Abstract
African trypanosomes are the only organisms known to use RNA polymerase I (pol I) to transcribe protein-coding genes. These genes include VSG, which is essential for immune evasion and is transcribed from an extranucleolar expression site body (ESB). Several trypanosome pol I subunits vary compared to their homologues elsewhere, and the question arises as to how these variations relate to pol I function. A clear example is the N-terminal extension found on the second-largest subunit of pol I, RPA2. Here, we identify an essential role for this region. RPA2 truncation leads to nuclear exclusion and a growth defect which phenocopies single-allele knockout. The N terminus is not a general nuclear localization signal (NLS), however, and it fails to accumulate unrelated proteins in the nucleus. An ectopic NLS is sufficient to reinstate nuclear localization of truncated RPA2, but it does not restore function. Moreover, NLS-tagged, truncated RPA2 has a different subnuclear distribution to full-length protein and is unable to build stable pol I complexes. We conclude that the RPA2 N-terminal extension does not have a role exclusive to the expression of protein-coding genes, but it is essential for all pol I functions in trypanosomes because it directs trypanosomatid-specific interactions with RPA1.
INTRODUCTION
African trypanosomes of the species Trypanosoma brucei are unicellular eukaryotic parasites that are responsible for human African trypanosomiasis (HAT, or sleeping sickness) and the wasting disease nagana in livestock in sub-Saharan Africa. The annual burden of HAT has been estimated at ∼70,000 cases, and more than a million disability-adjusted life years (DALYs) are lost (12) T. brucei is transmitted by fly bite to the host bloodstream where it proliferates extracellularly, evading the host immune response by the antigenic variation of surface protein (for a review, see reference 4). The cell surface of the bloodstream-form trypanosome is covered with a dense coat of monotypic variant surface glycoprotein (VSG). The T. brucei genome contains hundreds of VSG genes, many of which are pseudogenes (3). In addition, there are ∼20 telomeric bloodstream expression sites (BESs), each of which contains a single VSG gene toward its 3′ end (16, 49). Only one of these BESs is transcriptionally active at any time, such that no more than one VSG gene is ever expressed by any cell. By switching the expressed VSG, the parasite can escape the host immune response against previously exposed VSGs and hence maintain the waves of parasitemia that are characteristic of the disease (for a review, see reference 36).
The active BES, containing the expressed VSG in bloodstream-form T. brucei, is transcribed by RNA polymerase I (pol I) (14, 28, 37). This is also the case for the metacyclic ESs, which encode VSGs expressed during the initial transmission to the bloodstream and the genes coding for the major surface proteins of the procyclic (midgut) stage of the tsetse fly. This transcription of endogenous protein-coding genes by pol I apparently makes African trypanosomes unique. In all other eukaryotic organisms analyzed, pol I is restricted to the transcription of the 45S rRNA precursor. In trypanosomes, the transcription of the active BES is spatially distinct from the nucleolar transcription of rRNA, occurring instead from an extranucleolar transcription focus called the expression site body (ESB) (33). Alongside the VSG, BESs contain a number of expression site-associated genes (ESAGs). A BES is transcribed as a single long polycistron from which mature mRNA is produced by the trans-splicing of a spliced-leader mini-exon to the 5′ end and polyadenylation at the 3′ end. It has been postulated that the ability of pol I to transcribe through the BES requires the association of the polymerase with the transcription elongation and processing factors necessary for mRNA generation (36, 45), although no components that might facilitate this association have been identified thus far (43). This raises the question of whether there are specific differences in the composition of pol I compared to other organisms which might allow such interactions to occur, or which function solely in the transcription of mRNA and not rRNA.
Eukaryotic RNA polymerases are multisubunit enzymes. Trypanosomatids differ from other model systems in the composition and form of their polymerase subunits. In trypanosomes, there are two paralogues for RNA polymerase subunits RPB5, RPB6, and RPB10, which in other eukaryotes are single paralogues shared between all three RNA polymerases (20, 24, 34, 47). The canonical RPB5, RPB6, and RPB10 paralogues are associated with pol II and pol III but are not part of the pol I complex (8, 9, 32). Instead, variant RPB5z, RPB6z, and RPB10z paralogues, which contain sequence insertions relative to the sequences of the canonical set, are specifically substituted for their canonical counterparts in pol I (10, 34, 47). The RNA interference (RNAi)-mediated knockdown of RPB5 or RPB5z abolished either pol II/pol III or pol I transcription, respectively, demonstrating that the function of these two subunits is class specific (10). Trypanosomal pol I also contains a novel subunit, p31, which is essential for the transcription of rRNA (35, 47), but the complex lacks the pol I-specific RPA43/RPA14 heterodimer, which is not encoded in the genome (24). Hence, several trypanosomatid-specific features of pol I have been demonstrated, but the functional relevance, if any, of the insertions in the pol I-specific subunits is not yet understood.
The most striking difference between the pol I complexes of trypanosomes and other eukaryotes is the sequence of the N terminus of the second-largest subunit, RPA2 (RPA135). In a proteomic study of the T. brucei pol I complex, Schimanski et al. showed that the RPA2 protein is ∼50 kDa larger than orthologues in most other eukaryotes (38) and contains an apparently trypanosomatid-specific N-terminal domain of ∼250 amino acids. This RPA2 N-terminal extension contains a serine-rich region but is probably not a site for phosphorylation, since RPA2 was found not to be phosphorylated in bloodstream-form or procyclic T. brucei (47).
Given its conservation among trypanosomatids, it has been hypothesized that the RPA2 N terminus provides a crucial function for these parasites (38), but this function and its biological relevance have remained elusive to date. One interesting scenario is that the RPA2 N terminus is necessary for the residency of pol I in different parts of the nucleus, in particular endowing pol I the capability to occupy the ESB in addition to its canonical location in the nucleolus. In this work, we address this hypothesis by performing a molecular dissection of the N-terminal region of RPA2 in T. brucei cells. In so doing, we characterize the influence of the N-terminal region on the cell and subnuclear compartmentalization of RPA2, as well as on the stable assembly of the pol I complex and polymerase function.
MATERIALS AND METHODS
Bioinformatic analyses.
Using BLASTp (1), a data set of 28 eukaryotic RPA2 proteins was generated by querying 28 predicted eukaryotic proteomes of evolutionarily diverse organisms with proteins annotated as RPA2 in the NCBI databases. Multiple-sequence alignments were done with MAFFT (23). The profile-hidden Markov model of the trypanosome N terminus was built from a seed alignment of RPA2 amino acids 1 to 374 of T. brucei and 1 to 381 of Leishmania major with hmmer 2.3.2 (10, 11). As a control model, a seed alignment of T. brucei RPA2 amino acids 375 to 750 with L. major RPA2 amino acids 382 to 767 was generated. These models were then used to search the predicted proteomes of 28 eukaryotic, evolutionarily diverse organisms (see Table S1 in the supplemental material).
Cell lines and cell culture.
