Abstract
Dermatophytes are the most common cause of superficial mycoses in humans and animals. They can coexist with their hosts for many years without causing significant symptoms but also cause highly inflammatory diseases. To identify mechanisms involved in the modulation of the host response during infection caused by the zoophilic dermatophyte Arthroderma benhamiae, cell wall-associated surface proteins were studied. By two-dimensional gel electrophoresis, we found that a hydrophobin protein designated HypA was the dominant cell surface protein. HypA was also detected in the supernatant during the growth and conidiation of the fungus. The A. benhamiae genome harbors only a single hydrophobin gene, designated hypA. A hypA deletion mutant was generated, as was a complemented hypA mutant strain (hypAC). In contrast to the wild type and the complemented strain, the hypA deletion mutant exhibited “easily wettable” mycelia and conidia, indicating the loss of surface hydrophobicity of both morphotypes. Compared with the wild type, the hypA deletion mutant triggered an increased activation of human neutrophil granulocytes and dendritic cells, characterized by an increased release of the immune mediators interleukin-6 (IL-6), IL-8, IL-10, and tumor necrosis factor alpha (TNF-α). For the first time, we observed the formation of neutrophil extracellular traps against dermatophytes, whose level of formation was increased by the ΔhypA mutant compared with the wild type. Furthermore, conidia of the ΔhypA strain were killed more effectively by neutrophils. Our data suggest that the recognition of A. benhamiae by the cellular immune defense system is notably influenced by the presence of the surface rodlet layer formed by the hydrophobin HypA.
INTRODUCTION
Infections caused by the zoophilic dermatophyte Arthroderma benhamiae are increasingly being observed in humans. They are usually transmitted from the natural animal host, the guinea pig (13, 16). Recently, we reported the complete genome sequence of A. benhamiae (10). Because, in addition, A. benhamiae is amenable to genetic manipulation and grows well under laboratory conditions, this dermatophyte represents an excellent model organism for the elucidation of pathogenicity-associated traits. In the human host, A. benhamiae infections are characterized by inflammatory progression and are usually quickly eradicated by the host immune system. Besides the putative role of proteases and other enzymes abundantly secreted during infection or under other growth conditions (10, 17, 34), little is known about the molecular strategies which dermatophytes use to infect mammalian hosts and to counteract the initial immune response. On the host side, infected epithelial tissue reacts to fungal invasion by the secretion of a complex and species-specific cytokine pattern (32). The resulting inflammation includes the recruitment of neutrophil granulocytes to the site of infection (23).
During infection by pathogenic bacteria and fungi, a crucial factor for disease progression is the recognition of the pathogen by the host immune system. Since the cell wall of fungi consists of biopolymers not present in human cells, it displays structures that can be recognized by immune effector cells. Filamentous fungi such as Aspergillus fumigatus have been shown to be able to reduce this detection. A. fumigatus interferes with the host response, e.g., by inactivating complement components by proteolytic cleavage (5). It is conceivable that such a mechanism also exists in A. benhamiae due to its large number of secreted proteases (10). For A. fumigatus, it was also demonstrated that the rodlet layer on resting conidia formed by the hydrophobin protein RodA renders conidia immunologically inert, most likely by masking fungal immunogenic cell wall components. It was demonstrated previously that RodA is itself nonimmunogenic, e.g., not recognized by neutrophilic granulocytes, and does not stimulate the maturation of human dendritic cells (DCs) (1).
Here, we analyzed the rodlet layer of A. benhamiae, elucidated its genetic basis, and addressed the question of whether this rodlet layer helps the fungus to reduce recognition by the immune system. Therefore, we characterized the hydrophobin system of A. benhamiae and analyzed its function during the interaction with immune effector cells.
MATERIALS AND METHODS
Strains and cell culture conditions.
Arthroderma benhamiae LAU2354 (16) and the derived mutant strains were cultivated on Sabouraud 2% (wt/vol) glucose (SG) medium (Merck, Germany) or SG agar (15 g/liter select agar; Invitrogen, Germany). For conidiation, MAT agar (10% [vol/vol] SG medium, 1 g/liter MgSO4 · 7H2O, 1 g/liter KH2PO4, 15 g/liter agar) was used. Liquid cultures were shaken at 200 rpm at 30°C. The isolation of neutrophil granulocytes from fresh human blood was achieved by using Polymorphprep (Axis-shield, Scotland) according to the manufacturer's instructions. Cells were suspended in RPMI 1640 medium (Lonza, Germany)–5% (vol/vol) fetal calf serum (FCS) (Lonza) and used immediately for cocultivation experiments. To generate immature dendritic cells (imDCs), monocytes from human blood were isolated by using magnetically activated cell sorting (MACS) monocyte isolation kit II (Miltenyi Biotec, Germany). Cells were stimulated with 800 U/ml granulocyte-macrophage colony-stimulating factor (GM-CSF) and 1,000 U/ml IL-4 in RPMI medium for 6 days to differentiate into imDCs. Blood samples for the isolation of immune cells were taken from healthy volunteers after informed consent was obtained, as approved by the ethics committee of the University Hospital Jena (file reference number 2395-10/08).
Genetic manipulation of A. benhamiae.
Genetic manipulation was carried out essentially as described previously (18). For cloning, Escherichia coli strain DH5α was applied. The gene deletion construct used consisted of 1-kbp flanking regions of the hydrophobin gene hypA (NCBI accession number XP_003014413.1) cloned into pUC-hph up- and downstream of the hygromycin B resistance cassette (43). A. benhamiae was transformed with linearized vector DNA. The selection of transformants was based on resistance to 200 μg/ml hygromycin B (Roche Applied Science, Germany). Transformants were checked by Southern blotting (33) (Roche digoxigenin [DIG] system) for the replacement of the hypA gene by the resistance cassette. For the complementation of the hypA deletion mutant, the hypA gene including 1 kbp of the promoter region upstream and 0.5 kbp downstream was used. The construct contained a neomycin resistance marker and 1 kbp of the downstream flanking region for recombination. Transformants were selected with 300 μg/ml G418 (Roth, Germany) and analyzed for a single homologous recombination event that restored the hypA gene locus.
