Abstract
Sexual reproduction is essential for the maintenance of species in a wide variety of multicellular organisms, and even unicellular organisms that normally proliferate asexually possess a sexual cycle because of its contribution to increased genetic diversity. Information concerning the molecules involved in fertilization is accumulating for many species of the metazoan, plant, and fungal lineages, and the evolutionary consideration of sexual reproduction systems is now an interesting issue. Macrocyst formation in the social amoeba Dictyostelium discoideum is a sexual process in which cells become sexually mature under dark and submerged conditions and fuse with complementary mating-type cells. In the present study, we isolated D. discoideum insertional mutants defective in sexual cell fusion and identified the relevant gene, macA, which encodes a highly glycosylated, 2,041-amino-acid membrane protein (MacA). Although its overall similarity is restricted to proteins of unknown function within dictyostelids, it contains LamGL and discoidin domains, which are implicated in cell adhesion. The growth and development of macA-null mutants were indistinguishable from those of the parental strain. The overexpression of macA using the V18 promoter in a macA-null mutant completely restored its sexual defects. Although the macA gene encoded exactly the same protein in a complementary mating-type strain, it was expressed at a much lower level. These results suggest that MacA is indispensable for gamete interactions in D. discoideum, probably via cell adhesion. There is a possibility that it is controlled in a mating-type-dependent manner.
INTRODUCTION
Fertilization is an essential aspect of sexual reproduction, both to generate a new individual and to trigger its multicellular development. Although its molecular mechanisms have long been hidden, recent studies are successively uncovering the molecules involved in fertilization in a variety of organisms. For example, the requirement of CD9 (18, 20) on the egg and Izumo protein on the sperm surface (12) in mammalian sperm-egg fusions has been demonstrated, as well as many other proteins involved in sperm-egg coat interactions (11). Generative lineage-specific proteins, Hap2/GCS1, have been found to be indispensable for fertilization in Arabidopsis (21, 40) and other plants (41), as well as in malaria parasites (10). In the nematode, the products of spe and egg series genes are necessary for fertilization (33). Mechanisms for allorecognition or self-non-self discrimination have also been clarified in ascidians (8) and self-incompatible plants (28). Thus, it is now interesting to consider how the molecular mechanisms for sexual reproduction have evolved among the phyla.
The cellular slime mold Dictyostelium discoideum occupies a unique position in phylogeny, branching from the phylogenic trunk after plants but before the fungal lineages (4). D. discoideum cells proliferate by fission as unicellular amoebas, but they gather and initiate multicellular development upon starvation (15). However, under dark and submerged conditions, the formation of water-sensitive spores is not advantageous. Instead, the amoebas mature as gametes and fuse with appropriate mating-type cells to form zygote giant cells, which eventually develop into dormant structures called macrocysts. There are homothallic and heterothallic strains of D. discoideum (6, 29), and three mating types, types I, II, and III, are assigned to the latter group (37).
The molecular mechanisms of macrocyst formation are largely elusive (39). A previous study showed that a 138-kDa glycoprotein, gp138, was a target molecule of cell fusion-blocking antibodies (35). Although a member of the cell fusion-related (cfr; formerly GP138) gene family was suggested to encode it (1, 7, 43), the deletion of all 4 cfr genes in the KAX3 genome did not result in the complete disappearance of the gp138 protein (9). It also did not affect the sexual potency of the cells, leaving the actual gene for gp138 unknown. Genes expressed specifically in the gamete stage were screened by constructing a gamete-enriched gene pool (23). Although certain genes involved in the signal transduction pathway, such as ras and rac, were highly elevated in the gametes, their functions are unknown. The disruption of multiple gamete-enriched genes suppressed sexual potency (24), but only to a limited extent. Recently, a chromosomal region for mating-type determination has been identified (2); however, matA, a protein-coding gene in the mating-type locus of a type I strain, is predicted to be cytoplasmic and unlikely to function directly in gamete interactions.
In the present study, we generated sexually defective insertional mutants and identified the responsible gene. Its product was predicted to be expressed on the cell surface and possibly function in adhesion between gametes.
MATERIALS AND METHODS
Strains and culture conditions.