To facilitate the transfection of procyclic-form trypanosomes with RNAi and endogenous locus-tagging constructs, we made a single-marker procyclic line (SiMP1) expressing Tet repressor protein from the endogenous EP1 locus. Briefly, the T7 promoter was removed from the plasmid pBluescript II SK(+) (X52328; Stratagene) by digestion with NaeI and EcoRV, followed by self ligation to create the plasmid pSPR0. From genomic DNA (gDNA) purified from the single-marker bloodstream line of Wirtz et al. (48), we generated the following by PCR: a fragment of DNA corresponding to the region immediately upstream of the EP1 promoter (Ups), a fragment of DNA corresponding to the region downstream of the EP1 promoter (Downs), the EP1 promoter itself (EP1-prom), and a polycistron containing sequence encoding the Tet repressor protein and the neomycin resistance marker (Tet-Neo; see Table S2 in the supplemental material for all primer sequences). To create pSPR1, the Ups fragment was digested with AvaI/BglII, Downs was digested with SphI/SacI, and both were ligated, together with a linker made by annealing linker_F and linker_R, into pSPR0-cut AvaI/SacI. To create pSPR2, the EP1-prom fragment was digested with HindIII/BamHI, Tet-Neo was digested with BglII/SphI (partial, with selection for not cutting the internal SphI site), and both were ligated with pSPR1-cut HindIII/SphI. The identity of pSPR2 was confirmed by sequencing, and the insert was removed by SacI digestion and transfected into Lister 427 strain procyclic cells using standard electroporation protocols (48). Sequences and maps for all vectors are available at www.wicksteadlab.co.uk/.
SiMP1 cells were grown at 28°C in SDM79 medium (5) supplemented with 10% fetal bovine serum. Bloodstream-form Lister 427 and 90-13 (48) cells were grown in HMI-9 medium supplemented with 15% fetal bovine serum at 37°C and 5% CO2 (17). SiMP1 cells were cloned by limiting dilution on 96-well plates. Bloodstream-form growth was assessed by diluting the cells to 5 × 104 cells ml−1 and counting and rediluting them 24 h later during a period of 120 h. Procyclic cells were diluted to 2 × 105 cells ml−1, counted after 24 h, recounted after another 24 h, and rediluted during a period of 144 h. Tests were performed using the statistical programming package R (The R Project for Statistical Computing; www.r-project.org).
Endogenous locus tagging/truncation and ectopic expression of chimeric proteins.
The following strategy was used for the generation of cells with the N-terminal sequence of one endogenous allele of RPA2 replaced with a tag of the TY epitope (2) and enhanced yellow fluorescent protein (YFP). Fragments of 320 to 430 bp of the 5′ end of the targeted coding region and the 3′ end of the upstream intergenic region of the RPA2 gene (Tb11.03.0450) were amplified by PCR, and the fragments were ligated together into the endogenous locus-tagging vector pEnT5-Y (25) using the XbaI and BamHI restriction sites. Thus, the vectors pEnT5-Y:RPA2, pEnT5-Y:rpa2Δ1-113, pEnT5-Y:rpa2Δ1-172, and pEnT5-Y:rpa2Δ1-182 were generated. To add the T. brucei La protein NLS (RGHKRSRE) (31) to the N terminus of these chimeric proteins, DNA oligonucleotides with the relevant restriction sites, a start codon, codons coding for the La NLS optimal for T. brucei codon usage (18), and the TY epitope codons were designed. The oligonucleotides were annealed, phosphorylated, and ligated between the HindIII and SpeI sites of pEnT5-Y:rpa2Δ1-172, replacing the TY coding sequence with NLS-TY and generating pEnT5-Y:NLS:rpa2Δ1-172. pEnT5-Y:RPA2 and pEnT5-Y:NLS:rpa2Δ1-172 were then cut with SpeI and XhoI. The resulting fragment containing the YFP open reading frame (ORF) and the RPA2 targeting sequence was ligated between the SpeI and XhoI sites of the pEnT5-Y:NLS:rpa2Δ1-172 backbone fragment, thus generating pEnT5-Y:NLS:RPA2. These vectors were linearized at the XhoI restriction site and transfected into SiMP1 or bloodstream-form Lister 427 cells by following standard electroporation protocols (48). Stable transformants were selected for with 10 μg ml−1 (bloodstream form) or 50 μg ml−1 (procyclic form) hygromycin (Sigma).
For the generation of 90-13 bloodstream-form cell lines expressing the TY-YFP-RPA2N1-172 fusion protein, a DNA fragment coding for amino acids 2 to 172 of RPA2 followed by a stop codon was generated by PCR, and the product was ligated into pDEX577-Y (25) utilizing the XbaI and BamHI restriction sites and generating pDEX577-Y:RPA2N1-172. This vector and pDEX577-Y were linearized using NotI and transfected into 90-13 bloodstream-form cells by following standard procedures. Cells were selected for with 1 μg ml−1 phleomycin (Melford Laboratories). The expression of TY-YFP-RPA2N1-172 and TY-YFP-TY was induced by adding doxycycline (Sigma) to a final concentration of 1 μg ml−1.
Generation of single-allele knockouts and multiplex PCR.
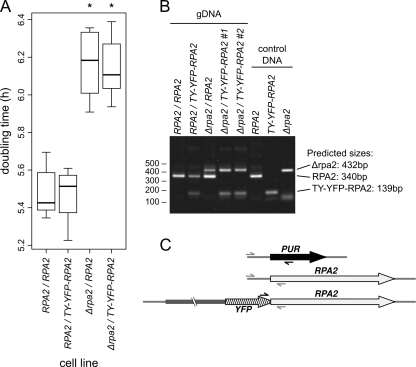
Amplicons encompassing 306 bp of the intergenic sequence directly upstream of the RPA2 coding sequence and 570 bp directly downstream were generated by PCR using the primer pairs GGAATTCCAGTTGTGAGGTTGTTGCGA and CTGGATCCGTTTGCACGCTTCACGC and also GCTCTAGA TAAGGAGGCAACTCAGA and TGGAGCTC TTTATTATATTTTATTTGCATTTTCCGG. These were digested with EcoRI/BamHI and XbaI/SacI, respectively, and ligated directly to BamHI/XbaI-cut puromycin resistance marker coding sequence amplified from pEnT6P-G (25) with CAGGATCCATGACCGAGTACAAGCC and CATCTAGATTAGGCACCGGGCTT. This DNA fragment was cloned into pBluescriptII SK(+) (Stratagene) and digested with EcoRI/SacI. The resultant vector was cut with EcoRI/SacI to release the puromycin resistance marker flanked by the RPA2 untranslated region (UTR), and this was transfected into Lister 427 bloodstream-form cells as described above. Multiplex PCR used the primers CAGTTGTGAGGTTGTTGCGA, AAGCCGAACCTACAACGG, GTCTGGATCGACGGTGT, and TCCGCCCTGAGCAAAGA, all at an equimolar nonsaturating concentration (100 μM).
Fluorescence microscopy and radial pixel intensity profiles.
