Abstract
The Candida albicans MYO5 gene encodes myosin I, a protein required for the formation of germ tubes and true hyphae. Because the polarized growth of opaque-phase cells in response to pheromone results in mating projections that can resemble germ tubes, we examined the role of Myo5p in this process. We localized green fluorescent protein (GFP)-tagged Myo5p in opaque-phase cells of C. albicans during both bud and shmoo formation. In vegetatively growing opaque cells, Myo5p is found at sites of bud emergence and bud growth, while in pheromone-stimulated cells, Myo5p localizes at the growing tips of shmoos. Intriguingly, cells homozygous for MTLa in which the MYO5 gene was deleted failed to switch efficiently from the white phase to the opaque phase, although ectopic expression of WOR1 from the MET3 promoter can convert myo5 mutants into mating-competent opaque cells. However, when WOR1 expression was shut off, the myo5-defective cells rapidly lost both their opaque phenotype and mating competence, suggesting that Myo5p is involved in the maintenance of the opaque state. When MYO5 is expressed conditionally in opaque cells, the opaque phenotype, as well as the mating ability of the cells, becomes unstable under repressive conditions, and quantitative real-time PCR demonstrated that the shutoff of MYO5 expression correlates with a dramatic reduction in WOR1 expression. It appears that while myosin I is not directly required for mating in C. albicans, it is involved in WOR1 expression and the white-opaque transition and thus is indirectly implicated in mating.
INTRODUCTION
Under different environmental conditions, the human pathogen Candida albicans is able to grow in different morphological forms. In response to changes in nutrient status, temperature, and pH, for example, Candida cells can switch from the yeast growth form to pseudohyphae or true hyphae, and this well-studied switch has led to the classification of C. albicans as dimorphic. However, C. albicans can exist in other morphological forms. A second well-studied morphological transition is the white-opaque switch. White and opaque cells are easily distinguished under the microscope; white-form cells are of the classic ovoid yeast shape, and opaque-form cells are much more elongated (2). On agar plates, white cells form domed colonies of a whitish color, while opaque cells form colonies that are flattened and greyish (2). Multiple environmental stimuli that slow cell growth lead to an increase in white-to-opaque switching in C. albicans (1). The opaque phase is stable at 25°C; however, upon shifting of the temperature to 30 to 37°C, cells switch en masse to the white phase (47).
More than 450 genes are modulated in expression during the white-opaque transition (26), and recently a master regulator of white-opaque switching, the WOR1 gene (22, 46, 54), has been identified. The data suggest that Wor1p is autoregulated. That means that once Wor1p expression is established, it tends to remain on, with a positive-feedback loop keeping the cells in the opaque state (54). In addition there is chromatin level regulation; several chromatin-modifying enzymes, such as Hst2p, Set3, and Hos2, are involved in the modulation of white-opaque switching (20).
The white-opaque transition plays a key role in the mating of C. albicans. The mating type locus of C. albicans controls the white-opaque transition. Only cells of the a or α mating type are permissive for switching, and only opaque-form cells are able to mate with high efficiency. C. albicans cells are diploid and have never been observed to undergo meiosis; however, the morphological changes that opaque cells undergo in response to pheromone are similar to those seen in Saccharomyces cerevisiae haploid cells. Mating-competent cells form mating projections in response to pheromone, and although these forms have been termed shmoos in both cell types, C. albicans can generate very long hypha-like conjugation tubes, while S. cerevisiae forms only short projections (29, 49, 53). Although the variety of cellular forms exhibited by C. albicans can be triggered by distinct cellular processes, in each instance polarized growth events must take place. Polarized growth is directed by the actin cytoskeleton, which consists of actin patches and actin cables (31, 42, 43). The myosin family proteins appear to interact with actin filaments in different ways to promote polarized growth in yeasts (8). In particular, the myosin type I protein encoded by MYO5 in C. albicans is required for the organization and polarized distribution of cortical actin patches. Myosin I is found to be colocalized with actin patches at the bud and hyphal tips, and a myosin I-null mutant shows impaired polarized growth and is incapable of hyphal formation (38). Genetic studies have revealed that myosin I functions in many actin-based processes, in particular endocytosis and exocytosis (17, 33, 37, 42, 43). In C. albicans, it has been observed that the ability to form hyphae is strictly correlated with MYO5 function in endocytosis (37).
Because shmoo formation in C. albicans involves long polarized mating projections, we were interested in the potential role of Myo5p in this process. Intriguingly, in our study we found that a simple assessment of the role of Myo5p in shmoo formation was not possible, because cells lacking MYO5 fail to switch from the white phase to the opaque phase. However, myo5-defective cells are able to switch to the opaque state and mate if the WOR1 gene is overexpressed. Opaque cells with MYO5 conditionally expressed rapidly convert to the white form and lose mating competence when MYO5 expression is shut off. The drop in MYO5 expression reduces WOR1 expression as well, suggesting that MYO5 operates upstream of WOR1 gene expression in the white-opaque switching pathway.
MATERIALS AND METHODS
Strain construction.