A clonal line of KAX3, a heterothallic type I strain of D. discoideum with high sexual potency and high blasticidin S sensitivity, was used throughout the experiments. Strains V12, WS2162, and AC4 represent type II, type III, and homothallic strains, respectively (29). All of the strains were maintained as fruiting bodies on nutrient SM agar plates with Klebsiella aerogenes as a food source. KAX3 and its transformants were also grown in HL5 medium containing 50 μg/ml of streptomycin sulfate.
Assay for sexual potency.
The ability to form macrocysts and zygote giant cells was determined using standard procedures (37). Growth-phase cells cultured in a shaken suspension with condensed K. aerogenes in darkness for 16 h at 22°C are normally ready for fusion with appropriate mating-type cells and are called fusion-competent (FC) cells. Cells cultured in the same way but in the light or on agar plates with bacteria are fusion incompetent; the former cells are designated ICL cells and the latter ICP cells.
Transformation of D. discoideum cells.
The transformation of KAX3 and its derivatives was performed by the standard electroporation procedure (16) using Transfector 800 (BTX, San Diego, CA). The selection of transformants was carried out in HL5 containing either 10 μg/ml of blasticidin S or 10 μg/ml of G418 depending on the transformation constructs used.
Isolation of insertional mutants.
The insertional mutagenesis was carried out by the method of restriction enzyme-mediated integration (REMI) (17). For this purpose, 50 μg/ml of pUCBsrΔBam (pBSR) (22) linearized with BamHI and 100 U/ml of BamHI were used to transform KAX3 cells. The transformants were selected in 10 μg/ml of blasticidin S (Funakoshi, Tokyo, Japan) for 7 days. The resultant colonies were harvested and plaque cloned on SM agar plates containing 20 μg/ml blasticidin S. The clonal transformants thus obtained were screened for defects in sexual development using the macrocyst assay, as described previously (32).
Reverse transcription-PCR (RT-PCR).
Total RNA was isolated using an RNeasy minikit (Qiagen, Düsseldorf, Germany) according to the manufacturer's instructions. This was followed by the removal of contaminating genomic DNA using an RNase-free DNase set (Qiagen, Düsseldorf, Germany). First-strand cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen Corporation). The concentration of the template was adjusted by amplification using primers for Ig7 (rnlA) (accession no. U21880). For quantification, real-time PCR was performed using an ABI 7900HT sequence detection system (Applied Biosystems). The amplifications were carried out using SYBR premix Ex Taq (Takara, Otsu, Japan). The primer sequences used in the present study are shown in Table S1 in the supplemental material.
Proteome analysis.
The crude membrane fraction was obtained by the repeated freeze-thawing of the cells (34) and dissolved in the sample loading buffer for SDS-PAGE. Proteins were separated in a 10 to 20% gradient gel, size fractionated by horizontally slicing the gel, and then digested with trypsin for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis as described previously (42). The digested peptides were analyzed using a capillary liquid chromatography system (Ultima3000; Dionex, CA) connected online to a mass spectrometer (LTQ-XL; Thermo Scientific, MA). Raw spectral data were processed using SEQUEST software to extract peak lists. The obtained peak lists were analyzed using the MASCOT program against the D. discoideum protein database extracted from dictyBase (http://dictybase.org) and supplemented with B and MatC (mat locus gene products in type II) and D, S, and MatT (mat locus gene products in type III) sequences (2).
Nucleotide sequence accession number.
The nucleotide sequence of macA in strain AC4 was deposited in GenBank under accession no. AB698458.1.
RESULTS
Sexually defective mutants and a new class of gamete fusion gene.
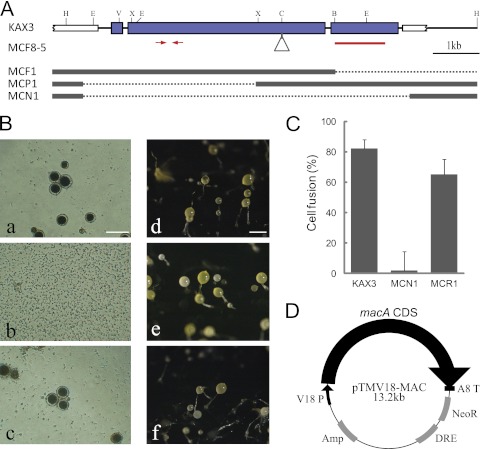
One BamHI-REMI mutant unable to produce macrocysts with V12 was obtained out of 7,000 independent transformants, and it was named macrocyst-free mutant 1 (MCF1). Since MCF1 appeared to have a large deletion downstream of the vector insertion site, the rescued plasmid harboring the upstream flanking region of the MCF1 genome was introduced into KAX3 to generate secondary mutants. Among the multiple transformants with the same sexual defects as MCF1, one clone, MCF8-5, was selected for further use. The vector insertion site of MCF8-5 was determined to be at the ClaI site in the middle of a long open reading frame on chromosome 5. The coding sequence extended in both directions with short interrupting introns, as confirmed by sequencing the RT-PCR products from FC cell RNA (Fig. 1A). This gene was named macA (accession no. AB259764.1), as it was responsible for macrocyst formation.