Northern blotting and protein analysis.
RNA was isolated from A. benhamiae by using a RiboPure Yeast kit (Ambion) according to the manufacturer's instructions. The expression of the hypA gene was performed by using standard Northern blot protocols (2) and the Roche DIG system for detection. HypA production and the cellular localization of HypA were analyzed by using two-dimensional (2D) gel electrophoresis of cell wall proteins. For this purpose, the mycelium from a liquid culture grown on SG agar for 2 days was harvested and freeze-dried. After thorough homogenization with a pestle and mortar, the cell wall fraction was washed with 1 M NaCl–phosphate-buffered saline (PBS) (8 g/liter NaCl [Roth, Germany], 0.2 g/liter KCl [Roth], 1.44 g/liter Na2HPO4 [AnalaR Normapur, Germany], 0.24 g/liter KH2PO4 [AnalaR Normapur]) and extracted twice for 5 min at 80°C with a preheated solution containing 2% (wt/vol) SDS (Serva, Germany), 0.28% (vol/vol) β-mercaptoethanol (Roth), and 100 mM EDTA (Roth) (pH 7.5). Each extraction was followed by sonication in an ultrasonic bath (Bandelin Sonorex Super RK 510 H) for 5 min. The cell wall extract was washed 5 times with cold water and lyophilized. The purified cell wall was treated with hydrogen fluoride (HF)-pyridine (Sigma, Germany) for 3 h on ice. After neutralization with 1 M ice-cold Tris base (Serva), proteins from the supernatant were precipitated with 10% (wt/vol) trichloroacetic acid (Roth)–100 mg/ml dithiothreitol (DTT; Roth) (end concentration). Proteins were purified as described elsewhere (25). Using this method, hydrophobin was the predominant protein present in the cell wall extract. To check the absence of hydrophobin in the ΔhypA mutant, lyophilized conidia were extracted directly with HF-pyridine without homogenization and SDS treatment. Protein samples were separated by using Bio-Rad Criterion precast Tris-HCl gels (8-to-16% gradient gels). Gels were stained with colloidal Coomassie brilliant blue G-250 (Roth). In cases where a higher level of sensitivity was required for the visualization of protein spots, gels were also silver stained (7). Protein identification by mass spectrometry was carried out as described previously (10).
Phenotype analysis of mutant strains.
To visualize the hydrophobicity of the sporulating mycelium, 100 μl bromophenol blue solution (0.005% [wt/vol]; Sigma, Germany) was carefully added to colonies on an agar surface. The distribution of conidia in an oil-water phase was tested as follows: spore suspensions in water at a concentration of 5 × 107 spores/ml were overlaid with the same volume of paraffin oil (2D cover fluid; GE Healthcare, Germany) and thoroughly shaken at 2,000 rpm for 2 min. Photographs were taken. For scanning electron microscopy (SEM) analysis, cultures were grown for 2 weeks on MAT agar. Conidia were transferred directly onto the sample mounting by stamping the colony. Samples were fixed for 24 h in a desiccator filled with 100 ml 25% (vol/vol) glutaraldehyde (Roth)–18.5% (vol/vol) formaldehyde (Roth). Further preparation and scanning electron microscopy were carried out as described elsewhere (37).
Preparation of fungal material for coincubation experiments.
Viable conidia were harvested from 14-day-old MAT agar plates, counted, and immediately used for coincubation experiments. To obtain hyphae, conidia were cultivated for 16 h in RPMI 1640-5% (vol/vol) FCS medium and washed two times with PBS. Fixed, inactivated fungal material was obtained by incubating conidial or hyphal suspensions overnight with 2.5% (vol/vol) (final concentration) formaldehyde. After neutralization with 100 mM NH4Cl (Merck), conidia or hyphae were washed twice with sterile PBS and stored in PBS.
Coincubation experiments with human cells.
Immature DCs were seeded into multidish wells at a density of 2 × 105 cells/cm2 and coincubated with fixed conidia at a multiplicity of infection (MOI) of 1:1 in RPMI medium containing 5% (vol/vol) FCS for 24 h at 37°C in 5% (vol/vol) pCO2 (partial pressure). The supernatant was obtained by a brief centrifugation at 300 rpm for 5 min. Experiments were repeated three times with imDCs prepared from different donors and at least two technical replicates. Freshly isolated neutrophil granulocytes were used at 2 × 105 cells/ml. Fungal preparations (viable or fixed conidia and hyphae) were used at an MOI of 1:1 or 1:5. The culturing time was 3 or 6 h at 37°C in 5% (vol/vol) pCO2 as indicated. The supernatant was obtained by a brief centrifugation at 300 rpm for 5 min.
ELISA, LDH assay, and NET quantification.
The cytokines interleukin-8 (IL-8), IL-6, IL-10, and tumor necrosis factor alpha (TNF-α) were assayed by using enzyme-linked immunosorbent assay (ELISA) kits from ImmunoTools GmbH and Maxisorp plates (Nunc) according to the manufacturers' instructions. Detection was achieved by using TMB substrate (3,3′,5,5′-tetramethylbenzidine; Kem-En-Tec Diagnostics, Denmark), and the reaction was stopped with 1% (wt/vol) sulfuric acid. The absorbance at 450 nm was measured by using a FLUOstar Optima plate reader (BMG Labtech). Lactate dehydrogenase (LDH) activity in coculture supernatants was assayed by using a cytotoxicity detection kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer's instructions. A readout was obtained at 490 nm with a FLUOstar Optima plate reader. The quantification of neutrophil extracellular traps (NETs) was carried out essentially as described previously by Bruns et al. (9). Propidium iodide (PI; Calbiochem, Germany) was added to the samples at a final concentration of 20 μg/ml following incubation at 37°C and brief shaking at 300 rpm for 10 min. The fluorescence was measured with excitation at 544 nm and emission at 612 nm and a signal amplification (gain) value of 2,000 in a FLUOstar Optima plate reader. Samples obtained from different donors showed a highly variable range in the cell response intensity, e.g., cytokine release. Moreover, the testing of the normal distribution of data (Shapiro-Wilk) failed for certain samples because of a low sample size. Therefore, significance testings were performed by using the nonparametric Wilcoxon signed-rank test, and significance thresholds in Fig. 4 and 5 refer to this test. However, a paired t test on the indicated data also resulted in at least the same significance thresholds. For the quantification of the IL-8 response and NETs against conidia (Fig. 5D and E), three replicates (3 plus 3 plus 3 donors) were analyzed. The quantification of the IL-8 response and NET formation against hyphae (see Fig. 5F and G) was based on data from four donors, and for the correlation between LDH release and NET production (see Fig. 5B and C), two replicates were measured (3 plus 3 donors).
