteria to survive in the hostile environments on the phyllosphere is questionable, because stress conditions on plant surfaces can restrict their survival (11, 42, 61). In the present study, when spray irrigation of parsley was performed with contaminated water, S. Typhimurium was detected in the phyllosphere, the rhizosphere, and the soil in which the plants were grown. The conditions on and around parsley plants were not optimal for Salmonella, since the quantity of Salmonella declined over time. Still a portion of the bacteria persisted and was recovered from the rhizosphere, the phyllosphere, and the soil even at the end of our sampling period (28 days). Several field studies have reported a decline in populations of enteric pathogens over time, but comparison of the reduction rates of Salmonella to reported reduction of other pathogens reveals that Escherichia coli and Listeria innocua decreased more rapidly (23, 25, 27, 45).
S. Typhimurium was not only recovered from leaves but also from surface-sterilized leaves. This may support the hypothesis that Salmonella cells invade the inner tissues of leaves (30, 38) or attach to sites that are inaccessible to sterilization treatments (13, 49). Our results are in agreement with reports about a small population of internalized E. coil O157:H7 in lettuce leaves after application through spray irrigation (25). It was suggested that Salmonella migrates from the surface of leaves to the inner tissues via open stomata (30, 38). Interestingly, the rates of internalization in these experiments, which were conducted in the lab with harvested leaves of parsley (1.9%), were very similar to the rates of internalization into leaves in growing plants in our experiment (1.5%). Nevertheless, internalization to the inner tissues of leaves is not the main strategy for persistence of Salmonella since, according to our results, the majority of the cells persisted as epiphytes.
Limited availability of good-quality water increases the need for the use of low-quality water with high microbial loads, including raw or partially treated wastewater, for irrigation of crops even in developed countries (26, 34, 41, 56, 58). The U.S. Environmental Protection Agency (EPA) observed that 40% of streams, 45% of lakes, and 50% of estuaries in the United States were not clean enough. Microbial contamination was also widespread in groundwater, especially in shallow aquifers, where mixing with surface water can occur (59, 60). Salmonella occurrence in various water bodies was reported worldwide with frequency of positive samples ranging from 3 to 100% (41), underlining the potential health risk associated with application for irrigation. We showed that the persistence of S. Typhimurium on the plant was dependent on its initial levels in irrigation water. Based on this, the quality of water applied in the production environment is linked to the potential for contamination. It was already shown that heavily contaminated water applied for irrigation resulted in extensive level of pathogens recovered from the plants (23, 25, 27, 33, 54). By applying sensitive methods (qRT-PCR with or without enrichment), we were able to show that even irrigation with water containing as little as ∼300 CFU/ml results in contamination and persistence of the pathogen in the plants for at least 48 h. To the best of our knowledge, quantification of pathogens from plants that were exposed to such a low Salmonella contamination through irrigation has not been described before. Quantification of naturally occurring Salmonella in environmental waters is seldom performed, but the existing reports reveal levels of up to 104 CFU/ml (41). Values of 300 CFU/ml are still higher than the levels of naturally occurring Salmonella in most investigated samples of water. However, it should be emphasized that Salmonella cells usually persist in water in a viable but nonculturable (VNC) state, meaning that the real viable counts might be 100- to 1,000-fold higher compared to counts obtained by culture-based methods (21).
In light of the observations that irrigation with water containing 300 CFU/ml results in contaminated leaves and in light of the reports that in many cases the gap between the last water application and harvest may be less than 24 h (58), the existence of Salmonella in irrigation water may lead to the harvesting of contaminated foods. Indeed, several outbreaks of salmonellosis from fresh produce were linked to contaminated irrigation water, including an outbreak of S. Newport in tomatoes and of S. Saintpaul in chili peppers (14, 31). Contaminated fresh produce can constitute a potential health risk even if contaminated with low levels of Salmonella, since the ingestion of as few as 10 to 100 cells has resulted in outbreaks associated with herbs, spices, and sprouts. In addition, there is a possibility of bacterial growth during the storage of cut produce. For instance, an outbreak of salmonellosis in Germany was traced to paprika and paprika-powdered potato chips. The estimated infective dose was as low as 4 to 45 salmonellae (40). Similarly, the infective dose of S. Newport on alfalfa sprouts was estimated to be <460 CFU (1).
