Abstract
The parabasalid protist Trichomonas vaginalis is a widespread parasite that affects humans, frequently causing vaginitis in infected women. Trichomonad mitosis is marked by the persistence of the nuclear membrane and the presence of an asymmetric extranuclear spindle with no obvious direct connection to the chromosomes. No centromeric markers have been described in T. vaginalis, which has prevented a detailed analysis of mitotic events in this organism. In other eukaryotes, nucleosomes of centromeric chromatin contain the histone H3 variant CenH3. The principal aim of this work was to identify a CenH3 homolog in T. vaginalis. We performed a screen of the T. vaginalis genome to retrieve sequences of canonical and variant H3 histones. Three variant histone H3 proteins were identified, and the subcellular localization of their epitope-tagged variants was determined. The localization of the variant TVAG_185390 could not be distinguished from that of the canonical H3 histone. The sequence of the variant TVAG_087830 closely resembled that of histone H3. The tagged protein colocalized with sites of active transcription, indicating that the variant TVAG_087830 represented H3.3 in T. vaginalis. The third H3 variant (TVAG_224460) was localized to 6 or 12 distinct spots at the periphery of the nucleus, corresponding to the number of chromosomes in G1 phase and G2 phase, respectively. We propose that this variant represents the centromeric marker CenH3 and thus can be employed as a tool to study mitosis in T. vaginalis. Furthermore, we suggest that the peripheral distribution of CenH3 within the nucleus results from the association of centromeres with the nuclear envelope throughout the cell cycle.
INTRODUCTION
The widespread human parasite Trichomonas vaginalis is a protist from the Parabasala group (5). The parasite causes a sexually transmitted disease, trichomoniasis, which is a common cause of vaginitis (30). In addition, trichomoniasis has been reported to be associated with an increased risk of human immunodeficiency virus infection, an increased risk of cervical cancer, and adverse outcomes of pregnancy (14). The strikingly large repetitive genome of T. vaginalis (∼160 Mb) is tightly packed into six chromosomes (4). Trichomonad nuclei divide by a specific form of mitosis called cryptopleuromitosis, during which the nuclear envelope is retained, the mitotic spindle is lateral, and its microtubules do not enter the nucleus or contact chromosomes directly (26). These features discriminate trichomonad mitosis from open mitosis, in which the nuclear envelope breaks down, from closed mitosis, which involves an intranuclear spindle, and from semiopen mitosis, in which microtubules of the extranuclear spindle penetrate through the nuclear envelope (25).
Kinetochore complexes mediate the capture of chromosomes by spindle microtubules and the migration of chromosomes to the cellular poles. These large proteinaceous structures are formed at the sites of centromeres. Kinetochores are transiently assembled during mitosis and display great structural diversity, while centromeres are present throughout the cell cycle. Centromeric chromatin is defined by the presence of the centromeric histone H3 variant (CenH3, also known as CENP-A), which replaces the core H3 histone in centromeric nucleosomes. CenH3 is crucial for recruitment of kinetochore proteins and thus serves as an epigenetic marker of the site where kinetochores assemble on the centromere (29).
Apart from CenH3, H3.3 is another variant of the core H3 histone. The protein sequence of histone H3.3 is almost identical to that of core H3, and both core H3 and H3.3 carry conserved lysines, which can be methylated. H3.3 is found in transcriptionally active chromatin and in pericentric heterochromatin and telomeres, as shown recently (35, 36). Unlike H3.3, CenH3 is a highly divergent variant of the core histone. In the C-terminal histone fold domain (HFD), the CenH3 variant shares 50 to 60% sequence similarity with core H3. Compared with core H3, CenH3s contain an N-terminal extension of variable length and primary structure (9). CenH3s have been identified in all eukaryotes studied to date, with the exception of Trypanosoma brucei (20). However, the origin of CenH3 is unclear. Phylogenetic analyses did not support CenH3 as a monophyletic group. Consequently, CenH3 function cannot be assigned to H3 variants without experimental characterization (9, 24). Other specific variants of histone H3 have evolved in the parasitic protists T. brucei and Giardia intestinalis. T. brucei histone H3V is enriched at telomeres and is a candidate variant that possibly serves as a substitute for CenH3 (20), while G. intestinalis histone H3B marks noncentromeric heterochromatin (9).