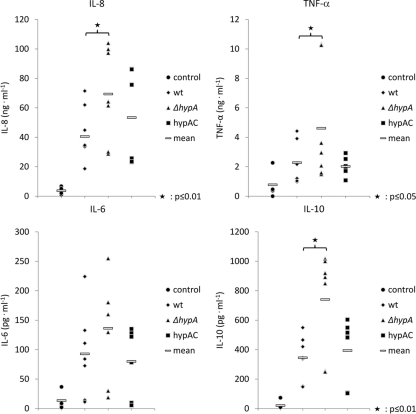
Fig 4.
Cytokine secretion of immature dendritic cells (imDC) during coincubation with A. benhamiae. The levels of the secreted cytokines IL-8, TNF-α, IL-6, and IL-10 were increased for the ΔhypA mutant. The hypAC strain showed wild-type (wt) levels.
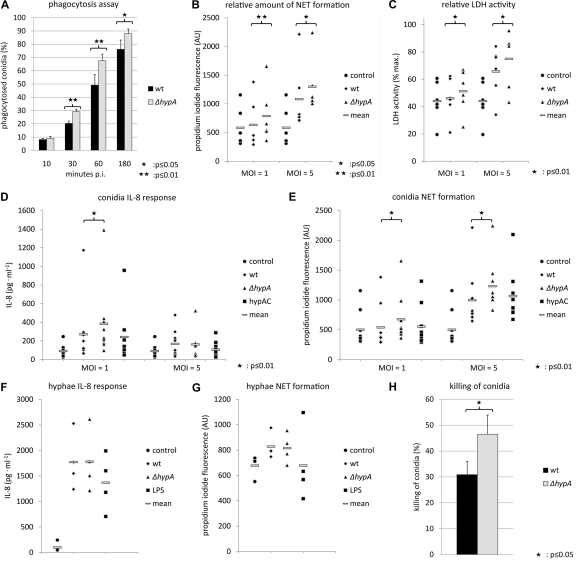
Fig 5.
Response of neutrophil granulocytes to A. benhamiae. (A) Phagocytosis of conidia at an MOI of 1 at different time points. (B and C) Correlation of NET formation and cell lysis. Samples used for NET quantification were tested for the release of lactate dehydrogenase (LDH). Relative values for cell lysis are given. Maximal cell lysis caused by treatment with 2% (vol/vol) Triton X-100 was set at a value of 100. AU, arbitrary units. (D and E) IL-8 secretion of neutrophils and induction of the formation of neutrophil extracellular traps (NETs) by conidia at MOIs of 1 and 5. DNA release was quantified by measuring propidium iodide fluorescence. Data are shown for the wild type, the ΔhypA hydrophobin deletion mutant, and the complemented strain (hypAC). (F and G) IL-8 secretion by neutrophils and NETs induced by hyphae. As a control, 10 ng/ml LPS was used. (H) Killing of A. benhamiae conidia after 6 h of coincubation with neutrophils at an MOI of 1.
Neutrophil granulocyte killing and phagocytosis assays.
To determine the number of conidia killed by neutrophils, conidia and neutrophils (2 × 105/ml) were coincubated at an MOI of 1:1. After incubation for 3 or 6 h at 37°C in 5% (vol/vol) pCO2, 1 U/ml DNase I (Epicentre) was added to the samples, followed by incubation for 10 min at 37°C. The samples were then diluted 10-fold in ice-cold PBS–0.002% (vol/vol) Tween 80 (Merck). The suspensions were streaked onto SG agar plates to obtain a maximum of 200 colonies, estimated based on the initial numbers of conidia (9). Killing values were calculated based on values for controls that were incubated without neutrophils. To assay phagocytosis, conidia were labeled with a prewarmed solution containing 100 μg/ml fluorescein isothiocyanate (FITC; Sigma) and 100 mM Na2CO3 (Sigma) at 37°C for 20 min. The conidia were then washed 3 times with sterile PBS to remove excess FITC. Coincubation was carried out as described above. Immediately after the incubation, cells were inactivated by the addition of formaldehyde at a final concentration of 2% (vol/vol). Experiments were carried out in two replicates using three and two donors, respectively. The colocalization of conidia and neutrophils was monitored microscopically (DMI 4000 B; Leica, Germany) by the counting of at least 600 neutrophils in bright fields and conidia using green fluorescent protein (GFP) filters (excitation at 470 nm and emission at 525 nm). Only conidia clearly covered by a phagocyte were counted as being colocalized, because in some cases, adhesion could not be differentiated from phagocytosis. From these data, the average number of conidia per neutrophil was calculated. Cells were also measured with a BD LSR II cytometer. Based on the average number of conidia per neutrophil counted by microscopy, the rate of phagocytosis was calculated for the cytometer experiment. Data obtained by both experimental techniques were consistent. Experiments were carried out in two replicates using three donors each time.
RESULTS
Identification of the hydrophobin of A. benhamiae.