T. brucei cells expressing fluorescent fusion proteins and control cells were washed in phosphate-buffered saline (PBS; 137 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) and allowed to settle onto glass (procyclic cells) or glutaraldehyde-derivatized silanized slides (bloodstream-form cells). Subsequently, cells were fixed for 10 min in 3% (wt/vol) formaldehyde. Cells then were permeabilized in −20°C cold methanol for at least 30 s. The slides were rehydrated in 0.25% (wt/vol) glycine in PBS and washed in PBS. Slides were then mounted in 1,4-diazabicyclo[2.2.2]octane (DABCO) medium containing 4′,6-diamidino-2-phenylindole (DAPI).
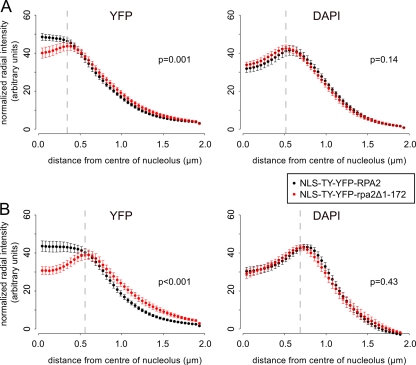
Radial pixel intensity profiles were extracted from images using the publicly available ImageJ software (National Institutes of Health; rsb.info.nih.gov) with the Radial Profile Plot plugin (rsb.info.nih.gov/ij/plugins/radial-profile.html). All profiles start from a pixel manually defined as the center of the nucleolus as assessed by eye from the DAPI signal. Plots shown in Fig. 8 show the mean intensities from several cells at increasing distances from the nucleolar center normalized to the total mean intensity for that cell line. The significance of divergence between curves was assessed by the Monte Carlo permutation (randomization) of the data sets using the sum square difference between the data as a test statistic. P values represent the proportion of 10,000 permutations that resulted in a sum square difference equal to or larger than that for the observed data. All tests were performed using the statistical programming package R.
Fig 8.
Analysis of radial fluorescence signal intensities of NLS-TY-YFP-RPA2 and NLS-TY-YFP-rpa2Δ1-172. Normalized radial intensities of YFP signal (left) and DAPI (right) measured from the center of the nucleolus in bloodstream-form (A) and procyclic (B) cells. Graphs represent means from 11 and 12 cells (A) and 10 and 11 cells (B) for NLS-TY-YFP-RPA2 and NLS-TY-YFP-rpa2Δ1-172, respectively. NLS-TY-YFP-rpa2Δ1-172 (red) is concentrated at the nucleolar periphery, while NLS-TY-YFP-RPA2 (black) is more homogeneously distributed in the nucleolus. Bars represent standard errors of the mean estimates. P values represent Monte-Carlo estimates from 10,000 random permutations. Dotted lines show the position of maximum intensity for the NLS-TY-YFP-rpa2Δ1-172 profiles.
Immunopurification of pol I complex.
For the purification of pol I complexes containing TY-YFP-tagged RPA2 derivatives, ∼8 × 109 procyclic-form cells expressing either NLS-TY-YFP-RPA2, NLS-TY-YFP-rpa2Δ1-172, or untagged RPA2 were harvested by centrifugation from mid-log-phase cultures. Cells were washed twice in ice-cold 300 mM glucose, 10 mM HEPES, pH 7.8, and then lysed by the addition of 2.4 ml of ice-cold HKMEG (150 mM KCl, 150 mM glucose, 25 mM HEPES, pH 7.8, 4 mM MgCl2, 1 mM EGTA) containing 1% (vol/vol) Nonidet P-40, 1 mM dithiothreitol, and protease inhibitors (2 mM 1,10-phenanthroline, 0.5 mM phenylmethanesulfonyl fluoride, 50 μM leupeptin, 7.5 μM pepstatin A, 5 μM E64-d), followed by vortexing for 2 min. Lysates were cleared by centrifugation at 20,000 × g for 30 min and then allowed to bind to affinity-purified rabbit anti-green fluorescent protein (GFP) polyclonal antibodies attached to paramagnetic beads (Dynabeads protein G; Invitrogen) for 2 h on ice. Beads were washed extensively in HKMEG containing 0.1% (vol/vol) Nonidet P-40, 0.5 mM dithiothreitol, and bound complex was subsequently eluted by incubating the beads in 100 mM glycine, pH 2.7. Eluates were separated by conventional SDS-PAGE and either stained with SYPRO ruby (Invitrogen) or transferred to nitrocellulose membranes for immunoblotting as described above. Immunoblottings were performed using either a mixture of mouse anti-GFP monoclonal antibodies 7.1 and 13.1 (Roche) or mouse monoclonal antibody 16B1a, which recognizes the largest subunit of pol I (33).
Mass spectrometry.
For the identification of isolated peptides, individual bands were excised from SYPRO ruby-stained gels. Gel fragments were washed with 50% acetonitrile in 50 mM NH4HCO3, pH 8.5, dehydrated in 100% acetonitrile, and air dried. Proteins were digested for 16 h with trypsin (Promega) at 20 μg ml−1 in 25 mM NH4HCO3, pH 8.5, at 37°C. Matrix-assisted laser desorption ionization tandem mass spectrometry (MALDI MS/MS) was performed on an ABI 4800 mass spectrometer at the University of Oxford Central Proteomics Facility (www.proteomics.ox.ac.uk). Masses were analyzed by the MASCOT search engine (Matrix Science) against a database of T. brucei predicted protein sequences (www.tritrypdb.org).
RESULTS
No homology between the trypanosome RPA2 N terminus and other proteins.
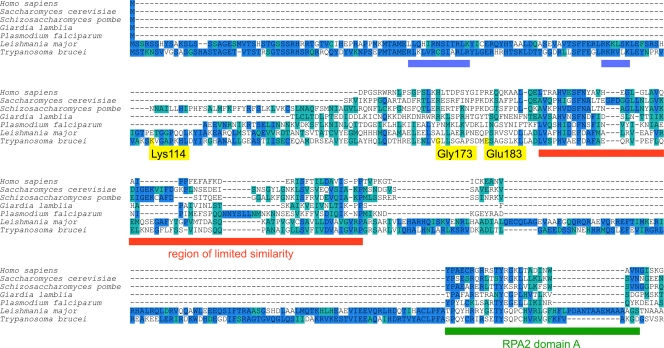
To test whether the extension of the trypanosome RPA2 N terminus is truly unique to these organisms, we built a multiple-sequence alignment from a data set of 28 eukaryotic RPA2 proteins. The alignment of N-terminal residues from a selection of organisms is shown in Fig. 1 (for a wider cross-section of organisms, see Fig. S1 in the supplemental material). The sequence N terminal to the conserved RPA2 domain A (marked with a green bar in Fig. 1) in T. brucei and Leishmania major contains >200 amino acids more than any of the other RPA2 proteins. Our alignment suggests that there follows a region of limited but identifiable similarity (marked with red bar in Fig. 1), implying that the sequence of the trypanosome RPA2 N terminus is the result of (i) an extension event (N terminal of the region of limited similarity) and (ii) an insertion event (between the region of limited similarity and domain A). Homologous RPA2 N-terminal extensions were found in the sequences of related trypanosomatids from publicly available sources (www.genedb.org; also see Fig. S2 and Table S3 in the supplemental material).