The mechanism of chromosome segregation and, in particular, the molecular basis of the interaction between spindle microtubules and chromosomes are virtually unknown in T. vaginalis. A set of spindle microtubules that contact the nuclear envelope has been observed; however, whether and how they interact with kinetochores is unclear. Several reports have suggested that spindle microtubules bind to kinetochores inserted into the nuclear membrane (3, 18, 25). However, this view was challenged by Ribeiro et al. (27). The investigation of this interaction is complicated by the lack of suitable centromeric markers. Therefore, the aim of this work was to identify T. vaginalis CenH3, to distinguish this protein from core H3 and possible other H3 variants, and to investigate its nuclear localization during the cell cycle. Our bioinformatic analysis of T. vaginalis H3 paralogs revealed three distinct H3 variants. These variants were expressed with a C-terminal hemagglutinin (HA) tag in T. vaginalis, and their deposition in chromatin during the cell cycle was examined. The first H3 variant (TVAG_087830) localized in a similar manner as H3.3, and it colocalized with an antibody against H3K4 methylation, which defines transcriptionally active sites. The second H3 variant (TVAG_185390) was deposited in bulk chromatin and resembled the canonical histone H3 (TVAG_270080) in distribution, although it has a rather divergent protein sequence. The third H3 variant (TVAG_224460), which possesses an N-terminal extension, localized mostly to 6 or 12 distinct spots at the periphery of the nucleus. We propose that this variant corresponds to the centromeric marker CenH3 and that centromeres are associated with the nuclear envelope during the cell cycle.
MATERIALS AND METHODS
Cell culture.
Trichomonas vaginalis strain T1 (kindly provided by J.-H. Tai, Institute of Biomedical Sciences, Taipei, Taiwan) was used in this study. Cells were grown in tryptone-yeast extract-maltose medium (pH 6.2) supplemented with 10% heat-inactivated horse serum at 37°C (11).
Sequences and variant histone H3_HA constructs.
Protein sequences of core histones and their variants found in selected members of main eukaryotic groups, including Opisthokonta, Amoebozoa, Plantae, Chromista, and Excavata (see Table S1 in the supplemental material), were used as queries to screen the trichomonad genome database TrichDB (http://trichdb.org/trichdb). For a complete list and copy number of T. vaginalis histone genes identified in the T. vaginalis genome, see Table S2 and Fig. S1 in the supplemental material. Sequences of H3 and variant H3 proteins from other organisms were downloaded from the NCBI Protein Database (http://www.ncbi.nlm.nih.gov). Protein sequences were aligned using the ClustalX program (37) and manually edited using BioEdit software (17). Secondary structures were inferred from published crystal structures of chicken core H3 (1tzy_C) and human CENP-A (3nqu_A) (31, 40) or predicted using the HHpred server (http://toolkit.tuebingen.mpg.de/hhpred) (32). The coding sequences of four selected H3 histones without their stop codons (TVAG_270080, TVAG_087830, TVAG_224460, TVAG_185390) were PCR amplified and cloned into the T. vaginalis expression vector TagVag (12) with a sequence coding for a double HA tag at the 3′ end. The following primers, including terminal 5′ NdeI and 3′ BamHI restriction sites (shown in italics), were used: TVAG_270080, 5′-CATATGGCTCGTACAAAGCAG-3′ and 5′-GGATCCGTTACGTTCTCCGCGGAT-3′; TVAG_087830, 5′-CATATGGCTCGTACTAAGCAA-3′ and 5′-GGATCCATTACGCTCTCCGCGGAT-3′; TVAG_224460, 5′-CATATGGCCAGTACCCGAATC-3′ and 5′-GGATCCCTCAGTAATTGAATCGCC-3′; and TVAG_185390, 5′-CATATGGAGGAGGAACCTCGG-3′ and 5′-GGATCCGCGATCTCCGCGGAGTCT-3′. Genomic DNA was isolated from trichomonads using a High Pure PCR template preparation kit (Roche) and was used as a template for PCR.
Selectable transformation of Trichomonas vaginalis.
Trichomonads were electroporated with the histone H3_HA constructs and maintained as described by Sutak et al. (34). Briefly, cells were pelleted by centrifugation, resuspended in fresh medium, and mixed gently with the TagVag constructs (300 μl of cell suspension, 50 μg DNA). A GenePulserXL apparatus (Bio-Rad) was used for electroporation (350 V; time constant, 175 ms). After 4 h, G418 (200 μg/ml; PAA Laboratories) was added to the transformants. Expression of tagged proteins was analyzed at 10 to 14 days after transformation.
Immunofluorescence microscopy.