Fig 1.
Generation of sexually defective mutants. (A) The genomic structure of the macA region for the parental KAX3 strain is shown on top. The thick blue and white bars indicate the exon of macA and neighboring genes, respectively. The triangle shows the vector insertion site in MCF8-5. For the other 3 mutants, substitutions to the vector sequences are shown by dotted lines. Red bars and arrows indicate the region used for the hybridization probe and the quantitative RT-PCR primers, respectively. Restriction sites are the following: B, BamHI; C, ClaI; E, EcoRI; H, HpaI; V, EcoRV; and X, XbaI. (B) Macrocyst (a, b, and c) and fruiting body formation (d, e, and f) of parental KAX3 (a and d), the macA-null mutant MCN1 (b and e), and the macA overexpression mutant in MCN1, MCR1 (c and f). For macrocyst formation, 100 cells were mixed with an equal number of V12 cells in 100 μl of BSS containing condensed K. aerogenes in a well of a 96-well plate and cultured for 4 days at 22°C. Bar, 100 μm. (C) Results of the cell fusion assays are shown. (D) Structure of the macA overexpression vector. The coding sequence of macA was inserted between the V18 promoter and the act8 terminator of pTMV18.
When 5′ partial (MCP1) and complete deletion mutants of macA (MCN1) were generated by homologous recombination (Fig. 1A), none were able to form macrocysts either with V12 (type II) (Fig. 1B) or WS2162 (type III) (data not shown). The deletion mutants were also defective in cell fusion with fusion-competent V12 cells, indicating that macA is required for cell fusion (Fig. 1C). To further confirm the relevance of macA to the mutant phenotype, the macA coding sequence was cloned in the expression vector pTMV18 (Fig. 1D) and introduced into MCN1. The recipient clone (MCR1) recovered from the sexual defects (Fig. 1B and C). The macA-null mutants formed fruiting bodies that were indistinguishable from those of the parental KAX3 cells (Fig. 1B, d to f). The growth of macA-null mutants in axenic and bacterial cultures was also the same as that for KAX3 (data not shown). It is not known whether macA is also necessary for the postfusion processes of macrocyst formation.
Structural analysis of the macA product.
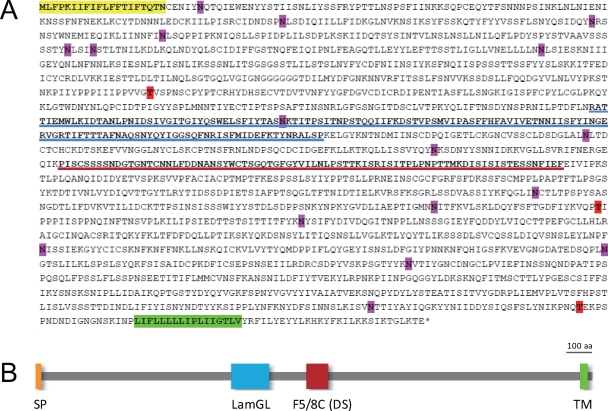
The deduced amino acid sequence of the macA product, MacA, was 2,041 amino acids long and estimated to have a molecular mass of ∼229 kDa. It possesses a signal peptide for membrane localization and a transmembrane region at its N and C termini, respectively, and was predicted to be a plasma membrane protein by two prediction programs, PSORT (25) (http://psort.hgc.jp/) and SOSUI (19) (http://bp.nuap.nagoya-u.ac.jp/sosui/). A very short cytoplasmic region was predicted (Fig. 2A and B). Although the overall sequence similarity to any nondictyostelid proteins was not significantly high, MacA contained two conserved protein domains related to cell adhesion, the LamGL (laminin G-like or ConA-like/glucanase subgroup) domain and the F5/8A (coagulation factor 5/8 type, C-terminal) domain, in its central region (Fig. 2B and C). The latter domain is also called a discoidin domain. Accordingly, MacA is weakly homologous to the type II discoidin of D. discoideum in this region. It was predicted to have 17 N-linked and 3 O-linked glycosylation sites (Fig. 2A).