Fig 1.
Multiple-sequence alignment of the RPA2 N terminus. A multiple-sequence alignment was generated from a data set of 16 eukaryotic RPA2 proteins, 7 of which were selected for display in this figure. Residues are shown with a blue or cyan background indicating either identity or similarity to the majority consensus, respectively. Yellow flags on the T. brucei sequence indicate the residues tagged in this study. Blue bars mark stretches rich in basic residues in the trypanosomatid homologues. The red bar marks the region of limited similarity; the green bar marks RPA2 domain A.
To test whether sequence homologous to the trypanosome N terminus can be found elsewhere in the trypanosome genome or in nontrypanosomatid organisms, a profile-hidden Markov model was built from the T. brucei and L. major N termini and used to search 28 predicted eukaryotic proteomes. This recovered only RPA2 orthologues in trypanosomatids. In contrast, a profile-hidden Markov model of similar length built from sequence elsewhere in RPA2 unambiguously identified 16 of the 28 homologues without iteration. These data suggest that the RPA2 protein N-terminal extension is specific to trypanosomatids and is not found in other sequences, including in trypanosomal RPB2 and RPC128, which are the pol II- and pol III-specific paralogues of RPA2.
Homology models built by aligning trypanosome RPA2 to the second-largest subunit of budding yeast RNA polymerases with published structures (7, 29) predict that the trypanosome RPA2 sequence N terminal to the region of limited similarity is likely to protrude from the pol I core structure (data not shown) and therefore is likely to be available for interactions with other complexes.
The RPA2 N terminus is required for nuclear localization.
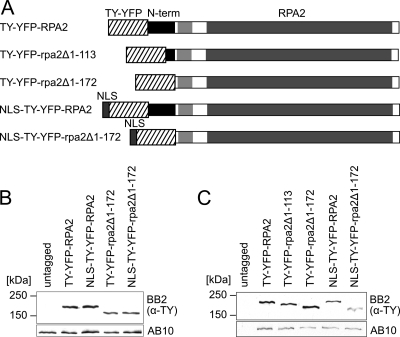
To test whether the T. brucei RPA2 N terminus is required for either nuclear or subnuclear localization, we designed a strategy to replace the N terminus of one endogenous RPA2 allele with a tag. We generated bloodstream-form cells expressing either full-length RPA2 or rpa2 missing residues 1 to 172 with an N-terminal TY epitope (2) and YFP tag (Fig. 1). Residues 1 to 172 were chosen because they are just N terminal of the region of limited similarity and therefore correspond to the first trypanosome-specific extension. The tagging of a similar position of Saccharomyces cerevisiae RPA2 (RPA135) has been used previously to isolate functional pol I complex (39, 41). A schematic representation of fusion proteins TY-YFP-RPA2 and TY-YFP-rpa2Δ1-172 is shown in Fig. 2A. Successful integration and expression was established by genomic PCR (see Fig. S2 in the supplemental material) and immunoblotting (Fig. 2B).
Fig 2.
Expression from endogenous loci of N-terminally truncated RPA2 tagged with TY-YFP or NLS-TY-YFP. (A) Schemes of chimeric proteins expressed. The region of limited similarity to nontrypanosomatid RPA2 homologues is indicated by a light gray box. Also shown are Western blottings of lysates from bloodstream-form (B) and procyclic (C) cells expressing RPA2 fusion proteins using monoclonal antibody BB2 (anti-TY-tag). Monoclonal antibody AB10 was used as a loading control.
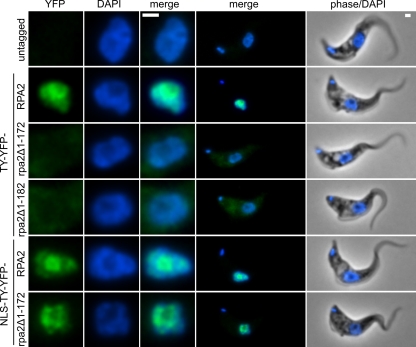
In bloodstream-form trypanosomes, TY-YFP-RPA2 localized to the nucleus with the major signal from the nucleolus, which can be clearly seen as a region of low DNA density of ∼0.5 μm in diameter in the nucleus (Fig. 3). A majority of cells (67% ± 5%; 68/102 nonmitotic cells) additionally displayed a single site of strong extranucleolar signal corresponding to the pol I-containing VSG ESB (Fig. 4A). This site often colocalized with a small nuclear region that was low in DAPI signal, most likely reflecting the transcriptionally permissive chromatin structure of the active ES (13, 42). As previously described (33), in a minority of cells it is not possible to unambiguously distinguish the ESB from the greater nucleolar pool of pol I because of the proximity of the two.
Fig 3.
Localization of N-terminal RPA2 truncations in bloodstream-form cells. While TY-YFP-RPA2 localizes to the nucleus, TY-YFP-rpa2Δ1-172 is excluded. NLS-TY-YFP-RPA2 is nuclear but has a subnuclear localization different from that of NLS-TY-YFP-rpa2Δ1-172. YFP fluorescence is pseudocolored in green, DAPI in blue. Bars, 1 μm.
Fig 4.
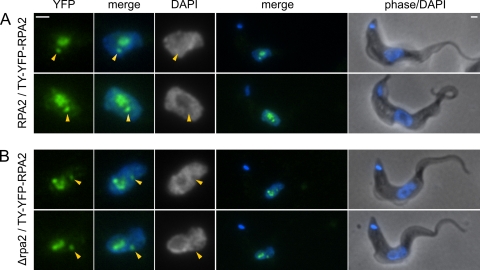
Localization of TY-YFP-RPA2 in bloodstream-form cells. Cells containing one tagged allele and a second which is either wild type (A) or a knockout (B) are shown. Localization is unaffected by the absence of untagged protein, showing incorporation into both the nucleolus and the ESB (arrows). YFP fluorescence is pseudocolored in green, DAPI in blue. Bars, 1 μm.
The fusion protein produced by the N-terminal TY-YFP tagging of full-length RPA2 is still competent for incorporation and function in the pol I complex, both in its nucleolar context and in the ESB. This was demonstrated by the knockout of the wild-type allele to produce cells containing only tagged protein (Fig. 4B and 5). The tagging of one allele of RPA2 has no detectable effect on the growth of bloodstream-form cells (Fig. 5A). However, heterozygous Δrpa2/RPA2 bloodstream-form cells show slowed growth relative to that of wild-type or tagged cells (Fig. 5A) (P < 0.01 by Student's t test), which was unchanged on the tagging of the remaining allele (Δrpa2/TY-YFP-RPA2) (P = 0.81 by Student's t test). The correct integration of constructs and the absence of extra copies of RPA2 was confirmed by diagnostic PCR from the genomic DNA of transformants (Fig. 5B and C). Importantly, the localization of TY-YFP-RPA2 is unchanged by the absence of untagged RPA2 (Fig. 4B), showing ESB structures in 72% ± 4% (81/113) of nonmitotic cells. Since trypanosomes cannot grow without a functional pol I enzyme and bloodstream-form cells require VSG mRNA (40), these data show that TY-YFP-RPA2 is a functional pol I subunit and that the localization seen for this tagged protein is representative of the wild-type complex.