The trichomonad culture was enriched in mitotic cells according to the protocol described by Torres-Machorro et al. (38). Trichomonads were cultivated with 1 mM colchicine (Sigma-Aldrich) for 6 h, followed by 5 min of hypotonic swelling in 75 mM KCl (42). Before immunodetection of HA-tagged histones, chromosomes were treated on microscope slides with 0.1% SDS for 15 min. Microscope slides were prepared according to the protocol described by Sutak et al. (34). Briefly, the cells were fixed with methanol and acetone on microscope slides and blocked in phosphate-buffered saline containing 0.25% bovine serum albumin, 0.25% gelatin, 0.05% Tween 20. HA-tagged histones were detected using a mouse anti-HA monoclonal antibody (Exbio, Prague, Czech Republic). A rabbit anti-monomethylated H3K4 polyclonal antibody (Millipore) was used for detection of transcriptionally active regions. Donkey anti-mouse Alexa Fluor-488 and anti-rabbit Alexa Fluor-546 antibodies (Molecular Probes) were used for immunostaining. The cells were mounted in Vectashield with DAPI (4′,6-diamidino-2-phenylindole; Vector Labs). Fluorescence microscopy was performed using an IX81 microscope with an IX2-UCB camera, and images were processed using Cell software (Olympus). Cell section images were analyzed using ImageJ software (NIH, Bethesda, MD).
RESULTS AND DISCUSSION
T. vaginalis genome database screen and analysis of H3 variant protein sequences.
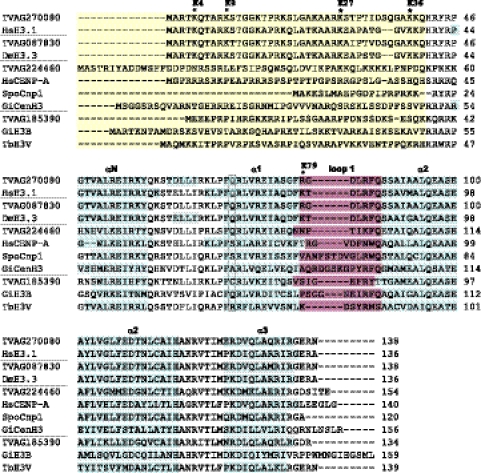
Although histones are among the most conserved proteins in eukaryotes, the core H3 and H4 histones from protists, including trichomonads, show remarkable sequence diversity compared with better-studied metazoan histones (22, 24). Consequently, current annotations of H3 histones and H3 variants are rather confusing in the T. vaginalis genome database, in which 12 histones are annotated as putative CenH3/CENP-A variants (http://trichdb.org/trichdb). Our screen of the T. vaginalis genome database revealed that all core histones are present in the T. vaginalis genome in multiple copies. In a haploid genome, there are 17 copies of histone H2A, 14 copies of H2B, and 21 copies of H4 (see Tables S1 and S2 in the supplemental material). The putative histone H3 is encoded by 23 genes, 20 of which are 100% identical at the amino acid sequence level and 88.7 to 99.7% identical at the nucleotide level (see Fig. S1 in the supplemental material). These sequences correspond to the core H3 histone of T. vaginalis described previously by Marinets et al. (22). The gene TVAG_270080 was randomly selected as a representative gene for the core H3 genes (Fig. 1). The three single-copy genes TVAG_087830, TVAG_185390, and TVAG_224460 displayed 95.7%, 48.9%, and 49.6% identity with T. vaginalis core H3, respectively. The genes coding for the core histones are commonly present in repeat arrays, while histone variants are found in single copies (36). Thus, these divergent H3 histones were obvious candidates for H3 variants.
Fig 1.
Analysis of protein sequences of H3 and H3 variants. Sequences are grouped with respect to putative function of the proteins (core histones, H3.3 variant histones, centromeric histones, histone variants with unknown function). Divergent N-terminal extensions (yellow) are not properly aligned between species. Positions of predicted α helices within the histone-fold domain are marked (blue). A conserved glutamine residue of core H3 (boxed) is substituted in all centromeric histones except the histone TVAG_224460. The loop 1 region (purple) shows amino acid insertions in the centromeric histones. Sites of conserved lysine residues known to play a role in gene activation or repression are marked by asterisks; the labels reflect the numbering commonly used for core H3. The TrichDB accession numbers for T. vaginalis sequences are as follows: TVAG_270080, core H3; TVAG_087830, H3.3 variant; TVAG_224460, CenH3; TVAG_185390, H3T (unknown function). NCBI protein database accession numbers are as follows: SpoCnp1, Schizosaccharomyces pombe Cnp1, NP_596473; HsCENP-A, Homo sapiens CENP-A, NP_001800; HsH3.1, Homo sapiens H3.1, NP_003520; GiCenH3, Giardia intestinalis CenH3, EDO81729; GiH3B, Giardia intestinalis H3B, EDO76497; DmH3.3, Drosophila melanogaster H3.3, CAA57080; TbH3V, Trypanosoma brucei H3V, EAN78895.