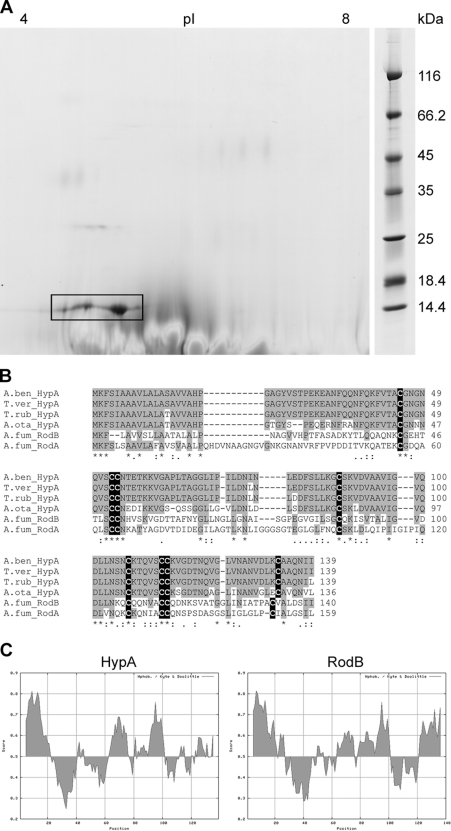
The cell wall is directly exposed to the fungal environment and thus of great importance for recognition by the immune system. To identify proteins potentially interfering with immune recognition, we depleted loosely bound proteins using an extensive extraction method (see Materials and Methods). The analysis of the cell wall extract by 2D PAGE indicated that the only protein present in considerable amounts was a protein exhibiting a conserved hydrophobin domain (Fig. 1A). This protein was identified by mass spectrometry (MS) and was shown to be encoded by a gene we designated hypA (NCBI accession number XP_003014413.1), the only gene of A. benhamiae encoding a putative hydrophobin with a typical structure of eight conserved cysteine residues (Fig. 1B). The deduced HypA protein belongs to hydrophobin class I (36). Previous hydropathy plot analyses (26, 41) indicated its amphiphilic character and gave a hydropathy pattern (Fig. 1C) (http://web.expasy.org/protscale/) similar to that of the A. fumigatus hydrophobins RodB (accession number XP_753093) and RodA (accession number XP_753681). The amino acid sequence of HypA is well conserved within the genera Arthroderma and Trichophyton but strongly diverges from sequences of other clades, including the A. fumigatus proteins mentioned above (Fig. 1B). In a previous study, we found that hypA transcript levels varied with the culture conditions; e.g., an increase was observed during keratinocyte cocultivation (10). The amino acid sequence of HypA contains an N-terminal secretion signal (6), but, in contrast to A. fumigatus RodA, no glycosylphosphatidylinositol (GPI) anchoring site (14) was detected. In addition, the programs NetNGlyc 1.0 and NetOGlyc 3.1 (22) failed to detect putative glycosylation sites, but this might be due to the data set (mammalian/human) which had been used to develop the prediction algorithms.
Fig 1.
The A. benhamiae hydrophobin HypA. (A) Separation of the HypA protein (ARB_06975) by two-dimensional polyacrylamide gel electrophoresis. HypA is marked by a black box. (B) Amino acid sequence comparison of A. benhamiae HypA with the homologous proteins from Trichophyton verrucosum, T. rubrum, and Arthroderma otae (Microsporum canis) and with two hydrophobins of Aspergillus fumigatus, RodA and RodB. Conserved amino acids are highlighted in gray, and cysteine residues are highlighted in black. (C) Hydropathy plot of A. benhamiae HypA and its bidirectional best hit (BBH) of A. fumigatus, RodB. Despite low sequence similarity at the amino acid level (see panel B), the distributions of hydrophilic and hydrophobic regions are similar.
Although for HypA, linkage to the cell wall glucan via a GPI anchor is unlikely due to the lack of a GPI anchoring site, the protein was neither present in culture supernatants nor isolated by concentrated (2 M) lithium chloride solution or SDS–β-mercaptoethanol extraction (data not shown). In contrast, HypA was released upon the treatment of resting conidia or fungal cell walls with hydrogen fluoride-pyridine and was shown to be the dominant extractable surface protein (Fig. 1A).
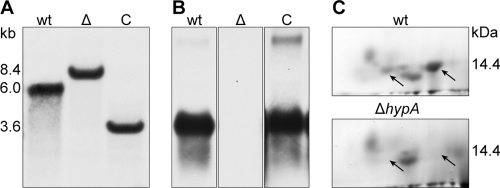
In contrast to many other filamentous fungi, A. benhamiae exhibits a rather efficient system of homologous recombination, making the generation of mutant strains feasible (18). To generate a genetically stable hydrophobin gene deletion mutant of A. benhamiae, protoplasts were generated and transformed with a gene replacement construct containing the hygromycin B resistance cassette (43). Nine of the 16 transformants analyzed showed a recombination event at the hypA locus. Six of these nine transformants showed the integration of a single copy of the gene deletion cassette at the hypA locus. One of these deletion strains was designated the ΔhypA strain (Fig. 2A). This mutant strain was complemented by the reintroduction of the hypA gene at its original locus using the neomycin resistance cassette and G418 (Geneticin) as a selective antibiotic (18). The screening of transformants was conducted based on the reversion of the hydrophobicity phenotype. As demonstrated by Southern blot analysis, three out of six analyzed transformants displayed a restored hypA locus. One of these transformants, designated the complemented hypA (hypAC) strain, showed the integration of a single hypA gene at the hypA gene locus (Fig. 2A). The lack of expression in the deletion strain as well as the reconstituted transcription of hypA in the hypAC strain was confirmed by Northern blot analysis (Fig. 2B). The absence of the hydrophobin protein on conidia of the ΔhypA mutant was shown by HF-pyridine extraction and subsequent 2D polyacrylamide gel electrophoresis (Fig. 2C).
Fig 2.
Molecular analysis of the A. benhamiae hypA mutant. (A) Southern blot analysis of the hypA deletion strain and complemented strain. The probe targeted 5′-upstream positions 51 to 1164 of hypA. DNA was cut by SfoI. Δ, ΔhypA strain; C, hypAC strain. Restriction fragment sizes are indicated in kilobases. (B) Northern blot analysis of the hypA gene in the wild type (wt), ΔhypA, and hypAC strains. Labeling is as described above for panel A. The probe is complementary to the hypA coding sequence. (C) 2D gel electrophoresis of proteins extracted from conidia using hydrogen fluoride-pyridine. The lower acidic region that contains the HypA protein is shown (Fig. 1A). The protein is absent in the ΔhypA strain (arrows).
Characterization of the hypA deletion mutant.