Fig 5.
Growth rates of T. brucei cells expressing only N-terminally tagged RPA2 protein. (A) Tukey boxplot of doubling times for bloodstream-form cells expressing TY-YFP-RPA2 from one locus in cells which are either wild type (RPA2) or knocked out (Δrpa2) at the other allele. Cells with haploinsufficiency caused by single-allele knockout grow more slowly (*, P < 0.01 by t test), but there are no significant differences between tagged and untagged cell lines; n = 6 for each boxplot. Bars indicate medians, boxes indicate interquartile ranges (IQR), and whiskers indicate ranges of measurements within 1.5× IQR. (B) Diagnostic multiplex PCR to confirm genotypes of cell lines. The presence of RPA2, TY-YFP-RPA2, or Δrpa2 results in a specific band of the size shown. Lanes labeled Δrpa2/TY-YFP-RPA2#1 and Δrpa2/TY-YFP-RPA2#2 are independent clones of these cell lines. (C) Schematic showing the amplicons generated from the multiplex PCR around the N terminus of the genes of interest.
In contrast to functional TY-YFP-RPA2 protein, TY-YFP-rpa2Δ1-172 was distributed throughout the cytoplasm and appeared to be completely excluded from the nucleus (Fig. 3). Equivalent results were obtained with cells expressing TY-YFP-rpa2Δ1-182, which lacks 10 more amino acids than TY-YFP-rpa2Δ1-172 (Fig. 3). These results showed that amino acids 1 to 172 of RPA2 are required for nuclear localization in T. brucei. To assess if this was an effect specific to bloodstream-form parasites, we also generated procyclic cell lines expressing tagged and truncated RPA2 fusion proteins from one of their endogenous alleles. We also generated a cell line expressing a fusion protein that had only 113 amino acids removed from the RPA2 N terminus, TY-YFP-rpa2Δ1-113 (Fig. 1). Successful integration was tested by genomic sequencing and immunoblotting (Fig. 2C; also see Fig. S3 in the supplemental material). In procyclic-form cells, as in bloodstream-form cells, TY-YFP-RPA2 localized predominantly to the nucleolus, whereas truncated forms TY-YFP-rpa2Δ1-113 and TY-YFP-rpa2Δ1-172 were entirely excluded from the nucleus (see Fig. S4 in the supplemental material). Hence, the N-terminal extension of RPA2 is required for the nuclear localization of this subunit in both bloodstream-form and procyclic cells.
The RPA2 N terminus is not sufficient for nuclear accumulation of a fusion protein.
One obvious explanation for the mislocalization of truncated RPA2 is that the N-terminal sequence contains a nuclear localization signal. Two small regions rich in basic residues are present within the N-terminal sequence of T. brucei RPA2 (blue bar in Fig. 1), potentially indicating an NLS in this region. However, these residues are poorly conserved in RPA2 sequences from other trypanosomatid species (see Fig. S2 in the supplemental material), which would not be expected if they form a necessary NLS.
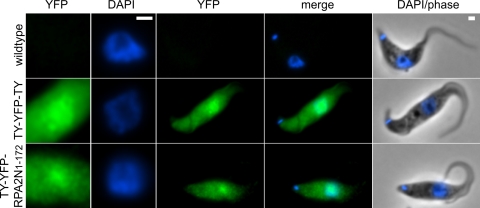
As a direct test of whether this sequence acts as a general NLS, we generated bloodstream-form cell lines conditionally expressing TY-YFP-RPA2N1-172 or TY-YFP-TY. Cells were induced for 48 h and then analyzed by fluorescence microscopy. TY-YFP-TY accumulated in cells to more than 10-fold the level of TY-YFP-RPA2N1-172. TY-YFP-TY was found throughout the cell body and also displayed a certain amount of accumulation in the nucleus (Fig. 6). The cellular distribution of TY-YFP-RPA2N1-172 was similar, but TY-YFP-RPA2N1-172 did not accumulate in the nucleus to any greater extent than TY-YFP-TY lacking the putative RPA2 NLS. Therefore, the N terminus of RPA2 is not an autonomous signal for nuclear localization.
Fig 6.
Cellular localization of ectopically expressed TY-YFP-RPA2N1-172 in procyclic cells. TY-YFP-RPA2N1-172 is not accumulated in the nucleus more than the control protein, TY-YFP-TY. Note that the YFP fluorescence of TY-YFP-TY was more than a magnitude higher than that for TY-YFP-RPA2N1-172, so they had to be processed differently for this figure. YFP fluorescence is pseudocolored in green, DAPI in blue. Bars, 1 μm.
Growth of bloodstream-form cells is slowed by N-terminal truncation of RPA2.
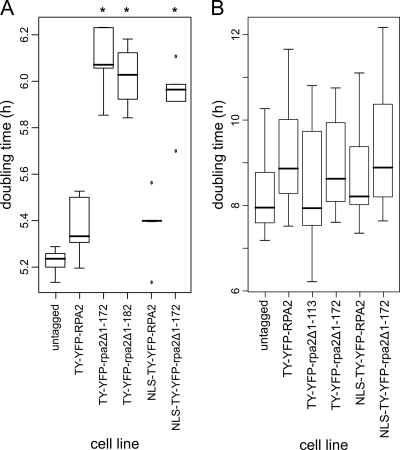
We showed above that the haploinsufficiency of RPA2 created by single-allele knockout caused slowed growth in bloodstream-form trypanosomes (Fig. 5). To assess the effect of the truncation of RPA2, we assessed the growth rate of bloodstream-form and procyclic cells expressing truncated proteins. While bloodstream-form cells expressing TY-YFP-RPA2 did not have a significantly different doubling time compared to that of wild-type cells (Fig. 5A and 6A), cells expressing either TY-YFP-rpa2Δ1-172 or TY-YFP-rpa2Δ1-182 grew with a significantly longer doubling time (∼0.8-h increase in doubling time; P < 0.01 by Student's t test) (Fig. 7A). Hence, the truncation of a single allele of RPA2 phenocopies haploinsufficiency created by complete gene knockout in bloodstream-form cells. In procyclic cells, no significant differences were seen between cells expressing wild-type, tagged, or truncated RPA2 from one of the two RPA2 loci (Fig. 7). This most likely reflects the different dependencies on pol I levels of these two life cycle stages (see Discussion).
Fig 7.
Growth rates of T. brucei cells expressing truncated RPA2 proteins. (A) Tukey boxplot of doubling times for bloodstream-form cells expressing truncated RPA2 proteins during 120 h. Cells expressing the truncated proteins grow at lower rates independently of the nuclear localization of the protein; n = 5 for each boxplot. Asterisks label cell lines with a doubling time significantly different from that of wild-type cells (P < 0.01 by t test). Bars, medians; boxes, interquartile ranges (IQR); whiskers, ranges of measurements within 1.5× IQR; dots, measurements outside the range of whiskers. (B) Tukey boxplot of doubling times for procyclic cell clones expressing tagged RPA2 proteins during 144 h. No significant differences between the growth rates of the cell lines were found (n = 6 for SiMP1 and TY-YFP-rpa2Δ1-172; n = 18 for others; t test was used to determine significance).