The H3 variant TVAG_087830 displayed hallmarks of H3.3. The protein sequences of H3.3 histones are almost identical with the sequence of core H3, except for 4 to 6 amino acids (19). In TVAG_087830, these substitutions occur at positions 11 (serine for threonine), 29 (serine for alanine), 30 (threonine for isoleucine), 33 (isoleucine for valine), 99 (serine for alanine), and 131 (glutamine for methionine) (Fig. 1). The secondary structure prediction for T. vaginalis H3 assigns four of these substitutions to the N-terminal tail of the protein (T11, A29, I30, and V33) and one of them to the α2 helix of the HFD (A99). This distribution is similar to what was observed in other species (Fig. 1) (19). An additional substitution occurs in T. vaginalis in the α3 helix of the HFD (M131), which was described in Tetrahymena thermophila as well (7). The core H3 and the H3.3 variant contain conserved lysine (K) residues that undergo modifications, such as acetylation and methylation, and are involved in the activation or repression of transcription (Fig. 1). Although the effects of specific histone modifications depend on the context in which they are presented, generally, methylated K4, K36, and K79 of H3 are considered to mark active genes, while methylation of K9 and K27 corresponds with gene silencing. Lysines, which can be acetylated, occur at positions K9, K14, K18, and K23 within the N-terminal tail and at position K56 within the histone fold; in this region, the DNA enters and exits the nucleosome (39). A complete set of conserved lysines is present at the N terminus of T. vaginalis H3 and H3.3 at positions corresponding to K4, K9, K14, K17, K23, K27, K36, and K37, as well as the conserved lysine within the HFD (K56) (Fig. 1). In T. vaginalis core H3 and H3.3, K79 is replaced by an arginine (R81). Protein arginine methyltransferases acting on histones have been described, and eight putative arginine methyltransferases were annotated in the T. vaginalis genome (15). Thus, methylation of R81 may be involved in the regulation of gene expression.
According to their protein sequences, either of the other two H3 variants (TVAG_185390, TVAG_224460) might serve as a marker of trichomonad centromeres (Fig. 1). Generally, centromeric H3 variants lack a conserved sequence motif, but their protein sequences share several specific features (1, 19). We found that TVAG_185390 and TVAG_224460 conform to most of the following criteria. (i) CenH3s have divergent N termini when aligned with canonical histone H3 and very often carry extensions of up to ∼150 amino acids (9). The N-terminal portions of both putative CenH3s of T. vaginalis are divergent. TVAG_224460 possesses an N-terminal extension of 14 amino acids, while the N-terminal sequence of TVAG_185390 is 3 amino acid residues shorter than that of core H3. (ii) Within the C-terminal HFD, CenH3s typically share only 50 to 60% identity with canonical H3 (6, 20). Putative CenH3s of T. vaginalis share 60% and 61% sequence identity with canonical H3 within the HFD. (iii) All experimentally validated centromeric H3 variants have at least 1 amino acid residue insertion in HFD loop 1. Insertion of a single amino acid is present in loop 1 of the variant TVAG_185390 as well as TVAG_224460. (iv) A conserved glutamine residue in the α1 helix of core histone H3 is often substituted in CenH3s (20). Accordingly, this substitution occurs in the TVAG_185390 variant; however, the glutamine is retained in the TVAG_224460 variant (Fig. 1).
In conclusion, our analysis of primary sequences suggested that TVAG_087830 was a candidate H3.3 histone, while TVAG_185390 and TVAG_224460 may represent CenH3 in T. vaginalis.
Localization of H3 variants in chromatin.