Fig 2.
Expected structure of MacA. (A) The deduced amino acid sequence is shown. The yellow and green highlights indicate the signal peptide (SP) and transmembrane (TM) sequences, respectively. Possible N-glycosylation (pink) and O-glycosylation (red) sites are also highlighted. Blue and red underlines represent LamGL and F5/8C (discoidin [DS]) domains, respectively. O-glycosylation sites were predicted using the NetOGlyc 3.1 server (http://www.cbs.dtu.dk/services/netoglyc/). (B) The overall structure of MacA is shown schematically.
Expression of macA.
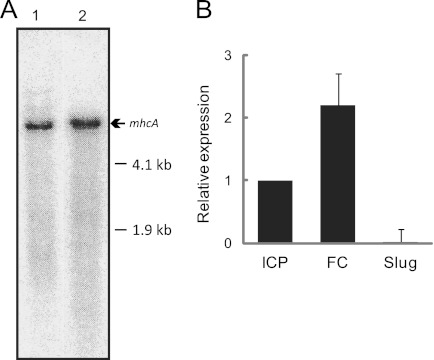
The detection of macA expression by Northern hybridization was possible only when poly(A)+ RNA (10 μg) was used, suggesting a very low level of its transcript. The macA signal was slightly lower in position than that of mhcA mRNA (6.3 kb plus untranslated regions) (Fig. 3A). According to quantitative RT-PCR, macA was approximately 2-fold enriched in FC cells compared to the level of ICP cells and was repressed in asexually developing cells at the slug stage (Fig. 3B).
Fig 3.
Detection of macA transcripts. (A) One hundred mg of poly(A)+ RNA prepared from FC cells of KAX3 was analyzed by Northern hybridization. The blot membrane was probed with the 32P-labeled macA fragment shown in Fig. 1 (lane 1) or clone SLH805 from the Tsukuba cDNA project, representing a 3′-portion of mhcA (lane 2). (B) Quantitative RT-PCR amplification was performed for RNA prepared from fusion-competent (FC) or -incompetent cells cultured on a bacterial plate (ICP) and slug-stage cells of KAX3.
For unknown reasons, we were unable to raise specific antibodies to MacA or to detect tag-fused MacA despite repeated trials. Therefore, we performed proteome analysis to confirm its translation. When the crude membrane fraction of FC and ICL cells of KAX3 were subjected to SDS-PAGE, followed by size fractionation and LC-MS/MS analysis, MacA-derived peptides were detected in a high-molecular-mass region of ∼280 kDa (see Fig. S1 in the supplemental material). The difference from the estimated molecular mass is probably due to heavy glycosylation, as suspected from Fig. 1A. The peptide spectra were obtained throughout the MacA sequences (see Table S2).
Specificity and conservation of macA.
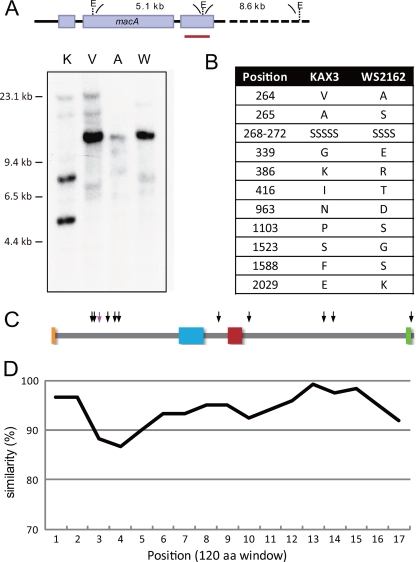
To see if macA is conserved among mating types, Southern hybridization was performed using EcoRI-digested genomic DNA from representative strains of each mating type. As shown in Fig. 4A, two bands of the expected size were obtained in KAX3. However, the other three strains gave rise to single bands that were similar in size (∼14 kbp). Since this suggested the possibility of the mating-type dependence of the macA structure, we analyzed the nucleotide sequences of macA in those strains. This was performed by PCR amplification and the cloning of 5 overlapping fragments of macA genomic regions, followed by sequencing and assembly. The primer sequences used for the PCR are listed in Table S1 in the supplemental material. The nucleotide sequence of V12 macA was different from that of KAX3 only at one position, G5403A, which happened to be the first nucleotide of the EcoRI site within the sequence used as the hybridization probe. Moreover, it was a synonymous substitution and the MacA sequence in V12 turned out to be exactly the same as that in KAX3, excluding the suspected possibility for a role of MacA in mating-type recognition. On the other hand, the macA coding sequence of the type III strain, WS2162, was different, having a single unit deletion containing a 3-nucleotide repeats and 13 nucleotide substitutions within the exons. These changes in nucleotide sequence resulted in the deletion of 1 amino acid and the alteration of 10 amino acids in the MacA sequence (Fig. 4B). Even so, these alterations did not fall within the two functional domains (Fig. 4C), nor was there any indication of extensive structural dissimilarities.
