Abstract
β-Arrestin proteins are critical for G-protein-coupled receptor desensitization and turnover. However, β-arrestins have recently been shown to play direct roles in nonheterotrimeric G-protein signal transduction. The Candida albicans β-arrestin-like protein Rim8 is required for activation of the Rim101 pH-sensing pathway and for pathogenesis. We have found that C. albicans Rim8 is posttranslationally modified by phosphorylation and specific phosphorylation states are associated with activation of the pH-sensing pathway. Rim8 associated with both the receptor Rim21 and the transcription factor Rim101, suggesting that Rim8 bridges the signaling and activation steps of the pathway. Finally, upon activation of the Rim101 transcription factor, C. albicans Rim8 was transcriptionally repressed and Rim8 protein levels were rapidly reduced. Our studies suggest that Rim8 is taken up into multivesicular bodies and degraded within the vacuole. In total, our results reveal a novel mechanism for tightly regulating the activity of a signal transduction pathway. Although the role of β-arrestin proteins in mammalian signal transduction pathways has been demonstrated, relatively little is known about how β-arrestins contribute to signal transduction. Our analyses provide some insights into potential roles.
INTRODUCTION
Seven-transmembrane-spanning-domain (7TM) receptors respond to a plethora of environmental conditions and are ubiquitous in the eukarya. In mammalian systems, ligand binding to 7TM receptors can promote activation of associated heterotrimeric G proteins, which transduce the environmental signal to a wide range of kinases and transcription factors. Ligand binding can also promote 7TM receptor phosphorylation via a G-protein-coupled receptor kinase (GRK), which promotes interaction with a β-arrestin protein and receptor internalization (8, 36). β-Arrestin-dependent 7TM receptor internalization was initially thought to function in desensitization by sequestering the receptor within endocytic vesicles away from additional ligands or by promoting the trafficking of the receptor to the vacuole for degradation. However, recent studies have demonstrated that β-arrestins also mediate G-protein-independent signaling from 7TM receptors (38).
While primarily studied in mammalian systems, β-arrestin-like proteins, also referred to as α-arrestins (2), have been identified in fungal systems. Saccharomyces cerevisiae encodes 10 β-arrestin-like proteins (Art1 to Art8, Art10, and Rim8). Art1 to Art6 and Art8 function in the endocytic uptake of specific transporter proteins (21, 33, 39); Rim8 functions in an environmental pH-sensing signal transduction pathway conserved throughout the fungi (14). Rim8, PalF in Aspergillus nidulans, interacts with the 7TM receptor Rim21/PalH (23, 24). Rim21/PalH is not known to associate with a heterotrimeric G protein but instead transmits extracellular information through the Rim8/PalF β-arrestin-like protein to promote activation of the Rim101/PacC transcription factor (23, 24, 32, 41).
From studies in S. cerevisiae and A. nidulans, the following model for Rim8/PalF in signaling has been developed. Neutral-alkaline pH is sensed by Rim21/PalH and Dfg16, another putative receptor located at the cell surface that is required for pH sensing (7). Upon receptor activation, the β-arrestin-like protein Rim8/PalF is ubiquitinated by the NEDD4 homolog Rsp5 (23, 24) and interacts with the endosomal sorting complex required for transport I (ESCRT-I) complex protein Vps23 (23). The ESCRT-I complex recruits the ESCRT-II complex, which recruits the ESCRT-IIIa Vps20-Snf7 heterodimer (4, 5, 27). Snf7 can then interact with Rim20/PalA, a scaffolding protein that interacts with the C-terminal inhibitory domain of Rim101/PacC, and Rim13/PalB, a protease that is predicted to proteolytically cleave the inhibitory C-terminal domain of Rim101/PacC (26, 28, 31, 57, 58). Proteolytically processed Rim101/PacC translocates into the nucleus and promotes changes in gene expression, promoting adaptation to neutral-alkaline environments.
We and others have identified homologs of the Rim101 pathway in Candida albicans, including Rim21, Dfg16, Rim8, Rim20, Rim13, and Rim101, and have shown that Rim101 activation in C. albicans requires the ESCRT pathway (7, 13, 28, 31, 42, 43, 53, 56, 58). C. albicans is a natural isolate of the human oral-pharyngeal, gastrointestinal, and urogenital tracks and primarily colonizes these mucosal surfaces as a commensal. However, in susceptible hosts, C. albicans can overgrow to cause mucosal infections as well as enter the bloodstream to cause systemic infections, which have an attributable mortality of 30 to 50% (15). As in other fungi, the C. albicans Rim101 pathway governs adaptation to neutral-alkaline environmental pH and is also required for pathogenesis in both mucosal and systemic models of infection (11, 12, 40, 50, 56, 61).
Although Rim8/PalF promotes Rim101 activation in S. cerevisiae, A. nidulans, and C. albicans, there are significant differences in Rim8 activity among these model organisms. For example, in S. cerevisiae Rim8 ubiquitination is pH independent; in A. nidulans PalF ubiquitination is pH dependent (23, 24). It is also unclear how the signaling complex affects the ESCRT machinery in order to promote Rim101 processing only at neutral-alkaline pH in any system. Here, we demonstrate that in C. albicans Rim8 is phosphorylated in response to neutral-alkaline pH and that this modification is correlated with Rim101 activation. Further, we find that Rim21, Rim8, and Rim101 coimmunoprecipitate, suggesting that Rim8 may directly bridge the receptor and transcription factor. Finally, our studies suggest that Rim8 represents a key step in negative feedback, allowing C. albicans cells to exhibit fine control of arrestin-dependent signaling.
MATERIALS AND METHODS
Media and growth conditions.
C. albicans strains were routinely grown at 30°C in YPD broth (2% Bacto peptone, 1% yeast extract, 2% dextrose). C. albicans transformants were selected on synthetic medium (0.67% yeast nitrogen base plus ammonium sulfate and without amino acids with 2% dextrose and supplemented with a dropout mix containing amino and nucleic acids except those necessary for the selection) (1).
Strains and plasmids.
All strains used in this study are listed in Table 1. The Rim8-hemagglutinin (HA) C-terminally tagged strains were generated by transformation with either the HA-URA3 and HA-HIS1 cassettes amplified in a PCR with primers 5′ Rim8-HA c-term and either 3′ Rim8-HA c-term URA or RIM8R2 (Table 2) and plasmid pMG1874 or pMG1921 as the template (18). Rim8-Myc C-terminally tagged strains were generated by transformation with the Rim8-Myc-HIS1 cassette amplified with primers RIM8FMyc and RIM8R2 and the plasmid pMG2093 as the template (18). Rim8-green fluorescent protein (GFP) C-terminally tagged strains were generated by transformation with the Rim8-GFP-HIS1 cassette amplified in a PCR with primers RIM8F1 and RIM8R2 and the plasmid pGFP-HIS1 (17). Correct integrations were identified via the PCR using primer seq4 rim8 in combination with primer 3′ HA detect, 3′myc detect, or GFP 3′detect.