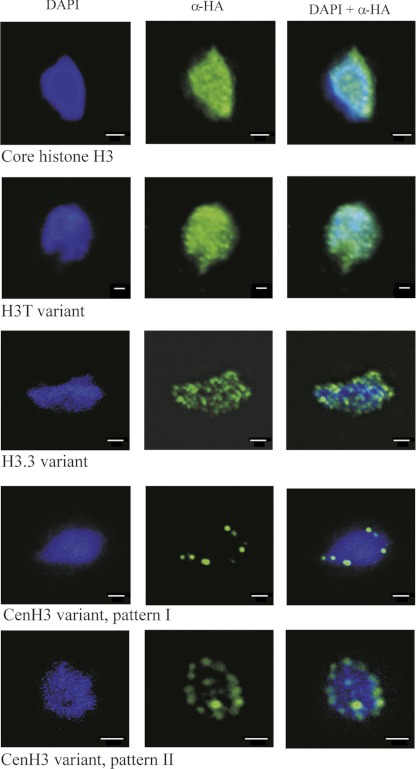
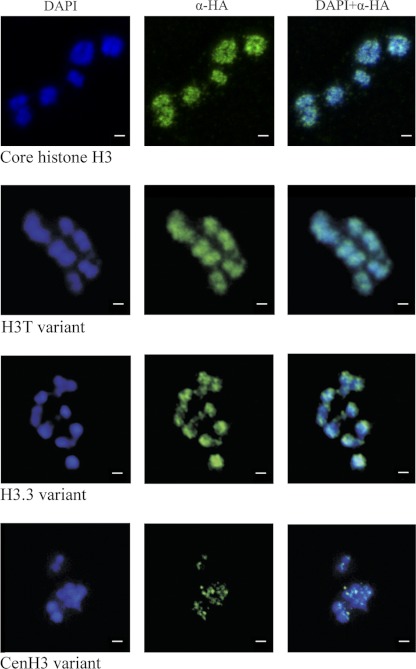
To examine the chromatin distribution of T. vaginalis histone H3 and H3 variants, we constructed plasmids encoding fusion proteins consisting of histones with a C-terminal HA tag. Expression of core H3-HA or its variants did not affect the viability or growth of the cultures (see Fig. S2 in the supplemental material). Western blot analysis of transformed cells using an anti-HA tag antibody revealed the presence of proteins of 20 kDa, a size that corresponds to the expected sizes of H3-HA histones (data not shown). Indirect fluorescent staining for core H3-HA showed a diffuse distribution throughout interphase nuclei and along the arms of metaphase chromosomes (Fig. 2 and 3). This distribution is consistent with the localization of core H3 observed in various eukaryotic cell lines (36).
Fig 2.
Localization of T. vaginalis core H3-HA and H3-HA variants in interphase nuclei. T. vaginalis mid-logarithmic-growth population was used for detection of tagged proteins. All cells displayed homogeneous labeling of all interphase nuclei in which core H3-HA (TVAG_270080) and H3T-HA (TVAG_185390) were expressed. The same punctate pattern was observed throughout the population that expressed H3.3-HA (TVAG_087830). Images are representative of tagged protein localization observed in over 200 cells per each strain; localization demonstrated in the figures was observed in 90% of examined cells. Two distinct patterns were observed for trichomonads that expressed CenH3 (TVAG_224460); patterns I and II were observed in 20% and 80% of examined cells, respectively. Bars, 1 μm.
Fig 3.
Localization of T. vaginalis core H3-HA and H3-HA variants in metaphase chromosomes. Images are representative of tagged protein localizations observed in over 200 cells per each strain. Core histone H3, TVAG_270080; H3T variant, TVAG_185390; H3.3 variant, TVAG_087830; CenH3 variant, TVAG_224460. Bars, 1 μm.
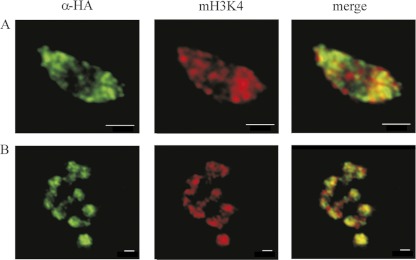
Expression of TVAG_087830-HA revealed a punctate pattern throughout interphase nuclei (Fig. 2). Numerous distinct foci were also observed on metaphase chromosomes (Fig. 3). This pattern indicated a distribution of the H3.3 variant whereby it is enriched at transcriptionally active sites (36). However, utilization of H3.3 is not universal, and in some eukaryotes, canonical H3-containing nucleosomes occur both in bulk chromatin and in transcriptionally active genes (1, 35, 36). An example of a species in which the H3.3 variant is missing is the diplomonad G. intestinalis, a close relative of T. vaginalis (9). This organism possesses a unique H3B variant. Similar to the results obtained with TVAG_087830-HA, H3B is also present in distinct foci. However, H3B does not define sites of active transcription, and its function is unknown (9). To distinguish whether TVAG_087830 is indeed H3.3 or rather an H3B-like variant, we assessed the localization of TVAG_087830-HA by immunostaining with an antibody specific for monomethylated H3 at K4 (mH3K4) as a marker for transcriptionally active sites. The pattern of TVAG_087830-HA distribution showed that it mainly colocalized with foci stained by the anti-mH3K4 antibody (Fig. 3 and 4). Thus, we assume that TVAG_087830 acts as an H3.3 variant and that methylated H3.3-containing nucleosomes are present in actively transcribed regions in T. vaginalis.