The strains used in this study are listed in Table 1. C. albicans strains were transformed by the one-step lithium acetate-polyethylene glycol transformation protocol (18). The strain containing MYO5-GFP (CaNK02) was constructed by transforming strain 3294 (MTLa/MTLa) (32) with BglII-cleaved pVEC-MYO5-GFP (Table 2) (38) and selecting for Ura+ recombinant clones. Strains CaNK03 (MTLα/MTLα CDC12-GFP) and CaNK01 (MTLα/MTLα MYO5-GFP) were derived from the MTLa/MTLα CDC12-GFP strain (U. Oberholzer, unpublished data) (created by inserting plasmid pVEC CDC12-URA3 into the CDC12 site of strain CaI-4) and the MTLa/MTLα MYO5-GFP strain, respectively (38), by growth on a medium containing 2% (wt/vol) sorbose as the sole carbon source at low density for 7 days at 30°C to select for the loss of one copy of chromosome 5, the chromosome containing the MTL locus (24). Homozygotes of specific mating types were verified by PCR with appropriate flanking primers.
Table 1.
C. albicans strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| 3294 | MTLa Δura3::imm34/Δura3::imm434his1/his1arg5,6/arg5 | 32 |
| 3315 | MTLα trp1/trp1 lys2/lys2 | 32 |
| CaNK01 | CaI-4 MTLα MYO5/myo5::pVEC MYO5-GFP-URA3 | This study |
| CaNK02 | 3294 MYO5/myo5::pVEC MYO5-GFP-URA3 | This study |
| CaNK03 | CaI-4 MTLαCDC12/cdc12::pVEC CDC12-GFP-URA3 | This study |
| CaNK06 | BWP17 MTLaNOP1/NOP1::YFP-HIS1 | |
| MYO5/myo5::pVEC MYO5-GFP-URA3 | This study | |
| CaNK07 | 3294 MYO5/myo5::hisG URA3 hisG | This study |
| CaNK08 | 3294 MYO5/myo5::hisG | This study |
| CaNK09 | 3294 myo5::hisG/myo5::hisG | This study |
| CaNK10 | 3294 myo5Δ/myo5ΔRp10/rp10::pRZ25 MET3-WOR1 | This study |
| CaNK11 | 3294 myo5::hisG/URA3-MET3-MYO5 | This study |
| CaNK12 | 3294 pVEC | This study |
| CaNK17 | 3294 sla2Δ871-1789::URA3/sla2Δ871-1789::HIS1 | This study |
Table 2.
Plasmids used in this study
Strain CaNK06, carrying both MYO5-GFP and NOP1-YFP, was generated from strain 6449, a BWP17 derivative containing a NOP1-YFP fusion (14). The MYO5-GFP allele was integrated into strain 6449 as described above. Positive transformants were screened by fluorescence microscopy and were confirmed by PCR. To obtain MTL isogenic derivatives, cells of strains containing fluorescent protein sequences were sorbose selected (see above). To create a mutant in which both copies of the MYO5 gene were deleted (CaNK09), strain 3294 was first transformed with pU21 (38), which had been digested with KpnI and SacI. Positive transformants derived from strain 3294 were then plated on a medium containing 5-fluoroorotic acid. The Ura− strains thus obtained were analyzed by PCR for the loop-out event that occurs by homologous recombination between the hisG direct repeats flanking the URA3 gene. The MYO5/myo5::hisG strain produced (CaNK08) was again transformed with pU21 digested with KpnI and SacI. Ura+ transformants were analyzed similarly for the correct integration event resulting in the disruption of the second MYO5 allele with the hisG::URA3::hisG blaster. Positive myo5::hisG/myo5::hisG::URA3::hisG transformants were analyzed for the correct loop-out event by plating on 5-fluoroorotic acid as described above. The Δmyo5/Δmyo5 (Δ/Δ myo5) mutant was transformed with pRZ25 (containing the MET3-WOR1 construct), which had been linearized with NcoI to direct integration to the RP10 locus (Table 2) (54).
A conditional knockout of MYO5 (CaNK11) was generated in the CaNK08 strain heterozygous for the deletion of MYO5 by insertion of the MET3 promoter before the ATG start codon of the remaining MYO5 sequence. Transformation cassettes were generated by PCR using a gene-specific primer pair from pFA carrying the MET3-URA3 cassette (Tables 2 and 3). A strain carrying the SLA2 loss-of-function mutation (sla2Δ871-1789) was constructed by the SLA2 disruption cassettes produced by PCR from pGEM carrying the URA3 and HIS1 cassettes, respectively (Tables 2 and 3). These disruption cassettes replace the SLA2 open reading frame (ORF) between nucleotides 871 and 1879.
Table 3.
PCR primers used in this study
| Primer | Sequence (5′–3′)a | Source |
|---|---|---|
| MYO5-Ca MET3F1 | TTTCTTCTTCTTTTATGACAAACATTTAAGATTTAATTCACATTTCCATTCATCAACTAATTAAACCTCTAGTCATTATCATTATTTATTTCATTtaatcgctctggcgcaggtgctt | This study |
| MYO5-CaMET3R1 | CGGCTTTTTTGATGCCACCACTACTACCACACCCAGAAGATTTGGCAGGTACTTGTTGTTGTTTTGTTTTGGTACGACCACCTCTTTTCACAATAGCCATtctgatatcatcgatgaattcgag | This study |
| SLA2 F1 | ACCATTACTCCCCCTAATATATTGTTAAACACTTTGCCCCCCCCCCCCCCCCTTAGCCCAtgtggaattgtgagcggata | This study |
| SLA2 F2 | AGAGAGACAGAAGAAATGAACACAACGAAAAAAAGAGCTCCGCCTATTAACTTATAACATACCATTACTCCCCCTAATA | This study |
| SLA2 R1 | GTGAATTATGGGTGGAAACAGAATATTATGATGAATGGAAACAAAGAATTCAAATACTTGgttttcccagtcacgacgtt | This study |
| SLA2 R2 | TCCCAAAATTGGCAAATTTATCATTAAAACAAGTACTTAATCCTAAAATTAGTGAATCTTGTGAATTATGGGGTGGAAACA | This study |
| ACT1 Fb | GAAGCCCAATCCAAAAGAGGT | 4 |
| ACT1 Rb | CTTCTGGAGCAACTCTCAATTC | 4 |
| WOR1 Fb | TGAACCCATGCCAATGACTA | This study |
| WOR1 Rb | GCTGCTGCAGTTGATGTTGT | This study |
| MYO5Fb | AACCGGCATATCCAATACCA | This study |
| MYO5Rb | TTGTAAGCTTGGTGCTGGTG | This study |
Capital letters indicate homology to genomic DNA. Lowercase letters indicate homology to plasmids pFA and pGEM.