Soil is considered a comparatively less hostile environment, although competition with native microorganisms can restrict the survival of pathogen populations (35). S. Weltevreden was detected in soil throughout a 28-day sampling period with only slight reduction in bacterial levels (4). In our study, culturable S. Typhimurium was present in soil samples even after irrigation challenge with water carrying as little as 4.5 log CFU/ml, but its levels declined over time. The quantity of Salmonella in the soil around the plants was always lower than its quantity on the leaves, probably because most water volume (with the bacteria in it) was retained in the phyllosphere. We presume that most bacteria arrived to the soil directly through drops of contaminated water. The rhizosphere has also been reported to serve as a reservoir for human pathogens (9). In our study, when contamination was mostly applied in the phyllosphere, we found bacteria attached to the roots, but their levels were lower compared to the other parts of the plants or to the soil. The decline of Salmonella associated with the roots was similar to the decline in the soil, indicating that in the planters used for the present study, the root environment does not provide better or worse conditions for Salmonella compared to the soil. Furthermore, whereas it was shown that S. enterica serotypes are able to move via chemotaxis toward root exudates in microcapillaries (37), in the conditions of our experiments the levels of bacteria associated with the roots were always similar or even lower than the levels in the soil. When irrigation was conducted by dripping directly to the soil, the presence of Salmonella and E. coli in the rhizosphere resulted in the contamination of the phyllosphere (39, 55). Here, when most bacteria were applied on the phyllosphere, the bacteria were also found in the rhizosphere, but further research is needed in order to understand whether the bacteria arrive to the roots through the soil or through the phyllosphere, since we have shown that ca. 1.5% of the bacteria are probably endophytic.
The physiology of the plant changes during the day and during the year, and it was hypothesized that these variations, together with variations in environmental conditions (such as sunlight, temperature, etc.) could influence the survival of Salmonella on the plants. Indeed, the levels of the pathogen recovered from the leaves were higher after the application of contaminated water during night hours. We suggest that variations in plant physiology that lead to different contents and concentrations of secondary metabolites in the infected tissues or impact the production of signals of plant immune response may play a role in the short-term adaptation of Salmonella in the leaf. Indeed, it was recently shown that the extent to which methyl salicylate is required for signaling systemic acquired resistance in plants, a defense response which is activated throughout a plant after local infection, is dependent on exposure to light during infection (44). In addition, it was shown that during photosynthesis, parsley cells may produce toxic oxygen species. These toxic oxygen species migrate outside the phytodetritus and potentially could affect the attached bacteria (47). With respect to field applications, our results indicate that irrigation of parsley during morning hours may have lower potential for contamination.
Presence of S. Typhimurium in the plant environment was subjected to changes throughout the seasons with the highest levels recorded during winter and the lowest levels during spring. The levels of S. Typhimurium in soil remained relatively constant between the seasons, while the levels of the pathogen in the phyllosphere and the rhizosphere varied considerably. A similar profile was reported for E. coli sprayed on lettuce (27). Interestingly, similar results (longer survival in the winter) were observed with Vibrio cholerae on stored parsley (29). This may suggest interactions of the pathogens with the plant or with the microflora. The quantity and diversity of microbial communities are subjected to seasonal changes (45, 52). Thus, indigenous bacteria may affect survival of S. Typhimurium, as shown for E. coli O157:H7 on lettuce (19). An increase in the logged relative humidity (RH) between spring and winter corresponded with the increase in Salmonella counts. Likewise, higher levels of Listeria spp. were recorded on artificially inoculated parsley leaves under high RH conditions (23). Lower counts of S. Typhimurium under low RH may be explained by the stress induced by limited available water, in spite the relative tolerance to dry conditions (12). Furthermore, low RH may induce a VNC state and result in lower recovery levels, as reported for L. monocytogenes on parsley leaves (24). Higher levels of persistence of enteric pathogens may lead to greater potential for food-borne illness outbreaks linked to fresh produce. Indeed, analysis of several high-profile outbreaks associated with fresh produce reveals that 17 of 25 outbreaks occurred during the autumn and winter (61), as opposed to the fact that most cases of human salmonellosis (originated from all types of foods) occur in summer (46).
Conclusions.
We have shown that spray irrigation of parsley with water containing a minimum of 300 CFU of S. Typhimurium/ml results in the persistence of detectable Salmonella on the leaves of parsley at least 48 h after the irrigation. The extent of contamination was affected mainly by the quantity of Salmonella in irrigation water and the time period between irrigation and harvest, seeing that the population levels of S. Typhimurium steadily declined during the field study. We furthermore showed that irrigation during the night versus during the day, and in the winter versus the other seasons, results in higher levels of pathogen on the phyllosphere. If parsley, which is used either as a dried spice or a fresh herb, is contaminated with pathogens such as Salmonella through exposure to polluted water, such pathogens might enter the food chain in a wide geographic distribution. Understanding the mechanisms underlying the seasonality of Salmonella will enable the implementation of effective irrigation and control strategies. Our results strengthen the paradigm that Salmonella is able to persist in soil and crops after application of contaminated water or compost for prolonged periods (7, 33, 39, 62), thus posing both health and environmental risks.