Table 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| JAD001 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG rim8::ARG4/RIM8-HA::URA3 | This study |
| JAD002 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG RIM8-HA::HIS1/RIM8 dfg16::ARG4/dfg16::URA3 | This study |
| JAD003 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG rim21::ARG4/rim21::URA3-dpl200 RIM8-HA::HIS1/RIM8 | This study |
| JAD004 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG rim20::ARG4/rim20::URA3-dpl200 RIM8-HA::HIS1/RIM8 | This study |
| JAD005 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG rim13::ARG4/rim13::URA3-dpl200 RIM8-HA::HIS1/RIM8 | This study |
| JAD006 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG rim101::ARG4/rim101::URA3 RIM8-HA::HIS1/RIM8 | This study |
| JAD007 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG snf::ARG4/snf7::URA3-dpl200 RIM8-HA::HIS1/RIM8 | This study |
| JAD008 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG vps4::ARG4/vps4::dpl200 rim8::ARG4/RIM8-HA::URA3 | This study |
| JAD009 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG doa4::ARG4/doa4::dpl200 rim8::ARG4/RIM8-HA::URA3 | This study |
| JAD010 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG bro1::ARG4/bro1::dpl200 rim8::ARG4/RIM8-HA::URA3 | This study |
| JAD011 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG rim8::ARG4/RIM8-HA::URA3 HIS1::RIM101-V5::his1::hisG/RIM101 | This study |
| JAD012 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG rim20::ARG4/rim20::dpl200 RIM8/RIM8-HA::URA3 HIS1::RIM101-V5::his1::hisG/RIM101 | This study |
| JAD013 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG rim21::ARG4/rim21::dpl200 RIM8/RIM8-HA::URA3 HIS1::RIM101-V5::his1::hisG/RIM101 | This study |
| JAD014 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG rim13::ARG4/rim13::dpl200 RIM8/RIM8-HA::URA3 HIS1::RIM101-V5::his1::hisG/RIM101 | This study |
| JAD015 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG vps4::ARG4/vps4::dpl200 RIM8/RIM8-HA::URA3 HIS1::RIM101-V5::his1::hisG/RIM101 | This study |
| JAD017 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG rim21::ARG4/RIM21-HA::URA3 RIM8-Myc::HIS1 | This study |
| JAD021 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG rim21::ARG4/RIM21-HA::URA3 | This study |
| JAD023 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG vps20::ARG4/vps20::URA3-dpl200 RIM8-HA::HIS1/RIM8 | This study |
| JAD025 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG vps27::ARG4/vps27::URA3-dpl200 RIM8-HA::HIS1/RIM8 | This study |
| JAD026 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG vps28::ARG4/vps28::URA3-dpl200 RIM8-HA::HIS1/RIM8 | This study |
| JAD027 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG vps36::ARG4/vps36::URA3-dpl200 RIM8-HA::HIS1/RIM8 | This study |
| JAD028 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG mvb12::ARG4/mvb12::URA3-dpl200 RIM8-HA::HIS1/RIM8 | This study |
| JAD029 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG vps22::ARG4/vps22::URA3-dpl200 RIM8-HA::HIS1/RIM8 | This study |
| JAD030 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG hse1::ARG4/hse1::URA3-dpl200 RIM8-HA::HIS1/RIM8 | This study |
| JAD031 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG rim21::ARG4/RIM21 RIM8-Myc::HIS1/RIM8 | This study |
| JAD032 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG mck1::ARG4/mck1::URA3-dpl200 RIM8-HA::HIS1/RIM8 | This study |
| JAD034 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG mkc1::hisG/mkc1::hisG RIM8-HA::HIS1/RIM8 | This study |
| JAD081 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG rim8::ARG4/RIM8-GFP-HIS1 | This study |
| JAD083 | his1::hisG/his1::hisG arg4::hisG/arg4::hisG ura3::hisG/ura3::hisG vps4::ARG4/vps4::dpl200 RIM8/RIM8-GFP-HIS1 | This study |
| DAY1145 | ura3::λimm434/ura3::λimm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG rim101::ARG4/rim101::dpl200 rim101-281::URA3::pDDB479::RIM101/RIM101 | 55 |
Table 2.
Primers used in the study
| Primer name | Sequence |
|---|---|
| 3′ HA detect | CGCATAGTCAGGAACATCGTATGGGTA |
| 3′ Rim8-HA c-term URA | TAATTTATATGTGTGTGTTTGTGTGTATATTTTAATTTTTTATATAATATGTAATGTTTATCTAGAAGGACCACCTTTGATTG |
| 3′myc detect | TAAATCTTCTTCAGAAATTAATTTTTGTTC |
| 5′ Rim8-HA c-term | TTGAACGTCAACAATGACCGTTTGATAGTTCCCCAGGATAATAACTCGAATTCAGAGACGCGGATCCCCGGGTTAATTAA |
| MCK1 5DR | AAACTAATTTGGTTTTTTTTTTTTGCCGAGTCCTGTTAATAAACTACTACCCTAGCAATGTTTCCCAGTCACGACGTT |
| MCK1 3DR | GTAGGTAGGTAGGTCAAGTAGTTTGGGTCTTAAAACTAACTACCTTTTTTTTAATTATGGGAATTGTGAGCGGATA |
| MCK1 5detect | TCAAACCTACAAGAGATACG |
| MCK1 3detect | ATTGATTTCTAGTGTCATGG |
| ACT1-F | AGAATTGATTTGGCTGGTAGAGAC |
| ACT1-R | AGAAGATGGAGCCAAAGCAGTAAT |
| GFP 3′detect | CCTTCAAACTTGACTTCAGC |
| RIM21+1397 | CAATAGCACAGCTCGTACCG |
| RIM21F1 | CAACCACAGACGAAGAAATGTTTATGTATACTCCAGAAAAGAAGTTATATTAGATGTATCTAGTGATGAAGGTGGTGGTTCTAAAGGTGAAGAATTATT |
| RIM21FHA | CAACCACAGACGAAGAAATGTTTATGTATACTCCAGAAAAGAAGTTATATTAGATGTATCTAGTGATGAACGGATCCCCGGGTTAATTAA |
| RIM21R1 | TCCAGGGTATGAAGAAACGTGTCTTGAAATAGCCAAAAGGGGTCATGAACATATTCAAGAATAATGAGATGTTCTAGAAGGACCACCTTTGATTG |
| RIM8F1 | GCCAAATTACTTGAACGTCAACAATGACCGTTTGATAGTTCCCCAGGATAATAACTCGAATTCAGAGACGGGTGGTGGTTCTAAAGGTGAAGAATTATT |
| RIM8FMyc | TTGAACGTCAACAATGACCGTTTGATAGTTCCCCAGGATAATAACTCGAATTCAGAGACGCGGATCCCCGGGTTAATTAA |
| RIM8R2 | CAACTTTATTTAGCTCAGAATCAATAATTTATATGTGTGTGTTTGTGTGTATATTTTAATTTTTTATATAATGAATTCCGGAATATTTATGAGAAAC |
| RTRIM8-F | GGGGTGTCCGTTAGTTTCATTCC |
| RTRIM8-R | CAGCGGCACAACATTTTCGTAAG |
| seq4 rim8 | GCAGCTATAGGAAACTCGTCG |
| PHR1-F | CCGCGGGCTCAGTTTCT |
| PHR1-R | GATTTGCCACACCATTCATACAT |
Rim101-V5-tagged strains were generated by transformation of NruI-digested pDDB322 as described previously (31).
Rim21-HA C-terminally tagged strains were generated by transformation of the HA-URA3 cassette amplified in a PCR with primers RIM21FHA and RIM21R1 and pMG1874 as the template (18). Correct integration was determined via the PCR using primers RIM21+1397 and 3′HA detect.
The mck1Δ/ΔRIM8-HA strain (JAD032) was generated as follows. BWP17 was transformed with the mck1::ARG4 cassette, which was amplified in a PCR using the MCK1 5DR and MCK1 3DR (Table 2) primers and the pRS-ArgΔSpe template (53), to generate the heterozygous mutant DAY399. DAY399 was then transformed with the mck1::URA3-dpl200 cassette, which was amplified in a PCR using the MCK1 5DR and MCK1 3DR primers and the pDDB57 template (52), to generate the homozygous mutant DAY406. Correct integration was demonstrated by the PCR using primers MCK1 5detect and MCK1 3detect, which flank the site of integration. RIM8-HA was introduced into DAY406 as described above to generate JAD032.
Protein preparation and Western blot analyses.
Overnight cultures were diluted 200-fold into fresh medium 199 (M199) buffered with 150 mM HEPES at the appropriate pH and grown at 30°C as indicated. Cell pellets were resuspended in ice-cold radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, pH 8, 150 mM NaCl, 1% NP-40, 3 mM EDTA, 0.5% deoxycholate, 0.1% SDS) containing 1 μg/ml leupeptin, 2 μg/ml aprotinin, 1 μg/ml pepstatin, 0.1 mM phenylmethylsulfonyl fluoride, and 10 mM dithiothreitol (DTT) and lysed by vortexing with acid-washed glass beads for 1 h at 4°C. Lysates were cleared by centrifugations at 4°C to remove cell debris, and the protein concentration was determined spectrophotometrically (19).
For Western blot assays, ∼0.7 mg of total protein was resuspended in 2× SDS gel-loading buffer (100 mM Tris-Cl, pH 6.8, 200 mM DTT, 4% SDS, 0.1% bromophenol blue, 20% glycerol), boiled at 95°C for 3 min, and run in either 6 or 8% SDS-polyacrylamide gels. Gels were transferred to a nitrocellulose membrane and blocked in 6% nonfat milk in TBS-T (50 mM Tris, pH 7.6, 150 mM NaCl, 0.1% Tween 20). Membranes were incubated with a 1:1,000 dilution of anti-HA (F-7 probe; Santa Cruz), a 1:5,000 dilution of anti-V5 or anti-Myc (Invitrogen), or a 1:15,000 dilution of antitubulin in 6% nonfat milk in TBS-T. Membranes were washed three times in TBS-T and then exposed to a 1:5,000 dilution of antimouse-horseradish peroxidase antibody (GE Healthcare) in 6% nonfat milk in TBS-T or a 1:10,000 dilution of antirat-horseradish peroxidase antibody (to detect antitubulin) in 6% nonfat milk in TBS-T. Membranes were washed three times in TBS-T incubated with enhanced chemiluminescence reagent (GE Healthcare) and exposed to film.