Quantitative PCR primer.
Media and growth conditions.
For conversion from the white to the opaque phase, starting strains in the white phase were grown overnight at 30°C in liquid yeast extract-peptone-dextrose (YPD), and a 2-μl aliquot was diluted with sterile water and was plated at low density on several synthetic complete (SC) (pH 4.5) plates (44) supplemented with 2% glucose as the carbon source (SCD) and containing 0.0005% (wt/vol) phloxine B; these plates were incubated at 24°C. Opaque-phase cells were purified from dark purple colonies, and the unique opaque-phase cell phenotype was verified microscopically and by mating tests.
For each experiment, strains were routinely restreaked from the original −80°C glycerol stock on SCD-phloxine B plates, and the cell morphology of a fresh opaque colony was reconfirmed by microscopy. For transformations, strains in the white phase were grown to stationary phase in YPD medium at 30°C. SCD liquid medium supplemented with 100 μg/ml uridine was used to maintain strains in the opaque phase at room temperature. For the MET3 induction experiments, cells were grown on solid SCD medium plus uridine (SCD+Uri medium) lacking Met and Cys (SCD-MC). To repress the MET3 promoter, SCD+Uri medium was supplemented with 2.5 mM (each) Met and Cys (SCD+MC). Spider medium containing 1% nutrient broth, 1% mannitol, 0.4% potassium phosphate, and 100 μg/ml uridine was used for mating and shmoo formation assays.
Pheromone treatment.
Opaque cells from phenotypically homogeneous 7-day colonies of the a mating type were individually inoculated in liquid SCD medium and were grown at 24°C for 48 h in a rotary shaker (240 rpm). Cells were harvested at mid-log phase (5 ×106 to 1 ×107 cells/ml). Harvested cells were first pelleted and then resuspended in an equal volume of fresh liquid SCD medium containing 1 μg/ml α pheromone (13-amino-acid [aa] version) and were incubated at 24°C in a rotary shaker (240 rpm) for 2 to 4 h.
Mating analysis.
Mating strains in the opaque phase were grown at 24°C overnight in liquid SCD medium, and cells of each mating type at an optical density at 600 nm (OD600) of approximately 3 were mixed and deposited onto 0.8-μm-pore-size nitrocellulose filters by using a Millipore 1225 vacuum sampling manifold. The filters were incubated on the surfaces of plates containing Spider medium at 24°C. Cells were collected from the filters in 5 to 7 and 16 h and were analyzed microscopically.
Patch mating assay.
Horizontal streaks of the strains under investigation were made using toothpicks on YPD plates. On separate plates, the tester α strain was streaked vertically. After 2 days' growth, the tester strains and the experimental strains were replica plated together on the surface of a fresh YPD plate and were incubated for 48 h at 24°C. After mating, this plate was again replica plated onto a selective medium so that only “tetraploids” would be able to grow. Confluent growth at any intersection was scored as efficient mating, while no growth at an intersection was scored as no mating (7).
Fluorescent staining.
For FM4-64 staining, 1 μl of a stock solution (1.64 mM in dimethyl sulfoxide [DMSO]) was added directly to 1 ml of culture, and the culture was then incubated for a further 10 min. Cells were harvested by brief centrifugation and were washed with 1× phosphate-buffered saline (PBS). Microscopic imaging was performed immediately (11).
Fluorescence microscopy.
For all types of fluorescence, cells were mounted on slides in PBS buffer (pH 6.5) or in SCD medium (pH 6.5) for visualization with the appropriate filter by epifluorescence microscopy using an upright Leitz Aristoplan microscope with a 100× immersion oil objective and a 10× projection lens.
Video microscopy.
Overnight opaque cell cultures of strains CaNK03 (MTLα CDC12-GFP) and CaNK06 (MTLa NOP1-YFP MYO5-GFP) were mixed at a ratio of 1:1, pelleted, and then placed in fresh SDC medium diluted 1:1 with water-agarose containing 2% agarose. The sample was mounted on the slide and was covered with a coverslip. Live video microscopy was carried out at room temperature using a Leica DM-IRB inverted microscope equipped with a Ludl motorized stage and a SenSys charge-coupled device camera. Pictures were taken with a 63× objective and a 10× projection lens every 10 min at 10 different locations.
Quantitative real-time PCR.
Cells of strain CaNK11 in the opaque phase were plated onto SCD-MC or SCD+MC plates. After 5 days of growth at 24°C, 3 opaque colonies from the −MC plate and 3 colonies each of wrinkled and smooth phenotypes from the +MC plate were selected and checked for the cell phenotype. Cultures were inoculated from a fresh colony and were grown overnight in SCD-MC or SCD+MC medium at 24°C. Cultures were then diluted to an OD600 of 0.1 in 100 ml of fresh SCD-MC or SCD+MC medium and were grown at the same initial temperature until an OD600 of 0.4 to 0.8 was reached. Cells were then centrifuged for 5 min at 3,500 rpm; the supernatants were removed; and the samples were quick-frozen and stored at −80°C. After that, cells were disrupted using acid-washed glass beads, and total RNA was extracted by a standard hot-phenol method (25).