ACKNOWLEDGMENTS
This study was supported in part by a CPS-BARD grant (CP-9036-09) and partially by a research grant from the Chief Scientist of the Israeli Ministry of Agriculture and Rural Development (grant 421-0177-09).
Footnotes
Published ahead of print 23 March 2012
REFERENCES
- 1. Aabo S, Baggesen DL. 1997. Growth of Salmonella Newport in naturally contaminated alfalfa sprouts and estimation of infectious dose in Danish Salmonella Newport outbreak due to alfalfa sprouts, p 425–426 Proceedings of Salmonella and Salmonellosis, Ploufragan, France [Google Scholar]
- 2. Ahmer BM, van Reeuwijk J, Watson PR, Wallis TS, Heffron F. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31:971–982 [DOI] [PubMed] [Google Scholar]
- 3. Andersen CR. 2012. Agriculture and natural resources. Home gardening series: parsley. Division of Agriculture, University of Arkansas System, Little Rock, AR: http://www.uaex.edu/Other_Areas/publications/PDF/FSA-6091.pdf [Google Scholar]
- 4. Arthurson V, Sessitsch A, Jaderlund L. 2011. Persistence and spread of Salmonella enterica serovar Weltevreden in soil and on spinach plants. FEMS Microbiol. Lett. 314:67–74 [DOI] [PubMed] [Google Scholar]
- 5. Barak JD, Gorski L, Naraghi-Arani P, Charkowski AO. 2005. Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl. Environ. Microbiol. 71:5685–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barak JD, Kramer LC, Hao LY. 2011. Colonization of tomato plants by Salmonella enterica is cultivar dependent, and type 1 trichomes are preferred colonization sites. Appl. Environ. Microbiol. 77:498–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barak JD, Liang AS. 2008. Role of soil, crop debris, and a plant pathogen in Salmonella enterica contamination of tomato plants. PLoS One 3:e1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Batz MB, Hoffmann S, Morris GJ. 2011. Ranking the risks: the 10 pathogen-food combinations with the greatest burden on public health. University of Florida, Emerging Pathogens Institute, Gainesville, FL: http://www.rwjf.org/files/research/72267report.pdf [Google Scholar]
- 9. Berg G, Eberl L, Hartmann A. 2005. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7:1673–1685 [DOI] [PubMed] [Google Scholar]
- 10. Berger CN, et al. 2010. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ. Microbiol. 12:2385–2397 [DOI] [PubMed] [Google Scholar]
- 11. Brandl MT. 2006. Fitness of human enteric pathogens on plants and implications for food safety. Annu. Rev. Phytopathol. 44:367–392 [DOI] [PubMed] [Google Scholar]
- 12. Brandl MT, Mandrell RE. 2002. Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl. Environ. Microbiol. 68:3614–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burnett SL, Beuchat LR. 2000. Human pathogens associated with raw produce and unpasteurized juices, and difficulties in decontamination. J. Ind. Microbiol. Biotechnol. 25:281–287 [DOI] [PubMed] [Google Scholar]
- 14. Centers for Disease Control and Prevention 2008. Outbreak of Salmonella serotype Saintpaul infections associated with multiple raw produce items–United States, 2008 MMWR Morb. Mortal. Wkly. Rep. 57:929–934 [PubMed] [Google Scholar]
- 15. Centers for Disease Control and Prevention 2012. Annual year review. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/outbreaknet/outbreaks.html [Google Scholar]
- 16. Centers for Disease Control and Prevention 2011. Reported foodborne disease outbreaks and illnesses by etiology and food commodities, United States. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/outbreaknet/surveillance_data.html [Google Scholar]
- 17. Centers for Disease Control and Prevention 2006. Salmonellosis: outbreak investigation, October 2006 Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/ncidod/dbmd/diseaseinfo/salmonellosis_2006/110306_outbreak_notice.htm [Google Scholar]
- 18. Charkowski AO, Barak JD, Sarreal CZ, Mandrell RE. 2002. Differences in growth of Salmonella enterica and Escherichia coli O157:H7 on alfalfa sprouts. Appl. Environ. Microbiol. 68:3114–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cooley MB, Chao D, Mandrell RE. 2006. Escherichia coli O157:H7 survival and growth on lettuce is altered by the presence of epiphytic bacteria. J. Food Prot. 69:2329–2335 [DOI] [PubMed] [Google Scholar]