Immunoprecipitation of HA-tagged proteins.
Twenty microliters of anti-HA agarose beads (Sigma) was added to 0.8 ml of crude extracts (∼12 mg of total protein), and the mixture was incubated overnight at 4°C. Beads were washed three times with TBS-T, and proteins were eluted by resuspension in 50 μl of 2× SDS gel-loading buffer and incubation at 95°C for 5 min.
Phosphatase assay.
HA-tagged proteins were immunoprecipitated, suspended in TBS, and incubated with and without calf intestine alkaline phosphatase (NEB) and with or without 50 mM EDTA. Reaction mixtures with beads were incubated at 37°C for 30 min. SDS gel-loading buffer (2×) was added, and the samples were boiled at 95°C for 5 min. The entire product was loaded in a 6% SDS-polyacrylamide gel to be analyzed by Western blotting.
RNA analyses.
Total RNA was extracted as described previously (13). Total RNA was further purified using an RNA purification kit (Qiagen). For reverse transcription-PCR (RT-PCR), RNA was treated with DNase I (Promega) and the RNA concentration was determined spectrophotometrically. Five micrograms of total RNA was reverse transcribed to cDNA using SuperScript II (Invitrogen) and random primers (Promega). Two microliters of cDNA was used as the template with either RIM8-specific primers (RTRIM8-F and RTRIM8-R) or ACT1-specific primers (ACT1-F and ACT1-R) as the loading control. PCR parameters were 94°C for 2 min, followed by 30 cycles, each consisting of 94°C for 30 s, 53°C for 30 s, and 72°C for 1 min.
Microscopy.
GFP-tagged Rim8 strains were grown overnight at 30°C to saturation in YPD. Cells were diluted 1/100 into fresh M199 at pH 4 and 30°C for 2 to 3 h. Cells were then shifted to 2 ml of fresh M199 at pH 4 or pH 8 containing 2.5 μl of 16 mM lipophilic dye FM4-64 (Invitrogen) and incubated at 30°C for 30 min. Cells were washed with phosphate-buffered saline, pH 4 or pH 7.2, and analyzed by fluorescence microscopy. Pictures were taken using a Zeiss Axio camera using either a GFP-specific or a rhodamine filter set.
RESULTS
Rim8 is posttranslationally modified.
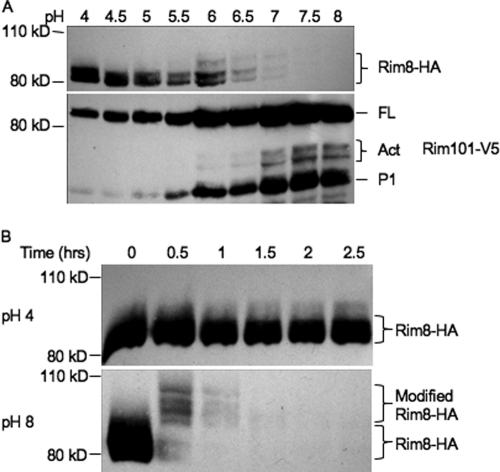
Rim8/PalF is posttranslationally modified in both A. nidulans and S. cerevisiae (23, 24). Thus, we asked if Rim8 is also posttranslationally modified in C. albicans. To address this question, we constructed a RIM8-HA/rim8Δ RIM101-V5 strain. The RIM8-HA/rim8Δ strain did not have the growth or filamentation defects associated with loss of Rim8, demonstrating that the Rim8-HA fusion was functional (data not shown). Using this strain background, we analyzed Rim8 expression across a range of pHs (Fig. 1A). At the most acidic pHs, pH 4 to 5.5, two distinct bands of Rim8-HA of ∼82 and 87 kDa were observed. However, at pH 6 to 7, a higher-molecular-mass band of ∼95 kDa was observed. We also noted a dramatic loss of Rim8-HA signal with increasing pH, as Rim8-HA levels were reduced beginning at pH 5.5 and were undetectable at pH 7.5 and 8 compared to Rim101-V5 levels. Thus, Rim8 is posttranslationally modified in response to environmental pH in C. albicans.
Fig 1.
Rim8 modification is correlated with Rim101 activation. (A) Rim8 levels are reduced concomitantly with processing of Rim101. The full-length (FL) and processed forms, both active (Act) and constitutive (P1), are indicated. The Rim101-V5 Rim8-HA fusion strain JAD011 was grown in M199 buffered at various pHs for 4 h. Total protein was extracted, separated by SDS-PAGE, probed with anti-HA, and stripped and probed with anti-V5. (B) Rim8 levels are rapidly reduced upon shift to alkaline conditions. The Rim8-HA strain JAD001 was grown in M199 at pH 4 for 4 h and shifted to either fresh M199 at pH 4 or M199 at pH 8. Cells were harvested every 30 min, total protein was purified, and Rim8-HA expression was determined by Western blot analysis with anti-HA.
The Rim101 transcription factor is proteolytically activated with increasing pH (31). Thus, we next asked whether Rim101 processing was correlated with Rim8-HA modification (Fig. 1A). As reported previously, Rim101-V5 was found in multiple forms (31). The full-length 85-kDa (FL) and 65-kDa processed (P1) forms were observed regardless of pH, although the levels of each varied depending on pH. Beginning at pH 6.0, two active forms (Act) of 72 and 74 kDa were observed. Thus, Rim101-V5 processing to the active 72- and 74-kDa forms occurs concomitantly with the modification and loss of Rim8-HA.
These initial analyses used samples that were grown for 4 h, which we reasoned represents steady state for Rim8-HA expression. To determine how quickly Rim8-HA modification occurs, we grew Rim8-HA-expressing cells in M199, pH 4, to logarithmic phase, shifted them to fresh M199 at pH 4 and pH 8, and monitored Rim8-HA expression over time (Fig. 1B). In fresh pH 4 medium, a broad band of ∼82 to 87 kDa was observed at all time points. In fresh pH 8 medium, by 30 min most of the Rim8-HA signal was found in at least 4 discrete bands of slower mobility between 95 and 105 kDa. However, we did note that some residual 82- and 87-kDa Rim8-HA protein remained after 30 min incubation. Furthermore, we observed that the Rim8-HA signal was drastically reduced within 30 min after the shift to pH 8 medium. At 1 and 1.5 h after the shift to M199 at pH 8, only the 95- to 105-kDa forms of Rim8-HA were apparent, although at even more reduced levels, and by 1.5 h after the shift to pH 8 medium, Rim8-HA was virtually undetectable. These results clearly demonstrate that Rim8 is rapidly modified and its expression is reduced in response to neutral-alkaline pH.
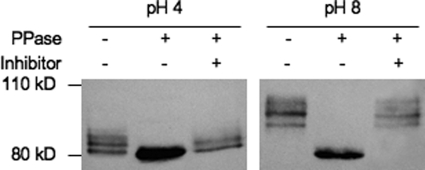
Rim8/PalF is posttranslationally modified by ubiquitination and phosphorylation in A. nidulans and S. cerevisiae (23, 24), and we predicted that the 95- to 105-kDa forms of Rim8-HA were due to ubiquitination. However, we could not detect ubiquitin-modified Rim8-HA in immunoprecipitates using a variety of antiubiquitin antibodies, although we readily detected other ubiquitinated proteins in the extracts (data not shown). Thus, we tested whether C. albicans Rim8 is phosphorylated. Rim8-HA-expressing cells were grown in M199 at pH 4 and shifted to fresh M199 at pH 4 and pH 8 for 30 min, and Rim8-HA was immunoprecipitated from crude extracts and incubated with or without phosphatase or with phosphatase plus a phosphatase inhibitor (Fig. 2). Cells grown in pH 4 medium showed Rim8-HA primarily in 2 distinct bands of 82 kDa and 87 kDa. However, only the 82-kDa band was apparent following phosphatase treatment, and both the 82-kDa and 87-kDa forms were present when EDTA was included as a phosphatase inhibitor. These results suggest that the 87-kDa form of Rim8 results from phosphorylation of the 82-kDa form. Cells grown in pH 8 medium showed Rim8-HA in several discrete bands ranging from 95 to 105 kDa. However, only the 82-kDa band was observed following phosphatase treatment and the 95- to 105-kDa bands were present if EDTA was included in the reaction mixtures. These results demonstrate that Rim8 is hyperphosphorylated in response to neutral-alkaline pH. Furthermore, these data suggest that all observed modifications of C. albicans Rim8 can be explained by phosphorylation.