Mycelial growth, conidiation, spore germination, and viability were not affected in the hypA deletion strain (data not shown). To check whether the lack of hydrophobin influences susceptibility to membrane-interfering detergents (Tween and SDS) and cell wall-disturbing compounds (Congo red and calcofluor white), the growth of the ΔhypA mutant was compared with that of the wild type on agar plates containing these compounds at concentrations affecting the growth of the wild-type strain. The levels of inhibition of growth turned out to be the same for the wild type and the ΔhypA mutant (data not shown). Thus, it is unlikely that HypA is a structural component necessary for cell wall integrity.
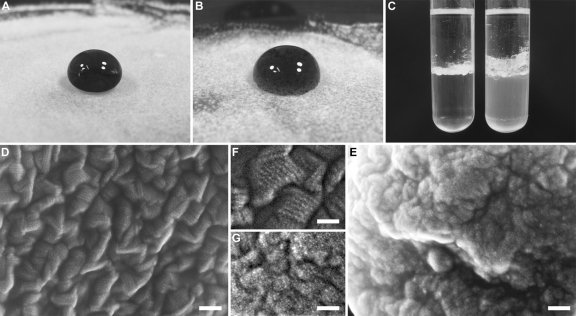
In contrast, a clear phenotype was observed with respect to mycelial hydrophobicity. The deletion mutant showed an “easily wettable” phenotype described previously for other rodlet layer-lacking mutants of filamentous fungi (27, 35). After the production of aerial mycelium and the initiation of sporulation, water dropped off the wild-type colonies, whereas drops of water attached to the mycelium of the hypA deletion strain (Fig. 3A and B, and see Fig. S1 in the supplemental material). Furthermore, conidia of the ΔhypA mutant strain showed a strongly reduced tendency to float at the water-air interface of conidial suspensions. When spore suspensions were overlaid and mixed with hydrophobic paraffin oil, conidia of the wild type were concentrated at the interface layer, whereas ΔhypA conidia remained in the water phase (Fig. 3C, and see Fig. S2 in the supplemental material). To confirm that this phenotype corresponds to the initial assumption that HypA is the main cause of hydrophobicity in A. benhamiae, high-resolution scanning electron microscopy (SEM) images of conidial surfaces were generated. A. benhamiae wild-type conidia displayed the well-known “rodlet” layer, which was completely absent in the hypA deletion strain (Fig. 3D to G).
Fig 3.
Characterization of the hypA deletion mutant. (A and B) Wettability of mycelium grown for 2 weeks. The contact angle of the wild-type mycelium (A) is significantly below 90°, whereas the ΔhypA mycelium (B) is easily wettable (angle, ≥90°). (C) Phase distribution at the water-oil interface. When conidial suspensions were mixed with paraffin oil, wild-type conidia (left tube) were concentrated at the phase boundary, resulting in the clearance of the water phase. The effect was not observed for the ΔhypA mutant. (D to G) High-resolution scanning electron microscopy (SEM) of conidial surfaces. The surface of the wild type displayed the typical rodlet structure (D and F), which is absent from the ΔhypA conidia (E and G). Scale bars, 100 nm (D and E) and 50 nm (F and G).
Recognition of A. benhamiae by human DCs.
During the onset of infection, dermatophytes are confronted with various immune effector cells. An important role played by DCs is as antigen-processing cells. Therefore, their response to A. benhamiae was tested. CD14+ blood monocytes isolated from the blood of healthy donors were stimulated to differentiate into immature dendritic cells (imDCs) in vitro and were coincubated with A. benhamiae. For this type of immune effector cell, the release of the cytokines IL-6, IL-8, IL-10, and TNF-α was observed upon coincubation with viable and formaldehyde-inactivated conidia and hyphae. In comparison with coincubations of imDCs with inactivated conidia, formaldehyde-fixed hyphae induced a stronger cytokine response. However, the level of the response of imDCs was the same for fixed hyphae of the wild type and the ΔhypA mutant strain (see Fig. S3 in the supplemental material). When imDCs were exposed to killed conidia, cytokine release was still detectable but lower than that with exposure to live conidia. Furthermore, formaldehyde-fixed conidia of the ΔhypA mutant strain triggered an increased release of proinflammatory cytokines, providing evidence that the absence of hydrophobin on the conidial surface enhances immune recognition by DCs (Fig. 4).
Recognition by neutrophilic granulocytes.
Neutrophilic granulocytes are the predominant cell type in inflammatory dermatophytic lesions. Due to their short life span, the time of their cocultivation with A. benhamiae was limited to 6 h. During this time period, viable conidia started to swell but did not germinate. Hence, the experimental results were comparable to those obtained from experiments with formaldehyde-inactivated conidia. Upon the addition of conidia, neutrophils showed binding and an extensive phagocytosis of conidia. After 3 h of coincubation, the majority of conidia colocalized with neutrophils. Fluorescence microscopy and cytometer measurements showed that nearly all conidia were taken up by or adhered to neutrophils. By measuring the uptake of conidia at various time points, it was observed that for the ΔhypA strain, the number of conidia colocalizing with neutrophils had increased faster than wild-type conidia. This difference was highest after 1 h of coincubation, with 49% of all wild-type conidia colocalizing with neutrophils, compared with 68% of all ΔhypA conidia (Fig. 5A).
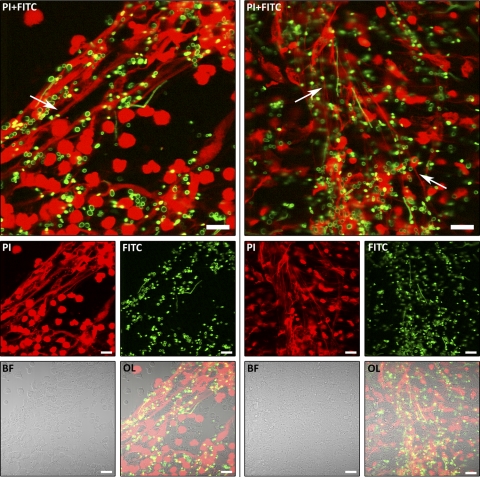
For the first time, we observed the formation of neutrophil extracellular traps (NETs) (8) during coincubation with A. benhamiae (Fig. 6). NET formation was quantified by the fluorescence intensity of propidium iodide (PI), which binds to extracellular DNA released during “NETosis.” Because there was no background fluorescence of conidia, the PI fluorescence originated from the DNA of lysed neutrophils. Neutrophil cell death was also quantified by the measurement of the lactate dehydrogenase (LDH) activity in the supernatant. No enzyme activity was found for A. benhamiae conidia, whereas PI fluorescence intensity values and LDH activity correlated well for samples, which were taken from coincubations (Fig. 5B and C). The response of neutrophils at the cytokine level was investigated by measurements of IL-8 levels.