- 20. Crook PD, et al. 2003. A European outbreak of Salmonella enterica serotype Typhimurium definitive phage type 204b in 2000. Clin. Microbiol. Infect. 9:839–845 [DOI] [PubMed] [Google Scholar]
- 21. Domingo JWS, Harmon S, Bennett J. 2000. Survival of Salmonella species in river water. Curr. Microbiol. 40:409–417 [DOI] [PubMed] [Google Scholar]
- 22. Doyle MP, Erickson MC. 2008. Summer meeting 2007: the problems with fresh produce: an overview. J. Appl. Microbiol. 105:317–330 [DOI] [PubMed] [Google Scholar]
- 23. Dreux N, Albagnac C, Carlin F, Morris CE, Nguyen-The C. 2007. Fate of Listeria spp. on parsley leaves grown in laboratory and field cultures. J. Appl. Microbiol. 103:1821–1827 [DOI] [PubMed] [Google Scholar]
- 24. Dreux N, et al. 2007. Viable but non-culturable Listeria monocytogenes on parsley leaves and absence of recovery to a culturable state. J. Appl. Microbiol. 103:1272–1281 [DOI] [PubMed] [Google Scholar]
- 25. Erickson MC, et al. 2010. Surface and internalized Escherichia coli O157:H7 on field-grown spinach and lettuce treated with spray-contaminated irrigation water. J. Food Prot. 73:1023–1029 [DOI] [PubMed] [Google Scholar]
- 26. FAO/WHO 2008. Microbiological hazards in fresh fruits and vegetables: meeting report. Microbiological risk assessment series. Food and Agriculture Organization of the United Nations/World Health Organization, Geneva, Switzerland [Google Scholar]
- 27. Fonseca JM, Fallon SD, Sanchez CA, Nolte KD. 2011. Escherichia coli survival in lettuce fields following its introduction through different irrigation systems. J. Appl. Microbiol. 110:893–902 [DOI] [PubMed] [Google Scholar]
- 28. Gandhi M, Golding S, Yaron S, Matthews KR. 2001. Use of green fluorescent protein expressing Salmonella Stanley to investigate survival, spatial location, and control on alfalfa sprouts. J. Food Prot. 64:1891–1898 [DOI] [PubMed] [Google Scholar]
- 29. Gerichter CB, Sechter I, Gavish A, Cahan D. 1975. Viability of Vibrio cholerae biotype El Tor and of cholera phage on vegetables. Isr. J. Med. Sci. 11:889–895 [PubMed] [Google Scholar]
- 30. Golberg D, Kroupitski Y, Belausov E, Pinto R, Sela S. 2011. Salmonella Typhimurium internalization is variable in leafy vegetables and fresh herbs. Int. J. Food Microbiol. 145:250–257 [DOI] [PubMed] [Google Scholar]
- 31. Greene SK, et al. 2008. Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiol. Infect. 136:157–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Islam M, et al. 2004. Fate of Salmonella enterica serovar Typhimurium on carrots and radishes grown in fields treated with contaminated manure composts or irrigation water. Appl. Environ. Microbiol. 70:2497–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Islam M, et al. 2004. Persistence of Salmonella enterica serovar typhimurium on lettuce and parsley and in soils on which they were grown in fields treated with contaminated manure composts or irrigation water. Foodborne Pathog. Dis. 1:27–35 [DOI] [PubMed] [Google Scholar]
- 34. Jacobsen CS, Bech TB. 2012. Soil survival of Salmonella and transfer to freshwater and fresh produce. Food Res. Int. 45:557–566 [Google Scholar]
- 35. Jiang X, Morgan J, Doyle MP. 2002. Fate of Escherichia coli O157:H7 in manure-amended soil. Appl. Environ. Microbiol. 68:2605–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kisluk G, Hoover DG, Kneil KE, Yaron S. 2012. Quantification of low and high levels of Salmonella enterica serovar Typhimurium on leaves. LWT-Food Sci. Technol. 45:36–42 [Google Scholar]
- 37. Klerks MM, Franz E, van Gent-Pelzer M, Zijlstra C, van Bruggen AH. 2007. Differential interaction of Salmonella enterica serovars with lettuce cultivars and plant-microbe factors influencing the colonization efficiency. ISME J. 1:620–631 [DOI] [PubMed] [Google Scholar]
- 38. Kroupitski Y, et al. 2009. Internalization of Salmonella enterica in leaves is induced by light and involves chemotaxis and penetration through open stomata. Appl. Environ. Microbiol. 75:6076–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lapidot A, Yaron S. 2009. Transfer of Salmonella enterica serovar Typhimurium from contaminated irrigation water to parsley is dependent on curli and cellulose, the biofilm matrix components. J. Food Prot. 72:618–623 [DOI] [PubMed] [Google Scholar]