Fig 2.
Rim8 is modified by phosphorylation. Rim8-HA-containing cells were grown in M199 at pH 4 for 4 h and then shifted to fresh M199 at either pH 4 or pH 8 for 30 min. Cells were harvested, total protein was purified, and Rim8-HA was immunoprecipitated. Immunoprecipitated Rim8-HA was untreated, treated with phosphatase (PPase), or treated with phosphatase and inhibitor for 30 min prior to Western blot analysis.
Requirements for Rim8 phosphorylation.
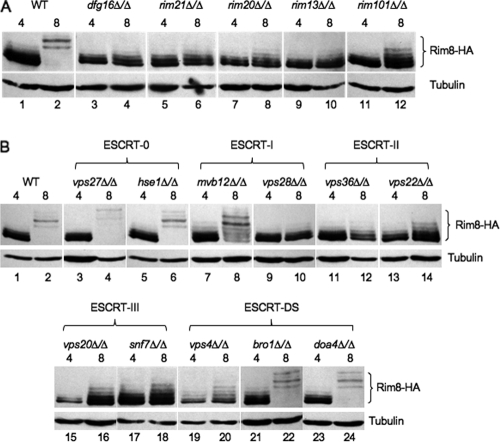
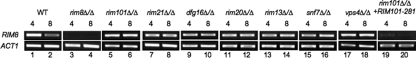
To determine the contribution of the Rim101 pathway for Rim8 phosphorylation, Rim8-HA was introduced into the Rim101 pathway mutants and Rim8-HA was monitored by Western blotting. After a 30-min shift to fresh pH 4 medium, wild-type cells showed the 82- to 87-kDa forms of Rim8-HA (Fig. 3A, lane 1). Similar results were observed in the Rim101 pathway mutants (Fig. 3A, lanes 3, 5, 7, 9, and 11), suggesting that this Rim8-HA phosphorylation is independent of the Rim101 pathway.
Fig 3.
Genetic requirements for Rim8 modification. Cells expressing Rim8-HA were grown in M199 at pH 4 for 4 h and shifted to fresh M199 at pH 4 and pH 8 for 30 min, and total was protein extracted. Protein was separated by SDS-PAGE and probed with anti-HA, followed by antitubulin as a loading control. (A) Rim8-HA was introduced into Rim101 pathway mutants to generate strains JAD001 (wild type [WT]), JAD002 (dfg16Δ/Δ), JAD003 (rim21Δ/Δ), JAD004 (rim20Δ/Δ), JAD005 (rim13Δ/Δ), and JAD006 (rim101Δ/Δ). (B) Rim8-HA was introduced into ESCRT pathway mutants to generate strains JAD001 (wild type), JAD025 (vps27Δ/Δ), JAD030 (hse1Δ/Δ), JAD028 (mvb12Δ/Δ), JAD026 (vps28Δ/Δ), JAD027 (vps36Δ/Δ), JAD029 (vps22Δ/Δ), JAD023 (vps20Δ/Δ), JAD007 (snf7Δ/Δ), JAD008 (vps4Δ/Δ), JAD010 (bro1Δ/Δ), and JAD009 (doa4Δ/Δ).
After a 30-min shift to fresh pH 8 medium, wild-type cells showed the 95- to 105-kDa forms of Rim8-HA (Fig. 3A, lane 2). However, the rim21Δ/Δ, dfg16Δ/Δ, rim13Δ/Δ, and rim20Δ/Δ mutants showed the 82- to 87-kDa forms of Rim8-HA and lacked the 95- to 105-kDa forms observed in wild-type cells grown at pH 8 (Fig. 3A, lanes 4, 6, 8, and 10). The rim101Δ/Δ mutant behaved similarly to the other Rim101 pathway mutants, although some faint higher-molecular-mass bands of ∼90 kDa could be detected (Fig. 3A, lane 12). Similar bands could also be observed in wild-type cells at pH 4 when the gel was overexposed (Fig. 3A, lane 1) and may represent a transient secondary modification that occurs prior to degradation (see below). Regardless, these results demonstrate that wild-type levels of Rim8 modification to the 95- to 105-kDa forms require both the signaling and processing components of the Rim101 pathway.
Rim101 activation requires the ESCRT-I, ESCRT-II, and ESCRT-IIIa components of the ESCRT pathway (28, 55, 56, 58). Thus, we asked if ESCRT components are required for Rim8 modification. To address this question, Rim8-HA was introduced into ESCRT pathway mutants (56). At pH 4, Rim8-HA modification in the ESCRT mutants was similar to that in wild-type cells (Fig. 3B). However, at pH 8, Rim8-HA modification was not modified to the 95- to 105-kDa forms in mutants lacking Vps28, Vps36, Vps22, Vps20, or Snf7 (Fig. 3B, lanes 10, 12, 14, 16, and 18). The ESCRT-0 components Hse1 and Vps27 as well as the ESCRT-DS components Bro1 and Doa4 were not required for wild-type-like Rim8-HA modification at pH 8 (Fig. 3B, lanes 4, 6, 22, and 24). Finally, the ESCRT-I mutant lacking Mvb12 had the 95- to 105-kDa bands at pH 8, but residual 82- to 87-kDa bands were still apparent, unlike the bands observed in the wild-type or the ESCRT-0 mutant background (Fig. 3B, lane 8). These results are in agreement with our previous studies that suggested that Mvb12 is not an essential member of the ESCRT-I complex for Rim101 signaling (56). These results demonstrate that ESCRT proteins required for Rim101 processing are also required for Rim8 phosphorylation.
Two kinases, Slt2 and Mck1, have reported genetic interactions with Rim8 in S. cerevisiae (45, 47). To determine if these kinases affect Rim8 phosphorylation, Rim8-HA was introduced into the mkc1Δ/Δ (the homolog of ScSlt2) and mck1Δ/Δ mutant backgrounds. Although MCK1 is reported to be an essential gene by the Candida Genomic Database, we were able to generate homozygous null strains using conventional disruption techniques (53). In both mkc1Δ/Δ and mck1Δ/Δ mutant backgrounds, Rim8-HA was modified similarly to wild type at pH 4 and pH 8 (Fig. 4). These results suggest that additional proteins phosphorylate Rim8, although it is possible that Mkc1 and Mck1 are redundant for Rim8 phosphorylation.
Fig 4.
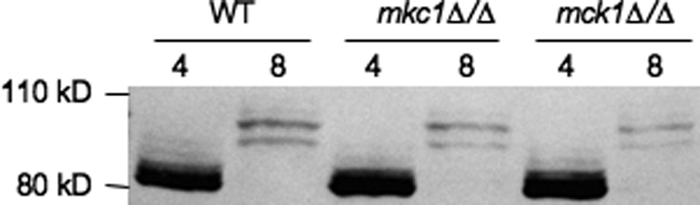
Wild-type (JAD001), mkc1Δ/Δ (JAD034), and mck1Δ/Δ (JAD032) strains expressing Rim8-HA were grown in M199 at pH 4 or M199 at pH 8, and total protein was extracted and separated by SDS-PAGE. Rim8-HA was detected using anti-HA.
Rim8 bridges the signaling and processing complexes.
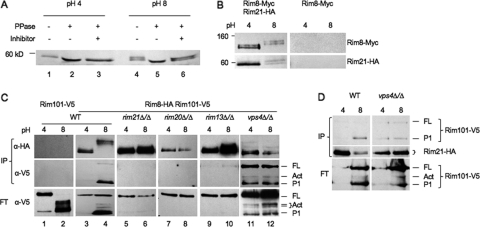
β-Arrestin proteins interact with plasma membrane receptors; Rim8/PalF interacts with the pH sensor Rim21/PalH in S. cerevisiae and A. nidulans (23, 24). To determine if this paradigm holds true in C. albicans, we constructed a strain coexpressing functional Rim21-HA and Rim8-Myc fusions. Our analysis of Rim21-HA expression showed that Rim21-HA ran at ∼59 kDa at pH 4 (Fig. 5A, lane 1) and at ∼59 kDa and ∼62 kDa at pH 8 (Fig. 5A, lane 4). We also noted that there was no apparent reduction in Rim21 intensity at pH 8 compared to pH 4, suggesting that Rim21 levels are not limiting for Rim101 signaling.
Fig 5.