Nuclear localization of truncated RPA2 does not restore function.
To test if the RPA2 N terminus is only required for nuclear import or provides additional functions, we created bloodstream-form and procyclic cell lines which expressed TY-YFP-RPA2 and TY-YFP-rpa2Δ1-172 from their endogenous loci with an ectopic NLS at their N termini (NLS-TY-YFP-RPA2 and NLS-TY-YFP-rpa2Δ1-172). As an NLS, we chose the T. brucei La protein sequence RGHKRSRE, which has been shown previously to be sufficient for the accumulation of exogenous protein in the trypanosome nucleus (31). Correct integration was confirmed by genomic PCR and immunoblotting (Fig. 2A and B; also see Fig. S3 in the supplemental material).
In both bloodstream-form and procyclic cells, the La protein NLS was sufficient for the accumulation of truncated RPA2 in the nucleus (Fig. 3; also see Fig. S4 in the supplemental material). As for TY-YFP-RPA2, in a majority of bloodstream-form cells, NLS-TY-YFP-RPA2 forms ESB structures which are visibly distinct from the nucleolus (see Fig. S5 in the supplemental material). There were, however, consistent differences between the nuclear distribution of the full-length and truncated NLS-TY-YFP-tagged proteins, with NLS-TY-YFP-rpa2Δ1-172 apparently being less present in the center of the nucleolus than NLS-TY-YFP-RPA2 (Fig. 3; also see Fig. S4 in the supplemental material). To investigate these differences, we measured the signal intensities of the YFP fusion proteins and also DAPI signal in a number of cells radially from the center of the nucleolus outwards (Fig. 8). The nucleolus forms a local minimum in DAPI signal, and its border can be seen from the position of the maximum in the radial DAPI distribution. The distribution of full-length NLS-TY-YFP-RPA2 is relatively homogeneous throughout the nucleolus, reflecting its incorporation into nucleolar pol I complexes. However, in both bloodstream-form and procyclic cells, truncated NLS-TY-YFP-rpa2Δ1-172 is significantly less present in the nucleolus (P < 0.001 in permutation test), instead having maximum intensity at the nucleolar periphery (Fig. 8). Moreover, while bloodstream-form cells expressing NLS-TY-YFP-RPA2 grew at rates similar to those of untagged controls, the doubling time of NLS-TY-YFP-rpa2Δ1-172-expressing cells was significantly longer for bloodstream-form parasites (P < 0.01 by Student's t test) (Fig. 7) but indistinguishable from cells in which RPA2 was excluded from the nucleus. These results show that the RPA2 N terminus not only is necessary for nuclear import but also is required for full pol I function and subnuclear distribution of the protein once inside the nucleus.
The RPA2 N-terminal extension is required for assembly of stable pol I complex.
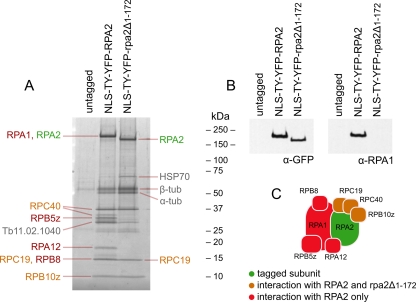
Since the N-terminal truncation of RPA2 phenocopies the effect of single-gene knockout (Fig. 5) with or without nuclear localization, we asked if this was a consequence of disruption in subnuclear localization or an inability to form functional polymerase complexes. To test this, we used the exogenous tag to immunoprecipitate the complexes formed by either full-length NLS-TY-YFP-RPA2 or NLS-TY-YFP-rpa2Δ1-172 and identified components by tandem mass spectrometry (Fig. 9). From cells expressing NLS-TY-YFP-RPA2, we isolated the 8-subunit core pol I complex isolated by Nguyen et al. by the tandem affinity purification of tagged RPA1 (34). This complex lacks RPB6z, RPB12, and p31, which are labile under the conditions used, but did show a strong interaction with the protein encoded by gene Tb11.02.1040. This protein is a predicted GTPase, and our analysis (not shown) suggests it is the trypanosome homologue of human GPN-loop GTPase 1 (GPN1/XAB1) and budding yeast NPA3, which have been found to interact with RNA pol II in these organisms (22, 44).
Fig 9.
Immunoprecipitation of complexes formed by tagged RPA2. (A) Purified complexes from cells expressing NLS-TY-YFP-RPA2, NLS-TY-YFP-rpa2Δ1-172, or untagged RPA2. Peptides have been separated by SDS-PAGE and stained with SYPRO ruby. The identity of the major component(s) of individual gel bands as determined by mass spectrometry is indicated next to each band. Colors indicate tagged subunit (green), pol I subunit interacting with tagged RPA2 and rpa2Δ1-172 (orange) or RPA2 only (red), or proteins that are not known components of pol I complex (gray). (B) Immunoblot of pol I precipitation showing the reaction of isolated peptides to antibodies raised against GFP or RPA1. (C) Schematic of the positions of the identified components of pol I complex on the S. cerevisiae complex (29). Colors are as described for panel A.
In contrast to the full-length protein, complexes formed by truncated NLS-TY-YFP-rpa2Δ1-172 contain only 3 other pol I subunits, RPC40, RPC19, and RPB10z (Fig. 9A). RPA1, RPB5z, RPA12, and RPB8, which are found associated with NLS-TY-YFP-RPA2, are absent from NLS-TY-YFP-rpa2Δ1-172 complexes. Levels of RPA1 in these truncation mutant complexes were so low that they were undetectable by Western blotting (Fig. 9B). Moreover, the missing subunits RPB5z, RPA12, and RPB8 all are predicted to interact directly with RPA1 based on the structure of yeast RNA polymerases (7, 29), while those still present interact directly with the RPA2 subunit. This suggests that the loss of the N-terminal region of RPA2 specifically disrupts the interaction between the largest and second-largest subunits of trypanosomal pol I.
DISCUSSION
Transcription by pol I in T. brucei is critical for the evasion of the mammalian immune response. Research during the last decade has described several trypanosome pol I-specific subunits which differ from their homologues in other systems by sequence insertions. Most dramatically, the second-largest subunit of pol I, RPA2, bears >200 additional residues at its N terminus, forming a region that is specific to trypanosomatids (38) and also to this polymerase complex (being absent from RPB2 and RPC128). The work described here tests whether this sequence is necessary for pol I function, either generally or in an ESB-specific context.
Our comparative analysis of trypanosome RPA2 sequences with a broad data set of eukaryotic RPA2 proteins suggests that the additional sequence is the result of both an insertion and an extension event near the N terminus. Homology modeling using the structural information available for the S. cerevisiae pol I and pol II complexes (7, 29) suggests that the RPA2 N terminus protrudes out of the pol I complex. It thus could provide a platform for interactions with other molecules, such as those involved in pol I biogenesis or transcription activation.