Fig 4.
Conservation of macA in other mating-type strains of D. discoideum. (A) Genomic DNA was prepared from KAX3 (K), V12 (V), WS2162 (W), and AC4 (A) cells, digested with EcoRI, and subjected to Southern hybridization using a 32P-labeled KAX3 macA fragment as a probe, which is shown by a red bar. (B) Amino acid alterations in WS2162 MacA. Due to an amino acid deletion, WS2162 MacA is 2,040 amino acids long. (C) Approximate positions of the deletion (pink arrow) and substitutions (black arrows) in WS2162 are mapped. (D) The similarity of AC4 MacA for every 120 amino acids is shown.
We also determined the nucleotide sequence of macA in AC4 (accession no. AB698458), a homothallic strain in D. discoideum. Although its sequence identity was much lower than that of heterothallic strains, the similarity of the deduced amino acid sequence to KAX3 MacA was >85% throughout. When the similarity value in a 120-bp window was plotted, the middle portion of the N-terminal region was found to be the most dissimilar (Fig. 4D). However, significant sequence homology of MacA was limited to the dictyostelid species. According to the BLASTP search results, only a short segment of a Caenorhabditis elegans hypothetical protein, C49C3.4, was weakly homologous (score, 109; E value, 0.097).
Cell type-dependent expression of macA.
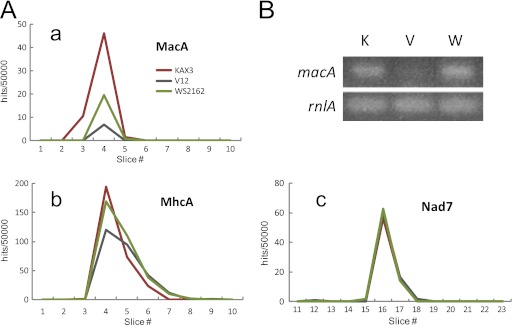
Despite the high sequence identity of MacA among the heterothallic strains of D. discoideum, which favors its involvement in membrane fusion rather than cell type recognition, proteome analysis revealed a significant difference in the detection level of MacA-derived peptides; it was approximately 11% of KAX3 in V12 and 33% in WS2162 (Fig. 5A; also see Table S2 in the supplemental material). It should be noted that even when the WS2162 MacA sequence was added to the database for peptide searching, no changes were observed. To rule out the possibility that this was caused by sample processing, we analyzed the mass spectrometry profiles of the NADH dehydrogenase subunit 7 (Nad7), which was unlikely to be mating type dependent. As shown in Fig. 5A, panel c, Nad7-derived peptides were detected at exactly the same level in the three strains. For the second example, myosin II heavy chain (MhcA) was examined, because its molecular mass was similar to that of MacA. Again, the MhcA-derived peptides were detected at nearly equal levels in the three strains; detection in V12 was 93% of that of KAX3, and that in WS2162 was 113%(Fig. 5A, panel b).
Fig 5.
Differential detection of macA expression with mating type. (A) The peptide hits from proteome analysis were plotted for MacA (a), Nad7 (b), and MhcA (c). (B) A portion of macA cDNA was amplified for FC cells of KAX3 (K), V12 (V), and WS2162 (W) using the primer set shown in Table S1 in the supplemental material.