Rim8 interactions with Rim101 pathway members. (A) Rim21-HA was immunoprecipitated from JAD021 cells grown in M199 at pH 4 and pH 8. Immunoprecipitated Rim21-HA was untreated, treated with phosphatase, or treated with phosphatase and inhibitor for 30 min prior to Western blot analysis. (B) Rim21-HA Rim8-Myc-expressing cells (JAD017) and Rim8-Myc-expressing cells (JAD031) were grown in M199 at pH 4 or pH 8, and total protein was extracted. Rim21-HA was immunoprecipitated with anti-HA beads and separated by SDS-PAGE. Blots were first probed with anti-Myc and then anti-HA. (C) Wild-type (JAD011) and rim21Δ/Δ (JAD013), rim20Δ/Δ (JAD012), rim13Δ/Δ (JAD014), and vps4Δ/Δ (JAD015) mutants expressing Rim8-HA and Rim101-V5 were grown and Rim8-HA protein was immunoprecipitated as for panel B. Blots were probed with anti-V5, stripped, and probed with anti-HA. (D) Wild-type (JAD016) and vps4Δ/Δ (JAD019) strains expressing Rim21-HA and Rim101-V5 were grown and Rim21-HA protein was immunoprecipitated using anti-HA. Blots were probed with anti-V5, stripped, and probed with anti-HA.
Since Rim21-HA migrated as two distinct bands at pH 8, we asked whether this was due to phosphorylation, as observed for Rim8. In extracts from cells grown at pH 4, Rim21-HA migrated at 59 kDa regardless of whether phosphatase was present (Fig. 5A, lanes 1 to 3). However, in extracts from cells grown at pH 8, addition of phosphatase abolished the presence of the 62-kDa band, which was restored when EDTA was included as a phosphatase inhibitor. These results demonstrate that Rim21 is phosphorylated in a pH-dependent manner correlated with activation of the Rim101 pathway.
To determine if Rim21-HA and Rim8-Myc interact, whole-cell extracts from cells grown at pH 4 or pH 8 were immunoprecipitated using anti-HA, separated by SDS-polyacrylamide gel electrophoresis (PAGE), and probed with anti-Myc and with anti-HA. Rim8-Myc protein was detected in Rim21-HA immunoprecipitates at both pH 4 and pH 8 (Fig. 5B, left lanes) but not from extracts lacking Rim21-HA (Fig. 5B, right lanes). We noted that Rim8-Myc was modified and that its levels were reduced at pH 8, and these findings are in agreement with our analyses demonstrating that Rim8 is rapidly modified and its levels are reduced at pH 8.
Since Rim8 modification is correlated with Rim101 processing (Fig. 1), we considered the possibility that Rim8 may be in complexes with Rim101. Thus, we conducted immunoprecipitations using cell extracts from a Rim8-HA Rim101-V5 strain grown at pH 4 or pH 8 (Fig. 5C, lanes 3 to 10). In cells grown at pH 4, no Rim101-V5 was detected in Rim8-HA immunoprecipitates, although Rim101-V5 was detected in the flowthrough (Fig. 5C, lane 3). In cells grown at pH 8, the 95- to 105-kDa forms of Rim8-HA were immunoprecipitated, and we detected the P1 form of Rim101-V5 and a faint signal of full-length Rim101-V5 (Fig. 5C, lane 4). No Rim101-V5 was detected from HA immunoprecipitates in the absence of Rim8-HA (Fig. 5C, lanes 1 and 2). These results suggest that Rim8 is in a complex that also contains Rim101.
To determine the genetic requirements for the Rim8-Rim101 interaction, we introduced Rim8-HA and Rim101-V5 into the rim21Δ/Δ, rim20Δ/Δ, and rim13Δ/Δ backgrounds and repeated the immunoprecipitations (Fig. 5C, lanes 5 to 10). In the absence of either the pH sensor Rim21 or the Rim101-recruiting protein Rim20, the 82- to 87-kDa forms of Rim8-HA were immunoprecipitated. No Rim101-V5 was coimmunoprecipitated, although Rim101-V5 was detected in the flowthrough. In the absence of the processing enzyme Rim13, low levels of full-length Rim101-V5 were detected in the immunoprecipitates. The Rim8-HA Rim101-V5 interaction in the rim13Δ/Δ mutant seems to be incomplete, as most of the Rim101-V5 is still found in the flowthrough. On the basis of these results, the pH sensor and Rim101-recruiting proteins, but not the processing enzyme, are required for Rim8-Rim101 interactions. Furthermore, these results indicate that Rim8 modification to the 95- to 105-kDa forms is not absolutely essential for interactions with Rim101.
Since Rim8 coimmunoprecipitates with Rim21 and Rim101 (Fig. 5B and C), we asked whether Rim21 and Rim101 interact. To address this question, we constructed a Rim21-HA Rim101-V5 strain and determined whether Rim101-V5 coimmunoprecipitates with Rim21-HA (Fig. 5D). At pH 8, when the Rim101 pathway is active, the P1 form of Rim101 was pulled down with Rim21-HA. In overexposed images, we also detected faint coimmunoprecipitation of the P1 form of Rim101 from pH 4-grown cells as well as full-length Rim101 at pH 4 and pH 8. These results support the idea that Rim21, Rim8, and Rim101 can be found within the same complex.
Vps4 limits Rim101 signaling.
Vps4, an AAA-ATPase, is required to form multivesicular bodies (MVBs) and disassemble ESCRT-III (6). In the absence of Vps4, ESCRT-III is stably localized at the endosomal membrane and vacuolar fusion is impaired. We and others have found that the vps4Δ/Δ mutant constitutively processes Rim101 (22, 56), suggesting that the stable association of ESCRT-III and specifically that of Snf7 with the endosomal membrane allows the Rim101-processing machinery to associate and process Rim101 in a pH-independent manner.
Thus, we predicted that Rim8-HA would be constitutively modified to the 95- to 105-kDa forms in the vps4Δ/Δ background. However, we found that while the vps4Δ/Δ mutant formed 82- to 90-kDa forms of Rim8-HA, it lacked the 95- to 105-kDa forms of Rim8-HA under pH 8 inducing conditions (Fig. 3B, lanes 19 and 20). At steady state, we could detect some Rim8-HA in the 95- to 105-kDa range, although the majority of Rim8-HA was still in the 82- to 90-kDa range (data not shown). Thus, cells lacking Vps4 do not promote wild-type-like modification of Rim8.
Since Vps4 affects Rim8-HA modification and constitutively processes Rim101, we asked how loss of Vps4 affects the association of Rim101 with Rim8 and Rim21. Immunoprecipitation of Rim8-HA in the vps4Δ/Δ mutant background did pull down Rim101-V5 (Fig. 5C, lanes 11 and 12). We found that in the absence of Vps4, the full-length, P1, and active forms of Rim101-V5 were coimmunoprecipitated with Rim8-HA regardless of pH. Similarly, Rim101-V5 coimmunoprecipitated with Rim21-HA regardless of pH, although only the full-length and P1 forms of Rim101-V5 were observed (Fig. 5D). These results demonstrate that in the absence of Vps4, Rim101 can associate with Rim21 and Rim8 independently of extracellular pH, suggesting an explanation for pH-independent Rim101 processing.
Rim8 appears to be degraded in the vacuole.
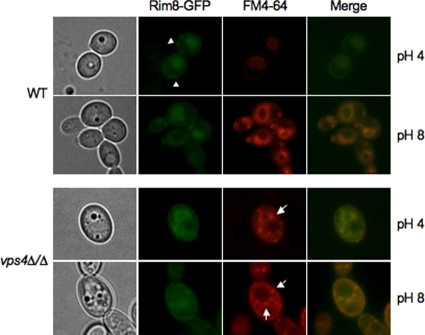
Rim8 immunoprecipitates with Rim21 and is modified in an ESCRT-dependent manner. Thus, we asked where Rim8 is localized. A functional Rim8-GFP C-terminal fusion was constructed, and localization was determined at pH 4 and pH 8 by fluorescence microscopy (Fig. 6). In acidic medium, Rim8-GFP was associated with the plasma membrane (Fig. 6, arrowheads), similar to results described for S. cerevisiae (23). However, we also noted that the lumen of the vacuole, identified with the lipophilic dye FM4-64 (49), had a robust signal. GFP is resistant to vacuolar proteases (39), suggesting that Rim8-GFP can be localized within the vacuole. In alkaline medium, we found little plasma membrane-associated localization and observed only vacuole-associated fluorescence. These results suggest that plasma membrane-localized Rim8-GFP may be transported to the vacuole.
Fig 6.