The trypanosomatid-specific RPA2 extension is required for the import of the protein into the nucleus. When we replaced the N-terminal 113, 172, or 183 residues of endogenous RPA2 with a tag, the fusion protein was excluded from the nucleus. This mislocalization was not an artifact of tagging, as tagged full-length RPA2 accumulated in the nucleus, localized to the nucleolus and ESB, and could fully replace the function of wild-type RPA2 with no detectable loss of fitness. Moreover, tags placed at the N terminus of RPA2 in budding yeast, which lacks the N-terminal extension found in trypanosomes, have been used to isolate functional pol I complex (39, 41).
In bloodstream-form cells, when one RPA2 allele is truncated, growth is significantly reduced. It is likely that TY-YFP-rpa2Δ1-172-expressing cells have only half the amount of functional RPA2 protein available in the nucleus that wild-type cells (or cells expressing tagged full-length RPA2) have. Studies of Schizosaccharomyces pombe pol II have shown that the three largest subunits are the least abundant pol II subunits (27). It seems plausible, therefore, that a 50% reduction of nuclear RPA2 crucially limits the number of pol I complexes and thus interferes with rRNA and/or VSG transcription. In agreement with this hypothesis, haploinsufficiency resulting from a clean knockout of a single allele of RPA2 resulted in the same growth rate decrease as N-terminal truncations. Interestingly, in S. cerevisiae the deletion of a short sequence (residues 5 to 15) in the N-terminal region of the second-largest subunit of pol III, RPC128, resulted in the exclusion of this protein from the nucleus and slow growth. As for trypanosomal RPA2 truncations, this functional impairment could not be cured by the addition of an ectopic NLS to the truncated sequence (15).
In contrast to the situation for bloodstream-form cells, no effect on growth was observed when RPA2 was truncated in procyclic cells, even though the resultant proteins still failed to access the nucleus. This difference might be explained by a lower requirement on pol I in procyclic cells, perhaps due to the higher levels of VSG required by bloodstream-form cells (21) compared to that for procyclin in insect-form cells (6). Bloodstream-form cells in which VSG production is depleted by RNAi are nonviable and very quickly arrest in their cell cycle (40), while culture-adapted procyclin-null procyclic mutants grow at wild-type rates (46). However, it may also be that the faster growth of bloodstream-form cells makes this life cycle stage more sensitive to reduced ribosome biosynthesis as a result of the depletion of nucleolar pol I.
Despite being required for nuclear localization, the RPA2 N-terminal sequence does not bear a classical NLS and does not act solely in nuclear import. The N-terminal 172 residues of RPA2 are not sufficient to confer nuclear localization on exogenous proteins. Our results cannot exclude the possibility of a complex nuclear localization signal that is only partially provided by RPA2N1-172. However, the nuclear import of the truncated protein by the addition of an ectopic NLS did not restore function or subnuclear localization, and the truncated protein was unable to form stable pol I complexes. This defect in complex formation suggests an alternative cause for mislocalization upon the removal of the N-terminal extension. If RPA2 accesses the nucleus not by an NLS in the N-terminal region but instead by virtue of an NLS on another pol I subunit with which it interacts, then the disruption of this interaction would also disrupt the nuclear import of RPA2. These two alternative scenarios are not separated by our data. It is clear, however, that the RPA2 N terminus does not act merely (if at all) as an NLS but is explicitly required to form functional pol I complexes in trypanosomes. This requirement identifies a trypanosomatid-specific component to either the assembly or stability of pol I in these organisms, as nontrypanosomatid organisms form pol I complexes without the need for an N-terminal extension to RPA2.
Since truncation disrupts the normal nucleolar localization of RPA2, it is difficult to unambiguously determine in these cells if truncated RPA2 is still recruited to the ESB. However, the truncation of RPA2 results in a complete failure to assemble stable pol I complexes with no detectable interaction with RPA1. This will disrupt the function of both nucleolar and ESB pol I regardless of potential recruitment to a particular subnuclear domain, and it is clear that the N-terminal extension does not function exclusively in ESB transcription.
Our data do not distinguish between defects in pol I assembly and defects in the stability of the assembled complex. In bacteria, the assembly of DNA-dependent RNA polymerase starts with the two α subunits and continues with the addition of first the β and then the β′ subunit (19). The stability of the subassembly of the pol II homologues of α2β in S. cerevisiae supported the view that the core assembly of the eukaryotic DNA-dependent RNA polymerases follows the same sequence (26). By extension, the equivalent assembly process for pol I would involve first RPC40 and RPC19 (the two α homologues), followed by RPA2 (β) and then the largest subunit RPA1 (β′) (30). Significantly, our RPA2 truncation mutants assemble complexes containing both RPC40 and RPC19, forming a pol I homologue of α2β. These complexes also contain RPB10z, suggesting that this subunit can also be incorporated directly into this subcomplex. Missing from the complexes are RPA1 and all subunits that interact directly with RPA1, indicating that it is the specific interaction between RPA2 and RPA1 that has a requirement for the RPA2 N-terminal extension in trypanosomes.
Our results with the N terminus of RPA2 provide the first evidence of functional significance for one of the sequence insertions seen in several pol I-specific subunits of trypanosomatids. They show that this protein region is required for nuclear import and subnuclear localization, either directly or through interaction with other subunits. They also demonstrate that this sequence is required for pol I function and for the stable interaction between RPA1 and RPA2 via a trypanosomatid-specific mechanism. However, the function of this region is not exclusive to the ESB but globally affects pol I complexes in trypanosomes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Wellcome Trust. J.-P.D. was supported by the EP Abraham Trust and the Studienstiftung des deutschen Volkes (German National Merit Foundation).
Predicted protein data sets were obtained from the sources specified in Table S4 in the supplemental material. We thank each of these organizations and the respective genome-sequencing projects for making sequence, gene model, and annotation data publicly available.