First-strand cDNAs were synthesized from 4 to 5 μg total RNA in a 20-μl reaction volume using the SuperScript First-Strand Synthesis system for reverse transcription-PCR (RT-PCR) (Invitrogen). Quantitative real-time PCR was performed in triplicate using the Corbett Rotor-Gene RG-3000A thermal cycler (Corbett Research, Sydney, Australia) with SYBR green fluorescence (Qiagen, Chatsworth, CA). Independent amplifications were performed using the same cDNA for both the gene of interest and the C. albicans ACTI ORF as a reference, using the SYBR green PCR master mix. The fold enrichment of the gene of interest was estimated using the comparative ΔΔCT method (45). The primer sequences used for this analysis are summarized in Table 3.
RESULTS
Myosin I localization in opaque cells during budding, shmooing, and mating in C. albicans.
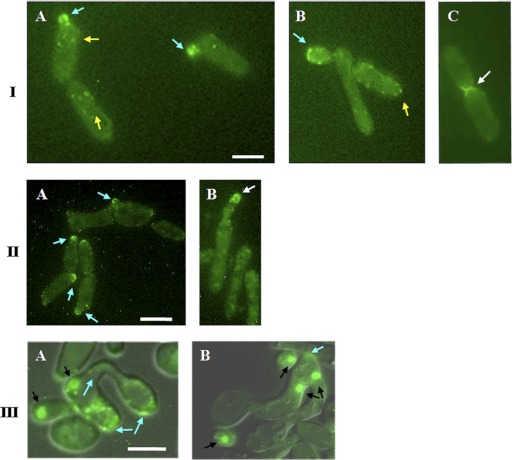
We investigated the role of Myo5p in the mating process of C. albicans. A MYO5-GFP fusion was integrated at the MYO5 locus of MTLa/MTLa MYO5/MYO5 strain 3294 (32) to generate strain CaNK02 (MYO5/myo5::pVEC MYO5-GFP-URA3). Cells bearing the fluorescent signal and verified for the correct site of the MYO5-GFP insertion were then screened for the mating-competent opaque form on SCD-phloxine B plates (3, 12). Fluorescence microscopy was used to detect the fusion protein in order to establish the localization of myosin I in vegetatively growing C. albicans opaque-form cells. Myo5p-green fluorescent protein (GFP) in exponentially growing opaque CaNK02 cells localized to patches that were dispersed in the mother cells and localized at the tips of emerging buds (Fig. 1IA and B); this localization was similar to that found in vegetatively growing white-form cells (38). As the bud grows, there is an alternation of highly polarized growth, where Myo5p-GFP localizes at the tip of the bud, and isotropic growth, where the Myo5p-GFP signal localizes more evenly over the entire bud periphery. These events function in a temporal sequence until nuclear division is completed, at which point the Myo5p-GFP signal is also observed at the mother-bud neck (Fig. 1IC).
Fig 1.
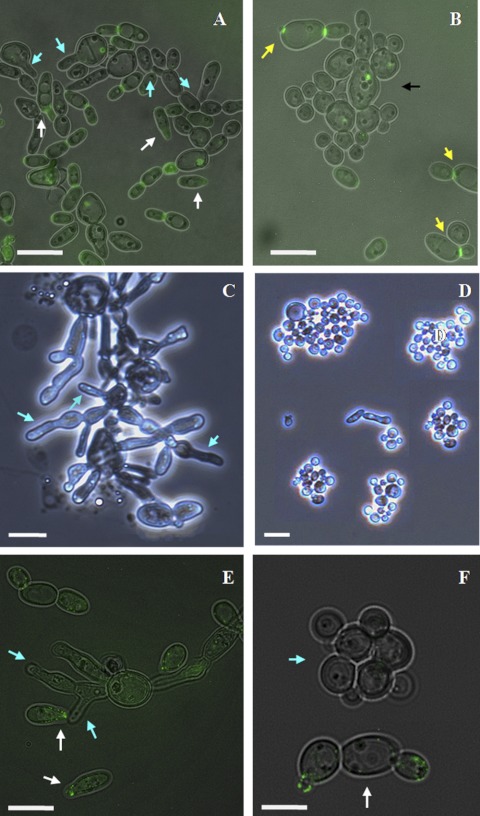
Localization of Myo5p-GFP in opaque C. albicans cells at different stages of the cell cycle and development. (I) (A and B) Myo5p-GFP is localized in cortical patches in MTLa/MTLa MYO5/MYO5-GFP mother cells (yellow arrows) and at the tips of emerging buds (blue arrows). (B) Subsequently, Myo5 redistributes evenly in an isotropic growing bud and then moves to the bud tip (blue arrow). (C) After nuclear division, Myo5p-GFP localization is observed at the mother-bud neck (white arrow). Bar, 3 μm. (II) Myo5p-GFP localizes in patches at the shmoo tips. C. albicans shmoos (blue arrows) (A) keep growing after emergence and continue to accumulate Myo5p at the shmoo apexes (white arrows) (B). Bar, 3 μm. (III) Mating between MTLa/MTLa NOP1/NOP1-YFP MYO5/MYO5-GFP and MTLα/MTLα MYO5/MYO5-GFP strains. (A) Myo5p patches (arrows) localize randomly in parent cells and in the conjugation bridge. (B) Concentration of Myo5p (arrows) at the site of first daughter cell formation. A mating mixture of the MTLa NOP1/NOP1-YFP MYO5/MYO5-GFP and MTLα MYO5/MYO5-GFP strains was cultivated O/N on solid Spider medium at 24°C. Black arrows point to the Nop1-GFP signal. Fluorescent images of Myo5p and Nop1p are superimposed on differential interference contrast images. Bar, 5 μm.