Rim8 localization in wild-type and vps4Δ/Δ cells. Rim8-GFP fusions were generated in a wild-type (JAD081) and vps4Δ/Δ (JAD083) background. Cells were grown in M199 at pH 4 or pH 8 and GFP and stained with FM4-64. Cells were analyzed and photographed by fluorescence microscopy.
Since the Rim8 signal is reduced at neutral-alkaline pH (Fig. 1) and appears to be localized to the vacuole, we considered the possibility that Rim8 may be degraded within the vacuole following activation of the Rim101 pathway. If true, Rim8-GFP localization to the vacuole should be Vps4 dependent. To address this idea, we introduced RIM8-GFP into the vps4Δ/Δ mutant background. In vps4Δ/Δ cells grown at either pH 4 or pH 8, Rim8-GFP was not observed within the vacuole but was colocalized with FM4-64 in class E-like structures around the presumptive vacuole (Fig. 6, arrows).
Rim8 is transcriptionally repressed.
RIM8 encodes a β-arrestin-like protein required for Rim101 activation at neutral-alkaline pH. However, RIM8 transcription is repressed at alkaline pH in a Rim101-dependent manner (9, 42). This led us to hypothesize that Rim8 is a target for negative-feedback regulation. To identify the genetic requirements for RIM8 repression, we analyzed RIM8 transcription at pH 4 and pH 8 by RT-PCR (Fig. 7). RIM8 mRNA levels were reduced by ∼40% at pH 8 compared to pH 4 in wild-type cells, no RIM8 product was observed at either pH in the rim8Δ/Δ mutant, and RIM8 was expressed similarly at either pH and at levels similar to those for wild-type cells at pH 4 in the rim101Δ/Δ mutant (Fig. 7, lanes 1 to 6). These results are in agreement with previous studies and establish the veracity of the RT-PCR approach.
Fig 7.
RIM8 transcription is repressed by activated Rim101. RT-PCR on total RNA purified from wild-type (DAY1), rim8Δ/Δ (DAY61), rim101Δ/Δ (DAY5), rim21Δ/Δ (JAD003), dfg16Δ/Δ (KBC033), rim20Δ/Δ (DAY23), rim13Δ/Δ (DAY349), snf7Δ/Δ (DAY534), vps4Δ/Δ (DAY537), and rim101Δ/Δ RIM101-281 (DAY1145) cells grown at pH 4 and pH 8. Controls lacking reverse transcriptase were performed, with no bands observed (data not shown). RT-PCR for ACT1 was included as a normalization control.
We next determined if other Rim101 pathway members were required for RIM8 repression. Indeed, the rim21Δ/Δ, dfg16Δ/Δ, rim20Δ/Δ, and rim13Δ/Δ mutants expressed RIM8 similarly to the rim101Δ/Δ mutant (Fig. 7, lanes 7 to 14), demonstrating that RIM8 repression requires Rim101 activation. We also found that the snf7Δ/Δ mutant failed to repress RIM8 (Fig. 7, lanes 15 and 16), as expected, since this mutant does not activate Rim101 (28, 55). However, we found that the vps4Δ/Δ mutant also failed to repress RIM8 (Fig. 7, lanes 17 and 18), which was unexpected since this mutant constitutively activates Rim101 (Fig. 5C) (56). Although the vps4Δ/Δ mutant constitutively activates Rim101, the levels of processed active Rim101 are reduced compared to those for wild-type cells (55). Thus, we considered either that the level of active Rim101 may not be sufficient to promote RIM8 repression or that processed Rim101 is necessary but not sufficient to promote RIM8 repression. To test the latter possibility, we analyzed RIM8 repression in a strain expressing only the constitutively active RIM101-281 allele (55), which bypasses the signaling and processing requirements of the Rim101 pathway. In the rim101Δ/Δ RIM101-281 background, RIM8, but not ACT1, transcription was greatly reduced at both pH 4 and pH 8 (Fig. 7, lanes 19 and 20). These results suggest that processed Rim101 is sufficient to promote RIM8 repression and suggest that there is insufficient active Rim101 generated in the vps4Δ/Δ mutant to promote repression.
DISCUSSION
β-Arrestin proteins function in receptor desensitization but also play a direct role in signal transduction. In fungi, the β-arrestin-like protein Rim8/PalF is essential for regulation of the Rim101 transcription factor in response to environmental pH (3, 13, 42, 48). Much like the β-arrestin proteins in mammalian systems, C. albicans Rim8 interacts with a receptor, Rim21, which senses environmental pH. Our studies further demonstrate that Rim8 can be found within complexes containing the transcription factor Rim101, which itself can be found in complexes containing Rim21. These results suggest that β-arrestin proteins can link a receptor with its cognate transcription factor. We also found that like some other β-arrestin-like proteins, Rim8 is posttranslationally modified by phosphorylation (34, 35). However, C. albicans Rim8 can be found in multiple phosphorylation states, which are linked with Rim101 activation. Finally, our results suggest a novel signal transduction regulation. When the Rim101 pathway is activated, the cytosolic Rim8 protein is delivered to the vacuole and degraded. Based on the similarities between fungal and mammalian β-arrestin-dependent signal transduction, we propose that lysosomal degradation may play an important role in β-arrestin regulation in mammalian systems as well. These analyses shed important insights into the evolving paradigm of the linkage between endocytosis and signal transduction pathways in eukaryotic cells.
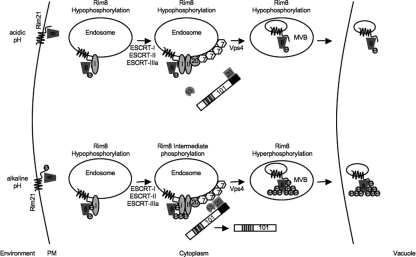
Model for Rim8 phosphorylation and activity.
Based on our analyses, we propose the following framework to consider Rim8 modification, localization, and function (Fig. 8). Rim8 was found in several phosphorylation states: a hypophosphorylation state (82- to 87-kDa forms), an intermediate phosphorylation state (87- to 95-kDa forms), and a hyperphosphorylated state (95- to 105-kDa forms). The hypophosphorylation state is observed at acidic pH, when Rim101 is not processed, and is thus considered inactive. The fact that Rim8 appears within the vacuole at pH 4 suggests that Rim8 is constitutively trafficked to the vacuole.
Fig 8.
Model of Rim8 modification and degradation. Under acidic conditions, Rim8 (8) is hypophosphorylated but can associate with Rim21 at the plasma membrane. Rim8 appears to be taken up via endocytosis and transported to the vacuole in a Rim21-dependent fashion. ESCRT-I (I), -II (II), and -IIIa components Vps20 (20) and Snf7 (7) associate with the endosome and promote MVB formation. The MVB then fuses with the vacuole. Under these conditions, Rim20 (20) does not associate with Snf7 and Rim101 (101) processing does not occur. Under neutral-alkaline conditions, hypophosphorylated Rim8 presumably associates with phosphorylated Rim21 and is taken up by endocytosis. ESCRT-I, -II, and -IIIa are recruited and Rim8 is phosphorylated to the intermediate 87- to 95-kDa forms. Via a currently unknown mechanism, Rim20 and Rim13 (13) can then associate with Snf7 to promote Rim101 processing. Rim8 is hyperphosphorylated, which may promote MVB formation and allow vacuolar fusion.
The intermediate and hyperphosphorylation states are observed at neutral-alkaline pH when Rim101 processing is defective or intact, respectively. This suggests that Rim8 hyperphosphorylation is directly linked to Rim101 processing. However, the vps4Δ/Δ mutant, which constitutively processes Rim101, does not promote Rim8 hyperphosphorylation. Thus, we are considering a model where hyperphosphorylation is a secondary modification occurring concomitantly with or after Rim101 processing that is linked to uptake into MVBs (Fig. 8). In mammalian systems, β2-adrenergic receptor stimulation promotes β-arrestin 1 association and clathrin-dependent endocytosis. Following receptor internalization, β-arrestin 1 can be phosphorylated by extracellular signal-regulated kinases, which inhibits β-arrestin 1 function (35). Thus, the hyperphosphorylated forms of Rim8 may be additional modifications that similarly inhibit Rim8 function or promote uptake of Rim8 into MVBs and transport to the vacuole. Although Rim8 does not appear to be localized to the vacuole in S. cerevisiae (23), a two-step modification system that differentiates internalization and vacuolar targeting has been demonstrated for yeast α-arrestin-dependent receptors, although this system uses ubiquitin instead of phosphorylation (16, 30).