Considering that the mass spectrometry analysis might not directly correspond to the actual protein level but rather could be affected by differential protein modifications, we compared the mRNA levels in those strains using RT-PCR. As is evident from Fig. 5B, the transcription level of macA was very low in V12. On the other hand, its level in WS2162 was similar to that of KAX3. Thus, macA expression seems to be regulated at the transcriptional and/or translational steps in a cell type-dependent manner.
DISCUSSION
In the present study, we generated sexually defective insertional mutants of D. discoideum strain KAX3 (mating type I) and identified the relevant gene, macA, which was indispensable for gamete interactions in this organism. Its product, the MacA protein, is a highly glycosylated cell surface protein with a molecular mass of ∼280 kDa that possesses two adhesion-related domains, the LamGL and discoidin domains. This is the first demonstration of a gene that is responsible for cell fusion in the sexual process of D. discoideum.
The actual molecular function of MacA in cell fusion is currently unknown. Its predicted localization to the plasma membrane suggests that it is involved in gamete interactions. Since the MacA sequences of KAX3 and V12 are exactly the same, it is less likely that MacA functions in mating-type recognition. It may function as a cell adhesion protein via the cell adhesion-related domains located in the central region of the protein. Greater variation in the N-terminal region of MacA might be involved in species recognition. Earlier studies on cell fusion mechanisms demonstrated the requirement for Ca2+ (3, 30) and the involvement of cell surface carbohydrates (13, 26). These findings coincide with the presence of a LamGL domain in MacA, which is known to be related to Ca2+-dependent cell adhesion (36) and to be heavily glycosylated. The discoidin domain protein reportedly participates in many adhesion-related processes in metazoan organisms (27, 31). It is noteworthy that porcine SED1/MFG-E8, a discoidin domain-containing protein originally called p47, is a sperm receptor for the zona pellucida (5). We suspect that MacA has a role in the establishment of sex-specific cell adhesion, which is possible after mating-type recognition and is a prerequisite for membrane fusion. The necessity for specific adhesion has been reported in mammalian fertilization. For example, CD9 on the mouse egg membrane generates the strong adhesion to sperm that is necessary for cell fusion (14).
Although the expression levels of the macA transcript were very low, the large size of MacA was an advantage for proteome analysis, so that appreciable numbers of peptide hits were obtained. The slightly higher number of hits in ICL cells than in FC cells (Fig. 4B) was apparently contradictory to the 2-fold abundance of mRNA in FC cells (Fig. 3B). One explanation for this is that peptide detection is sensitive to protein modifications that can cause charge and/or mass alterations of individual peptide fragments. This may also explain why peptides 5, 6, and 28 were not detected in FC cells despite having approximately 5 hits or more out of 5,000 normalized total hits in ICL cells. This observation suggests the occurrence of FC cell-specific modifications of MacA in the corresponding regions, although other explanations are equally possible. Differences in the structure and expression of macA among mating types may have some role, but the solution awaits further studies.
If MacA functions as a cell adhesion protein, there should be another component(s) responsible for membrane fusion itself as well as interactions with MacA. In light of this, we considered it interesting to see if macA is altered in the previously generated fusion-defective mutants XMC7 and XMC17 (38). These were generated by chemical mutagenesis, and the responsible genes have not yet been identified. We found that both the nucleotide sequence and expression level of macA were the same as those of KAX3, suggesting that other genes exist that are responsible for gamete fusion in type I strains of D. discoideum. It is worth pointing out that a homolog of Hap2/GCS-1 exists in the sequenced AX4 genome (4), which makes it a good candidate as another cell fusion protein in D. discoideum. Moreover, the relationship between MacA and MatA proteins should be clarified. Since MatA is predicted to be cytoplasmic, it most likely has a role in the intracellular regulatory or signaling cascades of the sexual process.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Miho Iijima, Shinichi Kawakami, and Akiko Ohtsuka for their assistance in some of the experiments described.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to H. Urushihara (06454683 and 22112502) and to L. Yamada (22112511).