The presence of α pheromone in the culture medium induces sexual differentiation in cells of the a/a mating type; this differentiation includes a pheromone-induced polarization termed shmoo formation (49). The localization of Myo5p-GFP in such shmoos was examined after treatment of strain CaNK02 (opaque; a/a MYO5/MYO5-GFP) with 1 μg/ml synthetic α-factor. Myo5p-GFP localized in patches at the shmoo tips (Fig. 1IIA), and throughout the shmoo elongation process, Myo5p-GFP was found in apically clustered patches (Fig. 1IIB).
To investigate the localization of Myo5p during the actual mating process, we created two strains: an a/a strain designated CaNK06 containing NOP1-YFP and MYO5-GFP that was derived from the a/α NOP1-YFP strain 6449 (a kind gift from J. Berman) and an α/α MYO5/MYO5-GFP strain designated CaNK01. Nop1p-YFP is a yellow fluorescent protein fusion to the C termini of the small nucleolar ribonucleoprotein Nop1p (14). This marker allows the visualization of nuclei in order to clearly differentiate the two potential parent cells and therefore distinguish a true mating fusion (Fig. 1IIIA) from a bud emerging at the end of a shmoo. Based on the Myo5p dynamics in shmoos, we would have expected to find a concentration of fluorescence at the point of fusion. However, after cell fusion, the Myo5p-GFP signal is found dispersed in the fusion bridge and parent cells, suggesting that the GFP signal relocalizes rapidly after mating. A concentration of Myo5p-GFP appears again in the fusion product only at the site of the formation of the first daughter cell (Fig. 1IIIB).
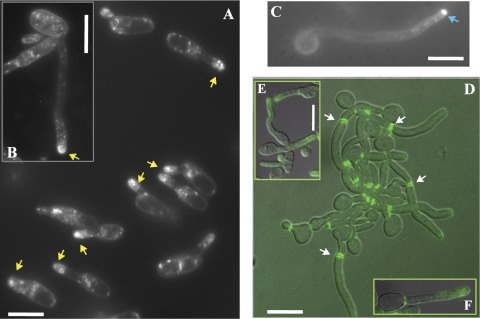
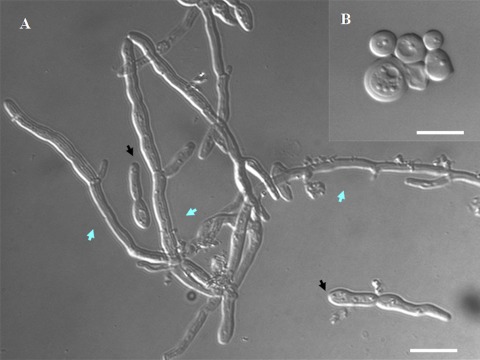
Interestingly, germ tubes and shmoos in C. albicans exhibit similar morphologies: each forms as an unconstricted projection growing out of the mother cell (Fig. 2) (10, 53). We plated cells from a mating mixture on Spider medium plates (Fig. 2A, B, and E); in this case, typically, cells generate very elongated shmoos that are highly reminiscent of standard germ tubes (Fig. 2C and D). We stained the shmoos with FM4-64, a dye capable of identifying the Spitzenkörper, a membrane-dense structure observed at the tips of growing hyphae (11, 15). Although FM4-64 staining is concentrated at the tips of the growing shmoos, the fluorescent signal is somewhat diffuse compared with the compact, solid signal in hyphal cells (Fig. 2A, B, and C). Recently, a detailed analysis of this Spitzenkörper-like structure in shmoos has been performed for both S. cerevisiae and C. albicans (10). In this work, a clear Spitzenkörper-like localization of FM4-64 was observed in C. albicans 1 h after induction by α-factor but was not detected in shmoos growing for 3 h. It is likely that we observed a less-compact FM4-64 staining, since we were observing shmoos forming by cellular stimulation rather than by artificial pheromone addition, and we were monitoring the cells at the later stages of shmoo formation.
Fig 2.
Shmooing (A, B, E, and F) and hyphal (C and D) cells of C. albicans. (A to C) Cells were stained with FM4-64. In shmooing opaque cells obtained from a mating mixture (MTLa/MTLa NOP1/NOP1-YFP MYO5/MYO5-GFP and MTLα/MTLα MYO5/MYO5-GFP strains) after 5 to7 h (A) and 16 h (B) of cultivation in liquid Spider medium at 24°C, we observed a diffuse bright spot (yellow arrows) at the tip of the mating projection. In hyphal cells (C) generated by overnight growth in YPD medium at 30°C, followed by dilution to an OD600 of 0.15 and cultivation in YPD medium supplemented with 10% (wt/vol) fetal calf serum at 37°C for 2, h we observed a tight fluorescent spot characteristic of a classic Spitzenkörper (blue arrow). (D to F) CDC12/CDC12-GFP cells were examined for Cdc12-GFP staining. Cdc-12p tagged with GFP localizes at the neck as bars parallel to the projection axis in both hyphal cells (D) and shmoos (E); however, although septin rings are evident in germ tubes (white arrows) (D), we never see similar structures inside mating projections (E and F). Fluorescent images of Cdc12p are superimposed on differential interference contrast images (D and E). Bars, 3 μm (A, B, C, D, and F) and 5 μm (E).