Fig 6.
NET formation of neutrophils upon coincubation with A. benhamiae. Neutrophils were coincubated with A. benhamiae microconidia at an MOI of 5 for 6 h. DNA was stained with propidium iodide (PI) (red pseudocolor). Conidia were labeled with FITC (green pseudocolor). Pictures where taken with a Zeiss LSM 5 instrument (magnification, ×630; scale bar, 20 μm). The large pictures (PI + FITC) show an overlay of the PI and FITC fluorescence signals. The small pictures show the signals individually; bright-field pictures (BF) and overlays of all signals together (OL) are shown. Staining by PI and FITC was carried out as described in Materials and Methods. Some NETs are indicated with arrows.
After 3 h of coincubation, the amount of DNA released into the culture supernatant and the concentration of IL-8 were low. In addition, no significant killing of conidia was detected after this incubation time (data not shown). In contrast, after 6 h of coincubation, a considerable amount of IL-8 was released by neutrophils (Fig. 5D). In addition, NETs were formed, in particular at a high MOI of 5 (Fig. 5E). The increase of the IL-8 level was accompanied by increases in the release of DNA and of LDH activity in the supernatant. Levels of both NET formation and LDH activity were higher during coincubations with the ΔhypA mutant strain. As expected, the hypAC strain exhibited levels similar to those of the wild type. The proportion of lysed neutrophils determined by DNA release and LDH activity was higher with a higher MOI. In contrast, IL-8 concentrations were lower at a high MOI. This finding implies that the level of production of IL-8 was reduced due to the extensive cell death of neutrophilic granulocytes (Fig. 5D and E). Hyphae of A. benhamiae induced IL-8 production, with cytokine levels that were comparable to those observed for positive controls treated with lipopolysaccharide (LPS). Moreover, the level of NET formation was low. Hyphae of the wild type and the ΔhypA mutant showed no difference with respect to cytokine release and NET formation (Fig. 5F and G). During coincubation, A. benhamiae conidia exposed to neutrophils were effectively killed. Whereas 30% of the wild-type conidia were killed within 6 h of coincubation with neutrophils, in the same period of time, almost 50% of ΔhypA conidia were killed (Fig. 5H).
Taken together, compared with the wild type, the recognition and killing of A. benhamiae ΔhypA mutant conidia by neutrophils were enhanced, most likely due to the lack of the hydrophobin rodlet layer on the conidial surface.
DISCUSSION
The intriguing characteristics of hydrophobins, which are unique to filamentous fungi, provoked much interest in analyses of their biological function. The physicochemical properties of hydrophobins include an amphiphilic character, resistance to all kinds of chemicals (1, 4), and a tendency to form aggregates (42), which cover both hydrophobic and hydrophilic surfaces under conditions of surface polarity reversion (21, 28). The aggregates formed are of interest for both material science and biotechnology. Moreover, since the RodA hydrophobin of A. fumigatus has been shown to render surfaces immunologically inert and prevent the adsorption of other proteins onto hydrophobin-covered surfaces (1, 39), it is the aim of research projects to establish long-term stable protective layers by using fungal hydrophobins. For conidia of dermatophytes, a rodlet layer was described in the 1970s. However, a detailed biochemical and molecular characterization was not possible at that time (20). Here, we provide evidence that the HypA protein of the dermatophyte Arthroderma benhamiae provides the unique structural basis for the hydrophobic rodlet layer of this fungus. Moreover, as shown for A. fumigatus, the rodlet layer of A. benhamiae reduces the intensity of the immune response and even enables conidia to prevent recognition by immune cells at the early stage of infection.
The presence of hydrophobic rodlet layers was found to be important for the dispersal of conidia. The eas mutant of Neurospora crassa lacking the rodlet layer is impaired in the release of conidia and the formation of protective coats on spores (3). Although, as shown here, the production and release of conidia from the mycelium of the A. benhamiae ΔhypA mutant were not impaired, dispersal and transmission through the air may be affected due to the reduced hydrophobicity of the ΔhypA mutant conidia and their tendency to remain in the aqueous phase (Fig. 3A to C). For some species, hydrophobins are even necessary for the development of microconidia (15), and their absence led to defects in morphogenesis and a reduced desiccation tolerance (24). In contrast, as shown here, in the A. benhamiae ΔhypA mutant, hyphal development and conidiation were not impaired. Moreover, conidia were produced abundantly and possessed the same viability as that of wild-type spores. Many basidiomycetes, such as Schizophyllum commune, produce several hydrophobins with distinct functions, e.g., for morphogenesis (29, 38, 40). However, it is likely that in ascomycetes lacking complex reproductive structures, the major function of hydrophobin, including the A. benhamiae hydrophobin HypA, is to coat conidia and hyphae with a protective layer. Hence, we propose that a major function of hydrophobins with respect to virulence is their ability to reduce recognition by the immune system. As shown previously for A. fumigatus, this immunoprotective effect is mediated by the rodlet layer but is abolished as soon as this protective layer is removed during the germination of conidia (9). The same observation was made for A. benhamiae: resting conidia of the wild type hardly triggered any proinflammatory response of DCs and neutrophils, whereas conidia of the ΔhypA mutant, lacking the rodlet layer, induced a strong immune response.