- 40. Lehmacher A, Bockemuhl J, Aleksic S. 1995. Nationwide outbreak of human salmonellosis in Germany due to contaminated paprika and paprika-powdered potato chips. Epidemiol. Infect. 115:501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Levantesi C, et al. 2011. Salmonella in surface and drinking water: occurrence and water-mediated transmission. Food Res. Int. doi:10.1016/j.foodres.2011.06.037 [Google Scholar]
- 42. Lindow SE, Brandl MT. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69:1875–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Little CL, Gillespie IA. 2008. Prepared salads and public health. J. Appl. Microbiol. 105:1729–1743 [DOI] [PubMed] [Google Scholar]
- 44. Liu PP, von Dahl CC, Klessig DF. 2011. The extent to which methyl salicylate is required for signaling systemic acquired resistance is dependent on exposure to light after infection. Plant Physiol. 157:2216–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moyne AL, et al. 2011. Fate of Escherichia coli O157:H7 in field-inoculated lettuce. Food Microbiol. 28:1417–1425 [DOI] [PubMed] [Google Scholar]
- 46. Ravel A, et al. 2010. Seasonality in human salmonellosis: assessment of human activities and chicken contamination as driving factors. Foodborne Pathog. Dis. 7:785–794 [DOI] [PubMed] [Google Scholar]
- 47. Rontani JF, Rabourdin A, Pinot F, Kandel S, Aubert C. 2005. Visible light-induced oxidation of unsaturated components of cutins: a significant process during the senescence of higher plants. Phytochemistry 66:313–321 [DOI] [PubMed] [Google Scholar]
- 48. Sagoo SK, et al. 2009. Assessment of the microbiological safety of dried spices and herbs from production and retail premises in the United Kingdom. Food Microbiol. 26:39–43 [DOI] [PubMed] [Google Scholar]
- 49. Sapers GM. 2001. Efficacy of washing and sanitizing methods for disinfection of fresh fruit and vegetable products. Food Technol. Biotechnol. 39:305–311 [Google Scholar]
- 50. Shirron N, Yaron S. 2011. Active suppression of early immune response in tobacco by the human pathogen Salmonella Typhimurium. PLoS One 6:e18855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sivapalasingam S, Friedman CR, Cohen L, Tauxe RV. 2004. Fresh produce: a growing cause of outbreaks of food-borne illness in the United States, 1973 through 1997. J. Food Prot. 67:2342–2353 [DOI] [PubMed] [Google Scholar]
- 52. Smalla K, et al. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sobel J, Griffin PM, Slutsker L, Swerdlow DL, Tauxe RV. 2002. Investigation of multistate food-borne disease outbreaks. Public Health Rep. 117:8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Solomon EB, Potenski CJ, Matthews KR. 2002. Effect of irrigation method on transmission to and persistence of Escherichia coli O157:H7 on lettuce. J. Food Prot. 65:673–676 [DOI] [PubMed] [Google Scholar]
- 55. Solomon EB, Yaron S, Matthews KR. 2002. Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl. Environ. Microbiol. 68:397–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Suslow TV. 2010. Produce Safety Project issue brief: standards for irrigation and foliar contact water. Produce Safety Project, Georgetown University, Washington, DC: http://www.producesafetyproject.org/reports?id=0007 [Google Scholar]
- 57. Teplitski M, Barak JD, Schneider KR. 2009. Human enteric pathogens in produce: un-answered ecological questions with direct implications for food safety. Curr. Opin. Biotechnol. 20:166–171 [DOI] [PubMed] [Google Scholar]
- 58. Tyrrel SF, Knox JW, Weatherhead EK. 2006. Microbiological water quality requirements for salad irrigation in the United Kingdom. J. Food Prot. 69:2029–2035 [DOI] [PubMed] [Google Scholar]
- 59. USEPA 2000. National water quality inventory report EPA-841-R-02–001. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 60. USEPA 2006. Occurrence and monitoring document for final ground water rule. EPA 815-R-06-012. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 61. Warriner K, Namvar A. 2010. The tricks learnt by human enteric pathogens from phytopathogens to persist within the plant environment. Curr. Opin. Biotechnol. 21:131–136 [DOI] [PubMed] [Google Scholar]
- 62. You Y, et al. 2006. Survival of Salmonella enterica serovar Newport in manure and manure-amended soils. Appl. Environ. Microbiol. 72:5777–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zweifel C, Roger S. 2012. Spices and herbs as source of Salmonella-related food-borne diseases. Food Res. Int. 45:765–769 [Google Scholar]