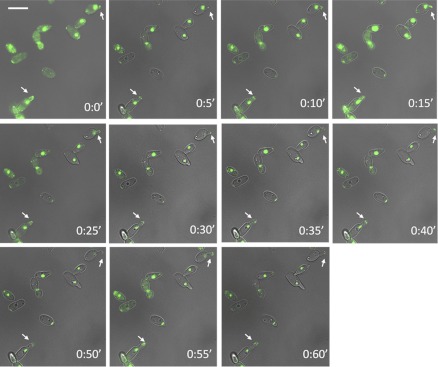
Another very specific characteristic of germ tubes and true hyphae, in contrast to structures such as pseudohyphae, is the localization of the septin ring. In germ tubes, the first septin ring is observed at a position distant from the mother cell-germ tube junction, while in buds and pseudohyphae, this ring is at the mother-bud neck. We examined the localization of GFP-tagged Cdc12p, one of the subunits of the septin ring, and observed that in C. albicans shmoos, the septins localize as side strips at the base of an emerging mating projection. This behavior is also seen in S. cerevisiae shmoos (30) and C. albicans germ tubes (48). However, in contrast to germ tubes, which ultimately form a septin ring at the future site of nuclear division, no septin ring forms in the shmoo projection, even in shmoos of significant length (Fig. 2D and E); this pattern is in agreement with that recently observed for a Cdc10-GFP fusion construct (10). However, the overall movement of Myo5p during shmoo growth is similar to that in hyphae (Fig. 3; see also time lapse movie S1 in the supplemental material) (38).
Fig 3.
Video monitoring of Myo5p localization during shmoo emergence. Shmooing (white arrows) was recorded in the mating mixture of MTLa NOP1/NOP1-YFP MYO5/MYO5-GFP and MTLα CDC12/CDC12-GFP cells. Bar, 5 μm.
Influence of myosin I on the white-opaque transition in C. albicans.
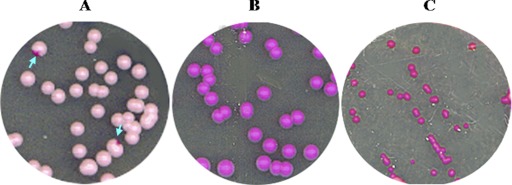
We created an a/a Δ/Δ myo5 mutant strain (strain CaNK09) to examine the role of myosin I in the shmooing of opaque cells and in the mating process. The white-form cells of this strain exhibited the classic myosin-defective phenotype: the cells looked abnormally round, were often enlarged, and were clumpy compared with wild-type (WT) cells (38). We screened this a/a Δ/Δ myo5 strain for colonies that had undergone the white-opaque transition, but after 7 days of incubation on plates containing phloxine B, we detected no colonies with dark purple sectors, which are a typical sign of the presence of opaque-phase cells (22, 35) (Fig. 4A and B). The screen was repeated in three replicas for four plates of both the myo5 mutant and the isogenic wild-type strain; each plate contained approximately 300 to 400 colonies. The control strain showed frequent opaque sectors, which ranged from 17 to 28 per plate, corresponding to an average frequency of 4.8 with a standard deviation of ±0.7 opaque sectors per 100 cells (Table 4). We found that all colonies of the mutant strain were isochromatic, though somewhat pinker than those of the control (Fig. 4B); inspection of these colonies did not identify any sign of opaque cells. Overall, it appears that the frequency of the white-opaque transition depends on the copy number of the MYO5 gene. Strain CaNK07, in which one of the MYO5 alleles is replaced with the URA3-blaster, has a 10-fold-lower frequency of switching than the wild type: on average, 0.36 ± 0.21 per 100 cells. The strain with an allele of MYO5 tagged with GFP showed a minor effect on white-opaque switching (2.4 ± 0.7 opaque sectors per 100 cells [Table 4]), suggesting that the GFP-tagged protein played an essentially normal role in the switching process.
Fig 4.
Identification of opaque-phase cells on SCD plates containing phloxine B. (A) MTLa/MTLa MYO5/Δmyo5 cells. Arrows point to colonies with sectors containing opaque cells. (B) MTLa/MTLa Δ/Δ myo5 cells. (C) MTLa/MTLa MET3-WOR 1 Δ/Δ myo5 cells grown on activating SCD-MC medium to induce WOR1 expression. This treatment generates colonies that are dark pink and smaller than MTLa/MTLa Δ/Δ myo5 and MTLa/MTLa MYO5/Δmyo5 colonies. Cells in these colonies have an abnormal morphology.
Table 4.
Induction of white-opaque switching in strains with different MYO5 contents
| Strain | No. of colonies |
Ratio (% ± SD) | |
|---|---|---|---|
| Total | With opaque sector | ||
| 3294 | 938 | 45 | 4.8 ± 0.7 |
| 3294+pVEC | 483 | 21 | 4.3 ± 0.9 |
| NK04 (MYO5-GFP) | 456 | 11 | 2.4 ± 0.7 |
| NK07 (Δmyo5/MYO5) | 838 | 3 | 0.36 ± 0.21 |
| CaNK09 (Δ/Δ myo5) | 1,469 | 0 | 0 ± 0.0 |
| CaNK17 (Δ/Δ sla2) | 1,211 | 0 | 0 ± 0.0 |
Ectopic WOR1 expression allows Δ/Δ myo5 cells to switch to the opaque phase.