Fig 4.
Active regions of transcription visualized by monomethyl H3K4 immunostaining and localization of the T. vaginalis H3.3 variant (TVAG_087830). (A) Interphase nuclei; (B) metaphase chromosomes. Images are representative of tagged protein localizations observed in over 200 cells per each strain. Bars, 1 μm.
Although the sequences of both trichomonad divergent histone variants (TVAG_185390 and TVAG_224460) show several features of CenH3s, expression of TVAG_185390-HA resulted in its uniform deposition in whole interphase nuclei and along entire arms of metaphase chromosomes, similar to core H3, and thus, TVAG_185390-HA did not conform to CenH3 (Fig. 2 and 3). This H3 variant was named H3T. In contrast, immunostaining of trichomonads expressing TVAG_224460-HA indicated a centromeric localization. CenH3 staining typically reveals multiple foci corresponding to chromosome number (8) or occurs exceptionally as a single dot, e.g., in the nuclei of Toxoplasma gondii, where all chromosomes are restrained close to one another in a specific region of the nucleus (2). In T. vaginalis, immunostaining of TVAG_224460-HA yielded two distinct patterns: (i) TVAG_224460-HA was observed as six discrete dots in 20% of examined cells, corresponding to the number of T. vaginalis chromosomes in the G1 phase (13), or (ii) TVAG_224460-HA appeared as six sets of double dots in about 80% of nuclei. This result is consistent with a duplicated chromatin state and monocentric chromosome appearance during the G2 phase of the cell cycle, which is a dominant phase in T. vaginalis cultures (10, 28). Importantly, single as well as double dots organized at the periphery of the interphase nucleus. This type of localization is consistent with an association of the centromere and/or kinetochores with the nuclear envelope, which allows their binding to spindle microtubules during mitosis. This localization also suggests that centromeres may associate with the nuclear membrane during the entire cell cycle. Specific foci of TVAG_224460-HA on parallel sister chromatids were also visible on metaphase chromosomes. However, to allow access of antibodies to the centromeric region for detection of TVAG_224460-HA in chromatids, the chromosomes were treated with SDS, which slightly affected their morphologies. In particular, chromosome constrictions were not clearly visible under these conditions; thus, the exact position of TVAG_224460-HA was not resolved. Apart from centromere-like staining, some signals for TVAG_224460-HA were observed in noncentromeric chromatin, indicating artifacts of CenH3 overexpression. The mammalian CenH3 homolog CENP-A was incorporated into chromatin of chromosome arms upon overexpression (33). Alternatively, the deposition of T. vaginalis CenH3 outside the centromeres is connected with its putative role in DNA repair, as previously suggested for CENP-A (41).
Our analysis of the pattern of CenH3 deposition into T. vaginalis chromatin complements earlier studies focusing on the T. vaginalis karyotype. The presence of distinct CenH3 foci corresponding to the number of chromosomes supports the view that trichomonad chromosomes are monocentric with point centromeres rather than holocentric with diffuse kinetochores spread along the entire length of chromatids (13). In holocentric chromosomes, CenH3 staining localizes as a band on the edge of each sister chromatid facing toward spindle microtubules (10, 23); we did not observe such a pattern in T. vaginalis. The presence of primary constrictions representing centromeres indicates the monocentric nature of chromosomes in T. vaginalis; furthermore, these structures are absent in holocentric chromosomes (1, 16, 21). A clearly visible constriction is found on four out of six chromosomes (chromosomes I, III, IV, V) during mitotic metaphase in T. vaginalis (13). Studies based on fluorescence in situ hybridization showed that the site of constriction of chromosome IV represents the site of a ribosomal DNA gene cluster, and thus, it is a secondary constriction (38, 43). Because all rRNAs are encoded on chromosome IV, the subtelomeric constrictions on chromosomes I, III, and V must be primary constrictions within the centromeric regions.