Antigen-presenting immature dendritic cells (imDCs) can be assumed to come into contact with the different morphotypes of A. benhamiae, including conidia. It was shown previously that pathogenic ascomycetes, including dermatophytes, bind to DCs via the C-type lectin DC-SIGN (dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin) (12, 30). A novel pattern recognition pathway to detect dermatophytes via binding to DC-HIL (dendritic cell-associated heparan sulfate proteoglycan-integrin ligand) (31) and the subsequent phosphorylation of an ITAM (immunoreceptor tyrosine-based activation motif) was suggested previously (11). Since binding and phosphorylation have been observed for Trichophyton rubrum but not for Candida albicans, it is conceivable that this mechanism is specific to dermatophytes. As described previously for A. fumigatus, an intact hydrophobin layer on conidia could prevent immune recognition until the onset of swelling and germination (1). As shown here, the response of imDCs to formaldehyde-inactivated ΔhypA conidia led to the same result: compared to the wild type, the level of release of the proinflammatory cytokines IL-8, IL-6, and TNF-α was shown to be elevated by the ΔhypA mutant (Fig. 4). Moreover, the release of IL-10 showed that the response of DCs to A. benhamiae is not exclusively proinflammatory but also appears to be important as an immunomodulatory signal. By releasing IL-10 at the site of infection, the differentiation of monocytes into macrophages that clear the site of infection without releasing inflammation mediators is triggered. This leads to a moderate inflammatory reaction (19). Here, we demonstrate that DCs show a complex response to A. benhamiae and that the rodlet layer formed by the HypA protein is able to delay the immune recognition by DCs.
Neutrophils immediately started to bind and phagocytose A. benhamiae conidia. Almost all conidia were taken up by neutrophils after 3 h of coincubation, and the killing of conidia occurred within the neutrophils. Probably, the phagocytosis of conidia is a prerequisite for the induction of NETosis but not a trigger for IL-8 release, since in experiments using hyphae, NET formation was almost completely abolished, whereas the level of cytokine secretion was high (Fig. 5D to G). Interestingly, this finding is in contrast to previous results obtained for A. fumigatus, where hyphae were shown to be a strong inducer of NET formation (9). The increased IL-8 signaling upon contact with hyphae demonstrates that A. benhamiae hyphae are still recognized by neutrophils. This suggests that the mechanism of hyphal recognition differs between these two fungal pathogens.
In conclusion, the immune recognition of A. benhamiae is strongly influenced by the presence of the conidial rodlet layer that is formed by the HypA protein. For hyphae, the presence of hydrophobin seems to play a minor role, since the expression of the hypA gene is absent and/or the protective effect of HypA is displaced by the exposition of cell wall components, which can be recognized by immune cells. Upon recognition, cytokine signaling is proinflammatory (IL-6, IL-8, and TNF-α), although the secretion of IL-10 by DCs implies a contribution of an immunomodulatory component. The defense reactions of neutrophils include cytokine signaling, phagocytosis, NET formation, and the killing of conidia. After the germination of conidia, the hyphae escape phagocytosis. The neutrophils then release IL-8 as an attractant for other neutrophils and immune cells, e.g., macrophages, but avoid cell death via NETosis.
Supplementary Material
ACKNOWLEDGMENTS
We thank the group of Peter F. Zipfel (Hans Knöll Institute, Jena, Germany) for help in performing experiments with human cell lines.
This research was supported by the Pakt für Forschung und Innovation of the Free State of Thuringia, the Federal Ministry of Science and Technology (BMBF) (Germany), and the International Leibniz Research School for Microbial and Biomolecular Interactions Jena (ILRS) as part of the DFG-funded excellence graduate school Jena School for Microbial Communication (JSMC). Work in the laboratory of Oliver Kurzai was supported by the BMBF within the Unternehmen Region program.
Footnotes
Published ahead of print 9 March 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Aimanianda V, et al. 2009. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460:1117–1121 [DOI] [PubMed] [Google Scholar]
- 2. Alwine JC, Kemp DJ, Stark GR. 1977. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc. Natl. Acad. Sci. U. S. A. 74:5350–5354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beever RE, Dempsey GP. 1978. Function of rodlets on the surface of fungal spores. Nature 272:608–610 [DOI] [PubMed] [Google Scholar]
- 4. Beever RE, Redgwell RJ, Dempsey GP. 1979. Purification and chemical characterisation of the rodlet layer of Neurospora crassa conidia. J. Bacteriol. 140:1063–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Behnsen J, et al. 2010. Secreted Aspergillus fumigatus protease Alp1 degrades human complement proteins C3, C4, and C5. Infect. Immun. 78:3585–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 7. Blum H, Beier H, Gross HJ. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93–99 [Google Scholar]
- 8. Brinkmann V, Laube B, Abu Abed U, Goosmann C, Zychlinsky A. 2010. Neutrophil extracellular traps: how to generate and visualize them. J. Vis. Exp. 2010(36):pii=1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bruns S, et al. 2010. Production of extracellular traps against Aspergillus fumigatus in vitro and in infected lung tissue is dependent on invading neutrophils and influenced by hydrophobin RodA. PLoS Pathog. 6:e1000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burmester A, et al. 2011. Comparative and functional genomics provide insights into the pathogenicity of dermatophytic fungi. Genome Biol. 12:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chung JS, et al. 2009. Binding of DC-HIL to dermatophytic fungi induces tyrosine phosphorylation and potentiates antigen presenting cell function. J. Immunol. 183:5190–5198 [DOI] [PubMed] [Google Scholar]