Footnotes
Published ahead of print 2 March 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Aiba K, Fang H, Yamaguchi N, Tanaka Y, Urushihara H. 1997. Isoforms of gp138, a cell-fusion related protein in Dictyostelium discoideum. J. Biol. 121:238–243 [DOI] [PubMed] [Google Scholar]
- 2. Bloomfield G, Skelton J, Ivens A, Tanaka Y, Kay RR. 2010. Sex determination in the social amoeba Dictyostelium discoideum. Science 330:1533–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chagla AH, Lewis KE, O'Day DH. 1980. Ca2+ and cell fusion during sexual development in liquid cultures of Dictyostelium discoideum. Exp. Cell Res. 126:501–505 [DOI] [PubMed] [Google Scholar]
- 4. Eichinger L, et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435:43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ensslin MA, Shur BD. 2003. Identification of mouse sperm SED1, a bimotif EGF repeat and discoidin-domain protein involved in sperm-egg binding. Cell 114:405–417 [DOI] [PubMed] [Google Scholar]
- 6. Erdos GW, Raper KB, Vogen LK. 1975. Mating types and macrocyst formation in Dictyostelium discoideum. Proc. Natl. Acad. Sci. U. S. A. 72:970–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fang H, Higa M, Suzuki K, Urushihara H, Yanagisawa K. 1993. Molecular cloning and analysis of the gene encoding gp138, a cell surface glycoprotein involved in the sexual cell fusion of Dictyostelium discoideum. Dev. Biol. 156:201–208 [DOI] [PubMed] [Google Scholar]
- 8. Harada Y, Sawada H. 2008. Allorecognition mechanisms during ascidian fertilization. Int. J. Dev. Biol. 52:637–645 [DOI] [PubMed] [Google Scholar]
- 9. Hata T, Takahashi M, Tanaka Y, Urushihara H. 2001. Total tetra knockout of GP138 multigene family implicated in cell interactions in Dictyostelium discoideum. Gene 271:33–42 [DOI] [PubMed] [Google Scholar]
- 10. Hirai M, et al. 2008. Male fertility of malaria parasites is determined by GCS1, a plant-type reproduction factor. Curr. Biol. 18:607–613 [DOI] [PubMed] [Google Scholar]
- 11. Ikawa M, Inoue N, Okabe M. 2008. Mechanisms of sperm-egg interactions emerging from gene-manipulated animals. Int. J. Dev. Biol. 52:657–664 [DOI] [PubMed] [Google Scholar]
- 12. Inoue N, Ikawa M, Isotani A, Okabe M. 2005. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 434:234–238 [DOI] [PubMed] [Google Scholar]
- 13. Ishikawa T, Urushihara H, Yanagisawa K. 1991. Involvement of cell surface carbohydrates in the sexual cell fusion of Dictyostelium discoideum. Develop. Growth Differ. 33:131–138 [DOI] [PubMed] [Google Scholar]
- 14. Jégou A, Ziyyat , et al. 2011. CD9 tetraspanin generates fusion competent sites on the egg membrane for mammalian fertilization. Proc. Natl. Acad. Sci. U. S. A. 108:10946–10951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kessin RH. 2001. Dictyostelium–evolution, cell biology, and the development of multicellularity. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 16. Knecht D, Pang KM. 1995. Electroporation of Dictyostelium discoideum. Methods Mol. Biol. 47:321–330 [DOI] [PubMed] [Google Scholar]
- 17. Kuspa A, Loomis WF. 1992. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc. Natl. Acad. Sci. U. S. A. 89:8803–8807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C. 2000. Severely reduced female fertility in CD9-deficient mice. Science 287:319–321 [DOI] [PubMed] [Google Scholar]
- 19. Mitaku S, Hirokawa T, Tsuji T. 2002. Amphiphilicity index of polar amino acids as an aid in the characterization of amino acid preference at membrane-water interfaces. Bioinformatics 18:608–616 [DOI] [PubMed] [Google Scholar]
- 20. Miyado K, et al. 2000. Requirement of CD9 on the egg plasma membrane for fertilization. Science 287:321–324 [DOI] [PubMed] [Google Scholar]
- 21. Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T. 2006. Generative cell specific 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8:64–71 [DOI] [PubMed] [Google Scholar]
- 22. Morio T, Adachi H, Sutoh K, Yanagisawa K, Tanaka Y. 1995. Bsr-REMI: an improved method for gene tagging using a new vector in Dictyostelium. J. Plant Res. 108:111–114 [Google Scholar]