Conclusion.
In this study, we identified three histone variants in T. vaginalis: H3T, which is distributed as core H3; an H3.3 variant that marks the sites of active transcription; and CenH3, which localizes to the centromeres. The protein sequence of HFD of T. vaginalis CenH3 is different from the sequences of experimentally validated centromeric histones in other species: similar to the canonical H3, T. vaginalis CenH3 retains a conserved glutamine in the α1 helix. We can only speculate whether it has any consequences with respect to DNA binding or interaction with other proteins. It is possible that this structural feature is involved in the specific mitosis of trichomonads. Mitosis occurs in a unique manner in trichomonads; during its course the nuclear envelope does not break down, and the mitotic spindle is extranuclear and does not penetrate inside the nucleus. Having a marker for centromeres provides us with a tool to elucidate whether the centromeres interact with the nuclear membrane and to study the possible involvement of nuclear pores in the attachment of chromosomes to spindle microtubules. Because several other members of the kinetochore complex were annotated in the T. vaginalis genome (e.g., CENP-B or CENP-C homologs), it would be interesting to define their roles in the formation of the trichomonad centromere/kinetochore complex during mitosis.
Supplementary Material
ACKNOWLEDGMENT
This study was supported by the Czech Ministry of Education (MSM 0021620858, LC07032).
Footnotes
Published ahead of print 9 March 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Ahmad K, Henikoff S. 2002. Histone H3 variants specify modes of chromatin assembly. Proc. Natl. Acad. Sci. U. S. A. 99:16477–16484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brooks CF, et al. 2011. Toxoplasma gondii sequesters centromeres to a specific nuclear region throughout the cell cycle. Proc. Natl. Acad. Sci. U. S. A. 108:3767–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brugerolle G. 1975. Étude de la cryptopleuromitose et de la morphogenèse de division chez Trichomonas vaginalis et chez plusiers de trichomonadines primitives. Protistologica 11:457–468 [Google Scholar]
- 4. Carlton JM, et al. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cepicka I, Hampl V, Kulda J. 2010. Critical taxonomic revision of Parabasalids with description of one new genus and three new species. Protist 161:400–433 [DOI] [PubMed] [Google Scholar]
- 6. Cervantes MD, Xi X, Vermaak D, Yao MC, Malik HS. 2006. The CNA1 histone of the ciliate Tetrahymena thermophila is essential for chromosome segregation in the germline micronucleus. Mol. Biol. Cell 17:485–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cui B, Liu Y, Gorovsky MA. 2006. Deposition and function of histone H3 variants in Tetrahymena thermophila. Mol. Cell. Biol. 26:7719–7730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dalal Y, Furuyama T, Vermaak D, Henikoff S. 2007. Structure, dynamics, and evolution of centromeric nucleosomes. Proc. Natl. Acad. Sci. U. S. A. 104:15974–15981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dawson SC, Sagolla MS, Cande WZ. 2007. The cenH3 histone variant defines centromeres in Giardia intestinalis. Chromosoma 116:175–184 [DOI] [PubMed] [Google Scholar]
- 10. Dernburg AF. 2001. Here, there, and everywhere: kinetochore function on holocentric chromosomes. J. Cell Biol. 153:F33–F38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diamond LS. 1957. The establishment of various trichomonads of animals and man in axenic cultures. J. Parasitol. 43:488–490 [PubMed] [Google Scholar]
- 12. Dolezal P, et al. 2005. Giardia mitosomes and trichomonad hydrogenosomes share a common mode of protein targeting. Proc. Natl. Acad. Sci. U. S. A. 102:10924–10929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drmota T, Král J. 1997. Karyotype of Trichomonas vaginalis. Eur. J. Protistol. 33:131–135 [Google Scholar]
- 14. Fichorova RN. 2009. Impact of T. vaginalis infection on innate immune responses and reproductive outcome. J. Reprod. Immunol. 83:185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fisk JC, Read LK. 2011. Protein arginine methylation in parasitic protozoa. Eukaryot. Cell 10:1013–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guerra M, et al. 2010. Neocentrics and holokinetics (holocentrics): chromosomes out of the centromeric rules. Cytogenet. Genome Res. 129:82–96 [DOI] [PubMed] [Google Scholar]