The hypothesis that Rim8 is taken up into MVBs also explains a previously unresolved observation. In C. albicans, Rim101 is processed to the small P1 form in a pH-independent manner (Fig. 1A) (31), although no function has been associated with this form of Rim101. In other fungi, Rim101/PacC processing does not occur at the more acidic pHs (29, 32, 41). Since Rim8 appears to be trafficked to the vacuole of cells grown at acidic pH, we propose that Rim101 may weakly associate with Rim8 on endosomes in the absence of an activating signal. Under these conditions, Rim101 is not processed but is taken up into MVBs with Rim8 and transported to the vacuole. In this model, the P1 form of Rim101 represents a partially stable degradation product. In S. cerevisiae, where no Rim101 processing is observed at acidic pH, Rim8 does not appear to be trafficked to the vacuole (23). It will be necessary to analyze Rim8/PalF localization in other fungal species to determine if vacuolar localization and degradation were lost in S. cerevisiae or gained in C. albicans.
Rim8 modification: phosphorylation versus ubiquitination.
In the related ascomycetes S. cerevisiae and A. nidulans, Rim8/PalF is posttranslationally modified by ubiquitination. In A. nidulans, this modification is linked to environmental pH and is required for PacC activation, but in S. cerevisiae it is not (23–25). We were not able to detect Rim8 ubiquitination in C. albicans but instead found robust phosphorylation (Fig. 2). A. nidulans PalF is also known to be phosphorylated in a pH-dependent manner (24), although the role that PalF phosphorylation plays in PacC activation has not been explored. Based on our findings, we suggest that the PalF phosphorylation observed in A. nidulans may also be critical for PacC pathway signaling.
The fact that we did not detect Rim8 ubiquitination in C. albicans suggests either that C. albicans Rim8 is not ubiquitinated or that Rim8 is ubiquitinated at levels too low to be detected using our assays. In S. cerevisiae, Rim8 ubiquitination occurs at residue K521 (23), which is located 12 residues C terminal to a PXY domain predicted to be critical for interacting with the WW domain of the Rps5 E3 ubiquitin ligase (20, 51). K521 is also located 13 residues N terminal to the SXP domain that interacts with the UEV domain of Vps23 (23). In C. albicans Rim8, the cognate lysine, K538, is similarly located 13 residues N terminal to an SXP domain. However, 12 residues N terminal to K538 is not a PXY motif but a divergent PXF sequence. This supports the idea that C. albicans Rim8 may not be ubiquitinated. However, a PXY domain is located 20 residues N terminal to K538, and there is not an absolute spacing requirement for Rsp5 function (44). It is noteworthy that C. albicans Rim8 has 64 potential phosphorylation sites (41 Ser, 5 Thr, and 12 Tyr), whereas S. cerevisiae has 42 potential phosphorylation sites (26 Ser, 7 Thr, and 9 Tyr). Of these sites, 28 and 15 have a high confidence (>0.950 from NetPhos software, version 2.0) of being phosphorylated in C. albicans and S. cerevisiae, respectively. Thus, C. albicans Rim8 has a greater potential to be phosphorylated than S. cerevisiae Rim8. Regardless, these analyses demonstrate several fruitful directions to elucidate the nature and role of specific Rim8 modifications.
Why does Rim8/PalF posttranslational modification vary between C. albicans, S. cerevisiae, and A. nidulans? While there is currently no definitive answer to this question, we suggest that environmental pH stresses found within the mammalian host may provide one explanation. C. albicans is a commensal of the oral and gastrointestinal tract. Thus, a C. albicans cell residing within the slightly alkaline oral cavity may find itself within a markedly acidic environment by the simple act of swallowing. Thus, it is possible that unique host environments colonized by C. albicans may require a more rapid and/or finely controlled response to extracellular pH.
Rim8 as a point of negative-feedback regulation.
Upon Rim101 pathway activation, Rim8 levels are rapidly reduced. We believe that our results best support a model where Rim8 levels are reduced in large part because Rim8 is taken up into MVBs and trafficked to the vacuole. We propose that reduction in Rim8 levels serves to negatively regulate the Rim101 pathway.
Upon exposure to neutral-alkaline pH, Rim101 is proteolytically processed to the active form, which promotes gene expression changes associated with growth at these environmental pHs. Active Rim101 promotes its own induction, leading to a positive-feedback loop (13, 43). This leads to the following questions: how is the positive-feedback loop limited, and what happens when cells are then exposed to acidic pH environments?
We propose that both questions are answered by the degradation of Rim8. Not only does active Rim101 induce its own expression, but it also represses RIM8 transcription ∼40% (Fig. 7) (9, 42). This reduces the message available to express new Rim8 protein. As Rim8 levels are reduced, less signaling from Rim21 can be transmitted. This serves to delay processing of newly translated Rim101, thus limiting the positive-feedback loop. If the environmental pH drops, Rim21 is no longer stimulated and newly translated Rim8 is not modified, leading to a rapid inhibition in continued Rim101 processing.
Activation of the Rim101 transcription factor by proteolysis of the inhibitory C-terminal domain has been compared to the activation of mammalian NFκB by the proteolysis of the inhibitory protein IFκB. Our work allows an extension of this comparison, as β-arrestin proteins can interact with IFκB and stabilize NFκB-IFκB (37, 54). This β-arrestin–NFκB–IFκB interaction inhibits activation of the transcription factor, which is the converse of our system, in which Rim8 promotes Rim101 activation. Regardless, the paradigm of arrestin-dependent regulation of a transcription factor is conserved and, we suggest, may be a more common mechanism for signal transduction than has previously been appreciated. Furthermore, the paradigm that we envision for Rim8 trafficking from the plasma membrane to the endosomal membrane and then within MVBs has also been suggested in higher eukaryotes. Glycogen synthase kinase 3, a cytoplasmic regulator of the Wnt signaling receptor frizzled, is also internalized into MVBs (46). Not surprisingly, like Rim101 activation, Wnt signaling requires endocytosis (10, 59, 60). Again, these results suggest that the intimate connection between the signal transduction and endocytic pathways is a conserved phenomenon throughout the eukarya.
ACKNOWLEDGMENTS
We thank Judith Berman for providing the antitubulin and antirat secondary antibodies. We also appreciate the kind gift of the mkc1Δ/Δ strain from Jesus Pla as well as the dfg16Δ/Δ strain from Aaron P. Mitchell. We thank all the members of the Davis laboratory and members of Kirsten Nielsen's laboratory for helpful discussion with this work. We also thank Julie Wolf and Lucia Zacchi for critical reading of the manuscript.
J.G.-R. was the recipient of a postdoctoral fellowship from Junta de Extremadura (ERDF, European Regional Development Fund). This research of Dana A. Davis was supported in part by an Investigators in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund and by NIH National Institute of Allergy and Infectious Diseases award R01-AI064054.