- 23. Muramoto T, et al. 2003. Construction of a gamete-enriched gene pool and RNAi-mediated functional analysis in Dictyostelium discoideum. Mech. Dev. 120:965–975 [DOI] [PubMed] [Google Scholar]
- 24. Muramoto T, Takeda S, Furuya Y, Urushihara H. 2005. Reverse genetic analyses of gamete-enriched genes revealed a novel regulator of the cAMP signaling pathway in Dictyostelium discoideum. Mech. Dev. 122:733–743 [DOI] [PubMed] [Google Scholar]
- 25. Nakai K. 1991. Predicting various targeting signals in amino acid sequences. Bull. Inst. Chem. Res. Kyoto Univ. 69:269–291 [Google Scholar]
- 26. O'Day DH, Rivera J. 1987. Lectin binding and inhibition studies reveal the importance of D-glucose, D-mannose and N-acelylglucosamine during early sexual development of Dictyostelium discoideum. Cell Differ. 20:231–237 [DOI] [PubMed] [Google Scholar]
- 27. Raymond A, Ensslin MA, Shur BD. 2009. SED1/MFG-E8: a bi-motif protein that orchestrates diverse cellular interactions. J. Cell Biochem. 106:957–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rea AC, Nasrallah JB. 2008. Self-incompatibility systems: barriers to self-fertilization in flowering plants. Int. J. Dev. Biol. 52:627–636 [DOI] [PubMed] [Google Scholar]
- 29. Robson GE, Williams KL. 1980. The mating system of the cellular slime mould Dictyostelium discoideum. Curr. Genet. 1:229–232 [DOI] [PubMed] [Google Scholar]
- 30. Saga Y, Okada H, Yanagisawa K. 1983. Macrocyst development in Dictyostelium discoideum. II. Mating-type-specific cell fusion and acquisition of fusion-competence. J. Cell Sci. 60:157–168 [DOI] [PubMed] [Google Scholar]
- 31. Schmidt S, Friedl P. 2010. Interstitial cell migration: integrin-dependent and alternative adhesion mechanism. Cell Tissue Res. 339:83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shimizu H, Morio T, Shimizu HD, Urushihara H. 1997. A mutation in the cAMP signaling pathway affects sexual development of Dictyostelium discoideum. Develop. Growth Differ. 39:227–234 [DOI] [PubMed] [Google Scholar]
- 33. Singson A, Hang JS, Parry JM. 2008. Genes required for the common miracle of fertilization in Caenorhabditis elegans. Int. J. Dev. Biol. 52:647–656 [DOI] [PubMed] [Google Scholar]
- 34. Sussman M, Boschwitz C. 1975. Adhesive properties of cell ghosts derived from Dictyostelium discoideum. Dev. Biol. 44:362–368 [DOI] [PubMed] [Google Scholar]
- 35. Suzuki K, Yanagisawa K. 1989. Identification of the cell surface molecule involved in sexual cell fusion of Dictyostelium discoideum. Differentiation 40:159–165 [DOI] [PubMed] [Google Scholar]
- 36. Tzu J, Marinkovich MP. 2008. Bridging structure with function: structural, regulatory, and developmental role of laminins. Int. J. Biochem. Cell Biol. 40:199–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Urushihara H. 2006. Cultivation and spore production, and mating. Methods Mol. Biol. 346:113–124 [DOI] [PubMed] [Google Scholar]
- 38. Urushihara H, Aiba K, Yanagisawa K. 1991. Isolation and characterization of Dictyostelium mutants defective in sexual cell fusion. Dev. Growth Differ. 33:517–524 [DOI] [PubMed] [Google Scholar]
- 39. Urushihara H, Muramoto T. 2006. Genes involved in Dictyostelium discoideum sexual reproduction. Eur. J. Cell Biol. 85:961–968 [DOI] [PubMed] [Google Scholar]
- 40. von Besser K, Frank AC, Johnson MA, Preuss D. 2006. Arabidopsis HAP2/GCS1 is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133:4761–4769 [DOI] [PubMed] [Google Scholar]
- 41. Wong JL, Johnson MA. 2010. Is HAP2-GCS1 an ancestral gamete fusogen? Trends Cell Biol. 20:134–141 [DOI] [PubMed] [Google Scholar]
- 42. Yamada L, Saito T, Taniguchi H, Sawada H, Harada Y. 2009. Comprehensive egg coat proteome of the ascidian Ciona intestinalis reveals gamete recognition molecules involved in self-sterility. J. Biol. Chem. 284:9402–9410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamaguchi N, et al. 1996. Targeted disruption of genes for gp138, a cell-fusion-related protein in Dictyostelium discoideum, revealed the existence of a third gene. Develop. Growth Differ. 38:271–279 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.