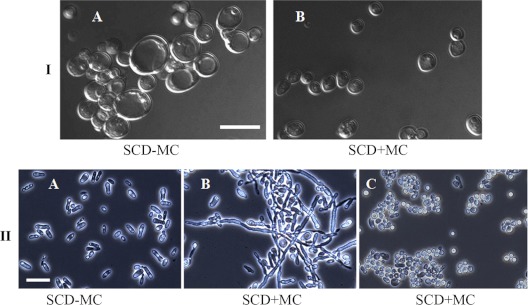
WOR1 is a master regulator of the white-opaque transition (22). We transformed the a/a Δ/Δ myo5 strain (CaNK09) with plasmid pRZ25, containing WOR1 expressed from the MET3 promoter (54), to create strain CaNK10 (a/a Δmyo5/Δmyo5 Rp10/rp10::pRZ25 MET3-WOR1). The expression of WOR1 would be expected to trigger the formation of opaque-form myosin mutant cells. When the transformants were incubated on plates containing SCD medium lacking Met and Cys (SCD-MC) to induce the expression of WOR1, they formed colonies characterized by a very dark pink color on phloxine B-containing plates (Fig. 4C). These colonies were also notably smaller than the Δ/Δ myo5 colonies grown on SCD medium supplemented with the dye. Cellular morphology was dramatically modified as well, with many cells exhibiting the classic elongated opaque cell shape, while other cells formed aberrant hypha-like structures (Fig. 5A). Thus, a high level of WOR1 expression in Δ/Δ myo5 cells leads to phenotypic changes similar to those caused by the white-opaque transition in wild-type cells and apparently reduces the growth rate. This suggests that the light pink colony color of the Δ/Δ myo5 mutants stained with phloxine B simply reflects the mutant genotype background (34, 36) and does not hide the presence of opaque cells; had Δ/Δ myo5 cells switched to opaque, we should have observed very dark pink opaque sectors within the lighter-staining colonies of white cells.
Fig 5.
Phenotype of the MTLa/MTLa Δ/Δ myo5 MET3-WOR1 mutant strain. (A) MTLa/MTLa MET3-WOR1 Δ/Δ myo5 cells growing on solid medium under MET3 inductive conditions (SCD medium lacking Met and Cys) can exhibit a morphology characteristic of opaque cells (black arrows) or can form aberrant hypha-like structures (blue arrows). (B) Cells are unable to sustain the opaque cell morphology under MET3 shutoff conditions (overnight growth in SCD liquid medium) and switch back to the white phenotype. Nomarski images are shown. Bars, 5 μm.
We were interested in determining whether the Δ/Δ myo5 MET3-WOR1 strain exhibited other phenotypic characteristics of the opaque state, so we checked cells grown on plates with SCD-MC medium for mating competence. Figure 6 shows that under inductive conditions, cells with ectopic expression of WOR1 but lacking both copies of MYO5 become mating competent and thus exhibit both the morphological and the physiological characteristics of opaque cells. This shows that Myo5p function is not essential for mating when the Wor1p circuit is activated.
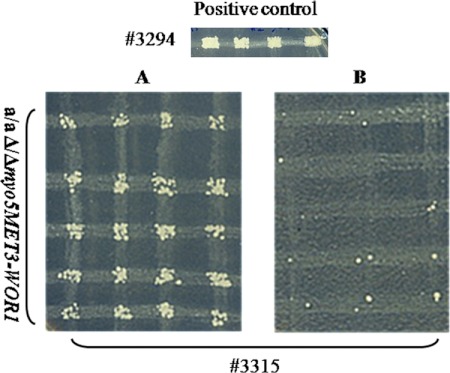
Fig 6.
Mating assay for the MTLa/MTLa Δ/Δ myo5 MET3-WOR1 strain. Cells were placed on plates containing SCD-MC or SCD+MC medium. After 4 to 5 days of growth at 24°C, cells from randomly chosen colonies were mated with tester strain 3315 (MTLα trp1/trp1 lys2/lys2) on SCD-MC or SCD+MC plates, respectively. The wild-type strain 3294 (MTLa Δura3::imm34/Δura3::imm434 his1/his1 arg5,6/arg5) was used as a control. Afterwards, plates were printed on selective SCD-MC medium (A) or on SCD+MC medium lacking histidine and lysine (B). Activation of WOR1 expression allowed all cells to mate.
Myosin I is required in order to maintain the opaque phenotype.
WOR1 expression is activated through the binding of the Wor1 protein to its own promoter (54), establishing a relatively stable epigenetic state. Thus, we expected to obtain stable opaque derivatives of the Δ/Δ myo5 MET3-WOR1 strain after ectopic WOR1 expression was shut off. However, the myo5 mutant cells that had been switched to the opaque state under MET3 inductive conditions failed to maintain the phenotype upon cultivation, even in SCD liquid medium, where the concentration of Met and Cys is too low (0.13 mM versus 2.5 mM) to completely suppress the expression of WOR1 from the MET3 promoter (Fig. 5B). Thus, the expression of WOR1 at a low level does not appear sufficient to maintain the opaque state in the absence of MYO5. We investigated the phenotype of 91 individual opaque Δ/Δ myo5 MET3-WOR1 cells placed on agar plates under WOR1 shutoff conditions. Within 24 h, 60 cells had generated morphologically white form segregants (Table 5), and when morphologically opaque cells were plated on SCD+MC medium, after 5 days all colonies (73 colonies) exhibited only the white phenotype. Thus, it appears that as residual Wor1p is depleted, the cells switch to the white state. Cells lacking the MYO5 gene retain the ability to switch to the opaque phase when WOR1 is ectopically expressed but are then unable to maintain the WOR1 circuit in the absence of continuous ectopic Wor1p expression.