- 17. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. (Oxf.) 41:95–98 [Google Scholar]
- 18. Heath IB. 1980. Variant mitoses in lower eukaryotes—indicators of the evolution of mitosis. Int. Rev. Cytol. 64:1–80 [DOI] [PubMed] [Google Scholar]
- 19. Henikoff S, Ahmad K. 2005. Assembly of variant histones into chromatin. Annu. Rev. Cell Dev. Biol. 21:133–153 [DOI] [PubMed] [Google Scholar]
- 20. Lowell JE, Cross GA. 2004. A variant histone H3 is enriched at telomeres in Trypanosoma brucei. J. Cell Sci. 117:5937–5947 [DOI] [PubMed] [Google Scholar]
- 21. Maddox PS, Oegema K, Desai A, Cheeseman IM. 2004. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 12:641–653 [DOI] [PubMed] [Google Scholar]
- 22. Marinets A, et al. 1996. The sequence and organization of the core histone H3 and H4 genes in the early branching amitochondriate protist Trichomonas vaginalis. J. Mol. Evol. 43:563–571 [DOI] [PubMed] [Google Scholar]
- 23. Nagaki K, Kashihara K, Murata M. 2005. Visualization of diffuse centromeres with centromere-specific histone H3 in the holocentric plant Luzula nivea. Plant Cell 17:1886–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Postberg J, Forcob S, Chang WJ, Lipps HJ. 2010. The evolutionary history of histone H3 suggests a deep eukaryotic root of chromatin modifying mechanisms. BMC Evol. Biol. 10:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raikov IB. 1994. The diversity of forms of mitosis in protozoa—a comparative review. Eur. J. Protistol. 30:253–269 [Google Scholar]
- 26. Ribeiro KC, Monteiro-Leal LH, Benchimol M. 2000. Contributions of the axostyle and flagella to closed mitosis in the protists Tritrichomonas foetus and Trichomonas vaginalis. J. Eukaryot. Microbiol. 47:481–492 [DOI] [PubMed] [Google Scholar]
- 27. Ribeiro KC, Pereira-Neves A, Benchimol M. 2002. The mitotic spindle and associated membranes in the closed mitosis of trichomonads. Biol. Cell 94:157–172 [DOI] [PubMed] [Google Scholar]
- 28. Riley DE, Krieger JN, Miner D, Rabinovitch PS. 1994. Trichomonas vaginalis: dominant G2 period and G2 phase arrest in a representative of an early branching eukaryotic lineage. J. Eukaryot. Microbiol. 41:408–414 [DOI] [PubMed] [Google Scholar]
- 29. Santaguida S, Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schwebke JR, Burgess D. 2004. Trichomoniasis. Clin. Microbiol. Rev. 17:794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sekulic N, Bassett EA, Rogers DJ, Black BE. 2010. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature 467:347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sullivan KF, Hechenberger M, Masri K. 1994. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 127:581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sutak R, et al. 2004. Mitochondrial-type assembly of FeS centers in the hydrogenosomes of the amitochondriate eukaryote Trichomonas vaginalis. Proc. Natl. Acad. Sci. U. S. A. 101:10368–10373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Szenker E, Ray-Gallet D, Almouzni G. 2011. The double face of the histone variant H3.3. Cell Res. 21:421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Talbert PB, Henikoff S. 2010. Histone variants—ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11:264–275 [DOI] [PubMed] [Google Scholar]
- 37. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Torres-Machorro AL, Hernandez R, Alderete JF, Lopez-Villasenor I. 2009. Comparative analyses among the Trichomonas vaginalis, Trichomonas tenax, and Tritrichomonas foetus 5S ribosomal RNA genes. Curr. Genet. 55:199–210 [DOI] [PubMed] [Google Scholar]
- 39. Turner BM. 2002. Cellular memory and the histone code. Cell 111:285–291 [DOI] [PubMed] [Google Scholar]
- 40. Wood CM, et al. 2005. High-resolution structure of the native histone octamer. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61:541–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zeitlin SG, et al. 2009. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc. Natl. Acad. Sci. U. S. A. 106:15762–15767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zubacova Z, Cimburek Z, Tachezy J. 2008. Comparative analysis of trichomonad genome sizes and karyotypes. Mol. Biochem. Parasitol. 161:49–54 [DOI] [PubMed] [Google Scholar]
- 43. Zubacova Z, Krylov V, Tachezy J. 2011. Fluorescence in situ hybridization (FISH) mapping of single copy genes on Trichomonas vaginalis chromosomes. Mol. Biochem. Parasitol. 176:135–137 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.