Footnotes
Published ahead of print 2 March 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bastin P, Bagherzadeh Z, Matthews KR, Gull K. 1996. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77:235–239 [DOI] [PubMed] [Google Scholar]
- 3. Berriman M, et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416–422 [DOI] [PubMed] [Google Scholar]
- 4. Borst P. 2002. Antigenic variation and allelic exclusion. Cell 109:5–8 [DOI] [PubMed] [Google Scholar]
- 5. Brun R, Jenni L. 1977. A new semi-defined medium for Trypanosoma brucei sspp. Acta Trop. 34:21–33 [PubMed] [Google Scholar]
- 6. Clayton CE, Mowatt MR. 1989. The procyclic acidic repetitive proteins of Trypanosoma brucei. Purification and post-translational modification. J. Biol. Chem. 264:15088–15093 [PubMed] [Google Scholar]
- 7. Cramer P, Bushnell DA, Kornberg RD. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863–1876 [DOI] [PubMed] [Google Scholar]
- 8. Das A, Li H, Liu T, Bellofatto V. 2006. Biochemical characterization of Trypanosoma brucei RNA polymerase II. Mol. Biochem. Parasitol. 150:201–210 [DOI] [PubMed] [Google Scholar]
- 9. Devaux S, et al. 2006. Characterization of RNA polymerase II subunits of Trypanosoma brucei. Mol. Biochem. Parasitol. 148:60–68 [DOI] [PubMed] [Google Scholar]
- 10. Devaux S, et al. 2007. Diversification of function by different isoforms of conventionally shared RNA polymerase subunits. Mol. Biol. Cell 18:1293–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14:755–763 [DOI] [PubMed] [Google Scholar]
- 12. Fèvre EM, Wissmann BV, Welburn SC, Lutumba P. 2008. The burden of human African trypanosomiasis. PLoS Negl. Trop. Dis. 2:e333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Figueiredo LM, Cross GAM. 2010. Nucleosomes are depleted at the VSG expression site transcribed by RNA polymerase I in African trypanosomes. Eukaryot. Cell 9:148–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Günzl A, et al. 2003. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell 2:542–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hardeland U, Hurt E. 2006. Coordinated nuclear import of RNA polymerase III subunits. Traffic 7:465–473 [DOI] [PubMed] [Google Scholar]
- 16. Hertz-Fowler C, et al. 2008. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One 3:e3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirumi H, Hirumi K. 1989. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75:985–989 [PubMed] [Google Scholar]
- 18. Horn D. 2008. Codon usage suggests that translational selection has a major impact on protein expression in trypanosomatids. BMC Genomics 9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishihama A. 1981. Subunit of assembly of Escherichia coli RNA polymerase. Adv. Biophys. 14:1–35 [PubMed] [Google Scholar]
- 20. Ivens AC, et al. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jackson DG, Owen MJ, Voorheis HP. 1985. A new method for the rapid purification of both the membrane-bound and released forms of the variant surface glycoprotein from Trypanosoma brucei. Biochem. J. 230:195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jeronimo C, et al. 2007. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell 27:262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kelly S, Wickstead B, Gull K. 2005. An in silico analysis of trypanosomatid RNA polymerases: insights into their unusual transcription. Biochem. Soc. Trans. 33:1435–1437 [DOI] [PubMed] [Google Scholar]
- 25. Kelly S, et al. 2007. Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol. Biochem. Parasitol. 154:103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kimura M, Ishiguro A, Ishihama A. 1997. RNA polymerase II subunits 2, 3, and 11 form a core subassembly with DNA binding activity. J. Biol. Chem. 272:25851–25855 [DOI] [PubMed] [Google Scholar]
- 27. Kimura M, Sakurai H, Ishihama A. 2001. Intracellular contents and assembly states of all 12 subunits of the RNA polymerase II in the fission yeast Schizosaccharomyces pombe. Eur. J. Biochem. 268:612–619 [DOI] [PubMed] [Google Scholar]
- 28. Kooter JM, Borst P. 1984. Alpha-amanitin-insensitive transcription of variant surface glycoprotein genes provides further evidence for discontinuous transcription in trypanosomes. Nucleic Acids Res. 12:9457–9472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuhn C, et al. 2007. Functional architecture of RNA polymerase I. Cell 131:1260–1272 [DOI] [PubMed] [Google Scholar]
- 30. Kwapisz M, Beckouët F, Thuriaux P. 2008. Early evolution of eukaryotic DNA-dependent RNA polymerases. Trends Genet. 24:211–215 [DOI] [PubMed] [Google Scholar]
- 31. Marchetti MA, Tschudi C, Kwon H, Wolin SL, Ullu E. 2000. Import of proteins into the trypanosome nucleus and their distribution at karyokinesis. J. Cell Sci. 113:899–906 [DOI] [PubMed] [Google Scholar]
- 32. Martínez-Calvillo S, Saxena A, Green A, Leland A, Myler PJ. 2007. Characterization of the RNA polymerase II and III complexes in Leishmania major. Int. J. Parasitol. 37:491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Navarro M, Gull K. 2001. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature 414:759–763 [DOI] [PubMed] [Google Scholar]
- 34. Nguyen TN, Schimanski B, Zahn A, Klumpp B, Günzl A. 2006. Purification of an eight subunit RNA polymerase I complex in Trypanosoma brucei. Mol. Biochem. Parasitol. 149:27–37 [DOI] [PubMed] [Google Scholar]
- 35. Nguyen TN, Schimanski B, Günzl A. 2007. Active RNA polymerase I of Trypanosoma brucei harbors a novel subunit essential for transcription. Mol. Cell. Biol. 27:6254–6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pays E. 2005. Regulation of antigen gene expression in Trypanosoma brucei. Trends Parasitol. 21:517–520 [DOI] [PubMed] [Google Scholar]
- 37. Rudenko G, Bishop D, Gottesdiener K, Van der Ploeg LH. 1989. Alpha-amanitin resistant transcription of protein coding genes in insect and bloodstream form Trypanosoma brucei. EMBO J. 8:4259–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schimanski B, et al. 2003. The second largest subunit of Trypanosoma brucei's multifunctional RNA polymerase I has a unique N-terminal extension domain. Mol. Biochem. Parasitol. 126:193–200 [DOI] [PubMed] [Google Scholar]
- 39. Schneider DA, Nomura M. 2004. RNA polymerase I remains intact without subunit exchange through multiple rounds of transcription in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 101:15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sheader K, et al. 2005. Variant surface glycoprotein RNA interference triggers a precytokinesis cell cycle arrest in African trypanosomes. Proc. Natl. Acad. Sci. U. S. A. 102:8716–8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shou W, et al. 2001. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell 8:45–55 [DOI] [PubMed] [Google Scholar]
- 42. Stanne TM, Rudenko G. 2010. Active VSG expression sites in Trypanosoma brucei are depleted of nucleosomes. Eukaryot. Cell 9:136–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stewart M, et al. 2010. Processing of a phosphoglycerate kinase reporter mRNA in Trypanosoma brucei is not coupled to transcription by RNA polymerase II. Mol. Biochem. Parasitol. 172:99–106 [DOI] [PubMed] [Google Scholar]
- 44. Tarassov K, et al. 2008. An in vivo map of the yeast protein interactome. Science 320:1465–1470 [DOI] [PubMed] [Google Scholar]
- 45. Vanhamme L, Pays E, McCulloch R, Barry JD. 2001. An update on antigenic variation in African trypanosomes. Trends Parasitol. 17:338–343 [DOI] [PubMed] [Google Scholar]
- 46. Vassella E, Bütikofer P, Engstler M, Jelk J, Roditi I. 2003. Procyclin null mutants of Trypanosoma brucei express free glycosylphosphatidylinositols on their surface. Mol. Biol. Cell 14:1308–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Walgraffe D, et al. 2005. Characterization of subunits of the RNA polymerase I complex in Trypanosoma brucei. Mol. Biochem. Parasitol. 139:249–260 [DOI] [PubMed] [Google Scholar]
- 48. Wirtz E, Leal S, Ochatt C, Cross GA. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- 49. Young R, et al. 2008. Isolation and analysis of the genetic diversity of repertoires of VSG expression site containing telomeres from Trypanosoma brucei gambiense, T. b. brucei and T. equiperdum. BMC Genomics 9:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.