- 12. Curtis BM, Scharnowske S, Watson AJ. 1992. Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc. Natl. Acad. Sci. U. S. A. 89:8356–8360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drouot S, Mignon B, Fratti M, Roosje P, Monod M. 2009. Pets as the main source of two zoonotic species of the Trichophyton mentagrophytes complex in Switzerland, Arthroderma vanbreuseghemii and Arthroderma benhamiae. Vet. Dermatol. 20:13–18 [DOI] [PubMed] [Google Scholar]
- 14. Eisenhaber B, Schneider G, Wildpaner M, Eisenhaber F. 2004. A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe. J. Mol. Biol. 337:243–253 [DOI] [PubMed] [Google Scholar]
- 15. Fuchs U, Czymmek KJ, Sweigard JA. 2004. Five hydrophobin genes in Fusarium verticillioides include two required for microconidial chain formation. Fungal Genet. Biol. 41:852–864 [DOI] [PubMed] [Google Scholar]
- 16. Fumeaux J, et al. 2004. First report of Arthroderma benhamiae in Switzerland. Dermatology 208:244–250 [DOI] [PubMed] [Google Scholar]
- 17. Giddey K, Favre B, Quadroni M, Monod M. 2007. Closely related dermatophyte species produce different patterns of secreted proteins. FEMS Microbiol. Lett. 267:95–101 [DOI] [PubMed] [Google Scholar]
- 18. Grumbt M, et al. 2011. Targeted gene deletion and in vivo analysis of putative virulence gene function in the pathogenic dermatophyte Arthroderma benhamiae. Eukaryot. Cell 10:842–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grütz G. 2005. New insights into the molecular mechanism of interleukin-10-mediated immunosuppression. J. Leukoc. Biol. 77:3–15 [DOI] [PubMed] [Google Scholar]
- 20. Hashimoto T, Wu-Yuan CD, Blumenthal HJ. 1976. Isolation and characterisation of the rodlet layer of Trichophyton mentagrophytes microconidial wall. J. Bacteriol. 127:1543–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hou S, et al. 2009. Surface modification using a novel type I hydrophobin HGFI. Anal. Bioanal. Chem. 394:783–789 [DOI] [PubMed] [Google Scholar]
- 22. Julenius K, Mølgaard A, Gupta R, Brunak S. 2005. Prediction, conservation analysis, and structural characterisation of mammalian mucin-type O-glycosylation sites. Glycobiology 15:153–164 [DOI] [PubMed] [Google Scholar]
- 23. Kahlke B, Brasch J, Christophers E, Schröder JM. 1996. Dermatophytes contain a novel lipid-like leukocyte activator. J. Invest. Dermatol. 107:108–112 [DOI] [PubMed] [Google Scholar]
- 24. Klimes A, Dobinson KF. 2006. A hydrophobin gene, VDH1, is involved in microsclerotial development and spore viability in the plant pathogen Verticillium dahliae. Fungal Genet. Biol. 43:283–294 [DOI] [PubMed] [Google Scholar]
- 25. Kniemeyer O, Lessing F, Scheibner O, Hertweck C, Brakhage AA. 2006. Optimisation of a 2-D gel electrophoresis protocol for the human-pathogenic fungus Aspergillus fumigatus. Curr. Genet. 49:178–189 [DOI] [PubMed] [Google Scholar]
- 26. Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132 [DOI] [PubMed] [Google Scholar]
- 27. Lauter FR, Russo VE, Yanofsky C. 1992. Developmental and light regulation of eas, the structural gene for the rodlet protein of Neurospora. Genes Dev. 6:2373–2381 [DOI] [PubMed] [Google Scholar]
- 28. Lumsdon SO, Green J, Stieglitz B. 2005. Adsorption of hydrophobin proteins at hydrophobic and hydrophilic interfaces. Colloids Surf. B Biointerfaces 44:172–178 [DOI] [PubMed] [Google Scholar]
- 29. Schuren FH, Wessels JG. 1990. Two genes specifically expressed in fruiting dikaryons of Schizophyllum commune: homologies with a gene not regulated by mating-type genes. Gene 90:199–205 [DOI] [PubMed] [Google Scholar]
- 30. Serrano-Gómez D, Leal JA, Corbí AL. 2005. DC-SIGN mediates the binding of Aspergillus fumigatus and keratinophylic fungi by human dendritic cells. Immunobiology 210:175–183 [DOI] [PubMed] [Google Scholar]
- 31. Shikano S, Bonkobara M, Zukas PK, Ariizumi K. 2001. Molecular cloning of a dendritic cell-associated transmembrane protein, DC-HIL, that promotes RGD-dependent adhesion of endothelial cells through recognition of heparan sulfate proteoglycans. J. Biol. Chem. 276:8125–8134 [DOI] [PubMed] [Google Scholar]
- 32. Shiraki Y, Ishibashi Y, Hiruma M, Nishikawa A, Ikeda S. 2006. Cytokine secretion profiles of human keratinocytes during Trichophyton tonsurans and Arthroderma benhamiae infections. J. Med. Microbiol. 55:1175–1185 [DOI] [PubMed] [Google Scholar]
- 33. Southern EM. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503–517 [DOI] [PubMed] [Google Scholar]
- 34. Staib P, et al. 2010. Differential gene expression in the pathogenic dermatophyte Arthroderma benhamiae in vitro versus during infection. Microbiology 156:884–895 [DOI] [PubMed] [Google Scholar]
- 35. Stringer MA, Dean RA, Sewall TC, Timberlake WE. 1991. Rodletless, a new Aspergillus developmental mutant induced by directed gene inactivation. Genes Dev. 5:1161–1171 [DOI] [PubMed] [Google Scholar]
- 36. Sunde M, Kwan AH, Templeton MD, Beever RE, Mackay JP. 2008. Structural analysis of hydrophobins. Micron 39:773–784 [DOI] [PubMed] [Google Scholar]
- 37. Thywißen A, et al. 2011. Conidial dihydroxynaphthalene melanin of the human pathogenic fungus Aspergillus fumigatus interferes with the host endocytosis pathway. Front. Microbiol. 2:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Wetter MA, Wösten HA, Wessels JG. 2000. SC3 and SC4 hydrophobins have distinct roles in formation of aerial structures in dikaryons of Schizophyllum commune. Mol. Microbiol. 36:201–210 [DOI] [PubMed] [Google Scholar]
- 39. von Vacano B, et al. 2011. Hydrophobin can prevent secondary protein adsorption on hydrophobic substrates without exchange. Anal. Bioanal. Chem. 400:2031–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wessels J, De Vries O, Asgeirsdottir SA, Schuren F. 1991. Hydrophobin genes involved in formation of aerial hyphae and fruit bodies in Schizophyllum. Plant Cell 3:793–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilkins MR, et al. 1999. Protein identification and analysis tools in the ExPASy Server. Methods Mol. Biol. 112:531–552 [DOI] [PubMed] [Google Scholar]
- 42. Wosten H, De Vries O, Wessels J. 1993. Interfacial self-assembly of a fungal hydrophobin into a hydrophobic rodlet layer. Plant Cell 5:1567–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu J, Gong ZZ. 2003. Intron requirement for AFP gene expression in Trichoderma viride. Microbiology 149:3093–3097 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.