Footnotes
Published ahead of print 16 March 2012
REFERENCES
- 1. Adams A, Gotschling DE, Kaiser CA, Stearns T. 1997. Methods in yeast genetics, 1997 ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 2. Alvarez CE. 2008. On the origins of arrestin and rhodopsin. BMC Evol. Biol. 8:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arst HN, Jr, Bignell E, Tilburn J. 1994. Two new genes involved in signalling ambient pH in Aspergillus nidulans. Mol. Gen. Genet. 245:787–790 [DOI] [PubMed] [Google Scholar]
- 4. Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3:271–282 [DOI] [PubMed] [Google Scholar]
- 5. Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3:283–289 [DOI] [PubMed] [Google Scholar]
- 6. Babst M, Wendland B, Estepa EJ, Emr SD. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barwell KJ, Boysen JH, Xu W, Mitchell AP. 2005. Relationship of DFG16 to the Rim101p pH response pathway in Saccharomyces cerevisiae and Candida albicans. Eukaryot. Cell 4:890–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benovic JL, et al. 1987. Functional desensitization of the isolated beta-adrenergic receptor by the beta-adrenergic receptor kinase: potential role of an analog of the retinal protein arrestin (48-kDa protein). Proc. Natl. Acad. Sci. U. S. A. 84:8879–8882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bensen ES, Martin SJ, Li M, Berman J, Davis DA. 2004. Transcriptional profiling in C. albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol. Microbiol. 54:1335–1351 [DOI] [PubMed] [Google Scholar]
- 10. Blitzer JT, Nusse R. 2006. A critical role for endocytosis in Wnt signaling. BMC Cell Biol. 7:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cornet M, et al. 2005. Deletions of endocytic components VPS28 and VPS32 affect growth at alkaline pH and virulence through both RIM101-dependent and RIM101-independent pathways in Candida albicans. Infect. Immun. 73:7977–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davis D, Edwards JE, Jr, Mitchell AP, Ibrahim AS. 2000. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 68:5953–5959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Davis D, Wilson RB, Mitchell AP. 2000. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20:971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davis DA. 2009. How human pathogenic fungi sense and adapt to pH: the link to virulence Curr. Opin. Microbiol. 12:365–370 [DOI] [PubMed] [Google Scholar]
- 15. Edmond MB, et al. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239–244 [DOI] [PubMed] [Google Scholar]
- 16. Erpapazoglou Z, et al. 2008. Substrate- and ubiquitin-dependent trafficking of the yeast siderophore transporter Sit1. Traffic 9:1372–1391 [DOI] [PubMed] [Google Scholar]
- 17. Gerami-Nejad M, Berman J, Gale CA. 2001. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18:859–864 [DOI] [PubMed] [Google Scholar]
- 18. Gerami-Nejad M, Dulmage K, Berman J. 2009. Additional cassettes for epitope and fluorescent fusion proteins in Candida albicans. Yeast 26:399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grimsley GR, Pace CN. 2004. Spectrophotometric determination of protein concentration. John Wiley & Sons, Inc., New York, NY: [DOI] [PubMed] [Google Scholar]
- 20. Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. U. S. A. 97:13871–13876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hatakeyama R, Kamiya M, Takahara T, Maeda T. 2010. Endocytosis of the aspartic acid/glutamic acid transporter Dip5 is triggered by substrate-dependent recruitment of the Rsp5 ubiquitin ligase via the arrestin-like protein Aly2. Mol. Cell. Biol. 30:5598–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hayashi M, Fukuzawa T, Sorimachi H, Maeda T. 2005. Constitutive activation of the pH-responsive Rim101 pathway in yeast mutants defective in late steps of the MVB/ESCRT pathway. Mol. Cell. Biol. 25:9478–9490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herrador A, Herranz S, Lara D, Vincent O. 2010. Recruitment of the ESCRT machinery to a putative seven-transmembrane-domain receptor is mediated by an arrestin-related protein. Mol. Cell. Biol. 30:897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Herranz S, et al. 2005. Arrestin-related proteins mediate pH signaling in fungi. Proc. Natl. Acad. Sci. U. S. A. 102:12141–12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hervas-Aguilar A, Galindo A, Penalva MA. 2010. Receptor-independent Ambient pH signaling by ubiquitin attachment to fungal arrestin-like PalF. J. Biol. Chem. 285:18095–18102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ito T, et al. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U. S. A. 98:4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katzmann DJ, Stefan CJ, Babst M, Emr SD. 2003. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162:413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kullas AL, Li M, Davis DA. 2004. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot. Cell 3:1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lambert M, Blanchin-Roland S, Le Louedec F, Lepingle A, Gaillardin C. 1997. Genetic analysis of regulatory mutants affecting synthesis of extracellular proteinases in the yeast Yarrowia lipolytica: identification of a RIM101/pacC homolog. Mol. Cell. Biol. 17:3966–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lauwers E, Jacob C, Andre B. 2009. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185:493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li M, Martin SJ, Bruno VM, Mitchell AP, Davis DA. 2004. Candida albicans Rim13p, a protease required for Rim101p processing at acidic and alkaline pHs. Eukaryot. Cell 3:741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li W, Mitchell AP. 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135:714–725 [DOI] [PubMed] [Google Scholar]
- 34. Lin FT, et al. 2002. Phosphorylation of beta-arrestin2 regulates its function in internalization of beta(2)-adrenergic receptors. Biochemistry 41:10692–10699 [DOI] [PubMed] [Google Scholar]
- 35. Lin FT, et al. 1997. Clathrin-mediated endocytosis of the beta-adrenergic receptor is regulated by phosphorylation/dephosphorylation of beta-arrestin1. J. Biol. Chem. 272:31051–31057 [DOI] [PubMed] [Google Scholar]
- 36. Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. 1990. Beta-arrestin: a protein that regulates beta-adrenergic receptor function. Science 248:1547–1550 [DOI] [PubMed] [Google Scholar]
- 37. Luan B, Zhang Z, Wu Y, Kang J, Pei G. 2005. Beta-arrestin2 functions as a phosphorylation-regulated suppressor of UV-induced NF-kappaB activation. EMBO J. 24:4237–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miller WE, Lefkowitz RJ. 2001. Expanding roles for beta-arrestins as scaffolds and adapters in GPCR signaling and trafficking. Curr. Opin. Cell Biol. 13:139–145 [DOI] [PubMed] [Google Scholar]
- 39. Nikko E, Pelham HR. 2009. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10:1856–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nobile CJ, et al. 2008. Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell. Microbiol. 10:2180–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Orejas M, et al. 1995. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 9:1622–1632 [DOI] [PubMed] [Google Scholar]
- 42. Porta A, Ramon AM, Fonzi WA. 1999. PRR1, a homolog of Aspergillus nidulans palF, controls pH-dependent gene expression and filamentation in Candida albicans. J. Bacteriol. 181:7516–7523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ramon AM, Porta A, Fonzi WA. 1999. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J. Bacteriol. 181:7524–7530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stamenova SD, Dunn R, Adler AS, Hicke L. 2004. The Rsp5 ubiquitin ligase binds to and ubiquitinates members of the yeast CIN85-endophilin complex, SlaI-Rvs167. J. Biol. Chem. 279:16017–16025 [DOI] [PubMed] [Google Scholar]
- 45. Su SS, Mitchell AP. 1993. Identification of functionally related genes that stimulate early meiotic gene expression in yeast. Genetics 133:67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taelman VF, et al. 2010. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell 143:1136–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tong AH, et al. 2004. Global mapping of the yeast genetic interaction network. Science 303:808–813 [DOI] [PubMed] [Google Scholar]
- 48. Treton B, Blanchin-Roland S, Lambert M, Lepingle A, Gaillardin C. 2000. Ambient pH signalling in ascomycetous yeasts involves homologues of the Aspergillus nidulans genes palF and paIH. Mol. Gen. Genet. 263:505–513 [DOI] [PubMed] [Google Scholar]
- 49. Vida TA, Emr SD. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Villar CC, Kashleva H, Nobile CJ, Mitchell AP, Dongari-Bagtzoglou A. 2007. Mucosal tissue invasion by Candida albicans is associated with E-cadherin degradation, mediated by transcription factor Rim101p and protease Sap5p. Infect. Immun. 75:2126–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang G, Yang J, Huibregtse JM. 1999. Functional domains of the Rsp5 ubiquitin-protein ligase. Mol. Cell. Biol. 19:342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wilson RB, Davis D, Enloe BM, Mitchell AP. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65–70 [DOI] [PubMed] [Google Scholar]
- 53. Wilson RB, Davis D, Mitchell AP. 1999. Rapid hypothesis testing in Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Witherow DS, Garrison TR, Miller WE, Lefkowitz RJ. 2004. beta-Arrestin inhibits NF-kappaB activity by means of its interaction with the NF-kappaB inhibitor IkappaBalpha. Proc. Natl. Acad. Sci. U. S. A. 101:8603–8607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wolf JM, Davis DA. 2010. Mutational analysis of Candida albicans SNF7 reveals genetically separable Rim101 and ESCRT functions and demonstrates divergence in bro1-domain protein interactions. Genetics 184:673–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wolf JM, Johnson DJ, Chmielewski D, Davis DA. 2010. The Candida albicans ESCRT pathway makes Rim101-dependent and -independent contributions to pathogenesis. Eukaryot. Cell 9:1203–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu W, Mitchell AP. 2001. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J. Bacteriol. 183:6917–6923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xu W, Smith FJ, Jr, Subaran R, Mitchell AP. 2004. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell. 15:5528–5537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yamamoto H, Komekado H, Kikuchi A. 2006. Caveolin is necessary for Wnt-3a-dependent internalization of LRP6 and accumulation of beta-catenin. Dev. Cell 11:213–223 [DOI] [PubMed] [Google Scholar]
- 60. Yamamoto H, Sakane H, Yamamoto H, Michiue T, Kikuchi A. 2008. Wnt3a and Dkk1 regulate distinct internalization pathways of LRP6 to tune the activation of beta-catenin signaling. Dev. Cell 15:37–48 [DOI] [PubMed] [Google Scholar]
- 61. Yuan X, Mitchell B, Hua X, Davis DA, Wilhelmus KR. 2010. The RIM101 signal transduction pathway regulates Candida albicans virulence during experimental keratomycosis. Invest. Ophthalmol. Vis. Sci. 51:4668–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]