Table 5.
Reversion of opaque-phase cells to white phase in strains with conditional expression of WOR1 or MYO5
| Strain genotype | No. of single opaque cells | No. of colonies in 24 h |
|
|---|---|---|---|
| White | Opaque | ||
| Δmyo5/MYO5 | 77 | 1 | 76 |
| Δ/Δ myo5 MET3-WOR1 | 91 | 60 | 31 |
| Δmyo5/MET3-MYO5 | 104 | 101 | 3 |
We further investigated the requirement for Myo5p in the maintenance of the opaque state by constructing a conditional allele of the MYO5 gene under MET regulation. The MET promoter was used to replace the promoter of the WT allele of MYO5 in the a/a MYO5/Δmyo5 strain designated CaNK8. Cells selected for the a/a MET3-MYO5/Δmyo5 (strain CaNK11) genotype exhibited phenotypes corresponding to a true MYO5 knockout (Fig. 7IA) under conditions repressive for MET3 promoter expression (SCD+MC), while cells grown on inductive medium (SC-MC) had the morphological phenotype characteristic of wild-type yeast cells (Fig. 7IB).
Fig 7.
Phenotype of MTLa/MTLa MET3-MYO5/Δmyo5 mutant cells. (I) (A) On SCD+MC medium, which suppresses the expression of MYO5, cells exhibit an abnormally round shape, with some cells enlarged, and the cells are clumped, characteristic of white-form myo5 mutants. (B) On SCD-MC medium, which allows MYO5 expression, the cells have a normal size and shape and are dispersed. An upright microscope was used. Magnification, ×90. Nomarski images are shown. Bar, 10 μm. (II) (A) MTLa/MTLa MET3-MYO5/Δmyo5 cells grown on SCD-MC medium can switch from the white to the opaque morphology. (B and C) MTLa/MTLa MET3-MYO5/Δmyo5 opaque cells grown on SCD+MC solid medium with phloxine B. (B) Opaque and hypha-like cells (from “wrinkled” colonies). (C) Clumped cells of abnormal shape (from “smooth” colonies). An inverted microscope was used. Magnification, ×40. Bar, 12.7 μm.
We could not find any colonies with sectors of opaque-phase cells among colonies obtained on the SCD repressive medium under conditions that normally support the white-opaque transition (data not shown). However, a/a Δmyo5/MET3-MYO5 cells plated on SCD inductive medium could be successfully switched to the opaque phase (Fig. 7IIA). We then examined whether the cell shape characteristic of opaque cells was stable under MYO5 loss-of-expression conditions. Opaque-phase cells were plated on SCD+MC solid medium (to repress MYO5 expression) supplemented with phloxine B to allow visualization of white and opaque colonies and were grown for 5 days at 24°C. Because even under repressed conditions the MET3 promoter can have residual activity in C. albicans (9), we expected to find a certain phenotypic variability if the opaque characteristics of Δmyo5/MET3-MYO5 cells became unstable under MYO5 loss-of-expression conditions. Under these conditions, we obtained two types of colonies, which were distinguishable phenotypically. One type of colony was dark pink and exhibited a wrinkled morphology. These colonies, as a rule, contained opaque-like and hypha-like cells similar to those of colonies derived from Δ/Δ myo5 MET3-WOR1 cells (Fig. 7IIB and 5A). It is intriguing that repression of MYO5 gene expression in opaque cells can lead to the appearance of cells with highly polarized projections, considering that polarized hyphal growth in white cells is blocked in the absence of Myo5p. The other type of colony was pink and had a smooth surface. These colonies contained mainly round and abnormally enlarged clumped cells with characteristics of the white form of Δ/Δ myo5 cells (Fig. 7C). As well, individual opaque cells placed on the solid SCD medium under MYO5 shutoff conditions typically generated a white morphology cell as the initial bud and in 24 h had formed a white-phase colony (101 of 104 cells picked generated white colonies [Table 5]).
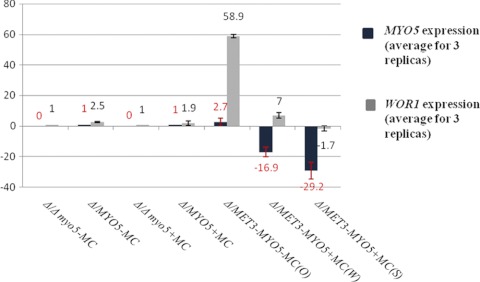
The inability of cells with a conditional knockout of the MYO5 gene to maintain a stable opaque phenotype when MYO5 expression is shut off supports the idea that Myo5p may be needed for proper WOR1 expression. We analyzed the expression of WOR1 and MYO5 by quantitative RT-PCR. Average data for the experiment carried out in triplicate are shown in Fig. 8. In spite of between-replica variation, it is apparent that when the MET3 promoter is repressed, both MYO5 expression and WOR1 expression decrease (compare the 59-fold gain of WOR1 expression in the Δmyo5/MET3-MYO5 strain [opaque] on SCD-MC medium with the 7-fold gain in the Δmyo5/MET3-MYO5 strain [wrinkled] on SCD+MC medium and the 1.7-fold decrease in the Δmyo5/MET3-MYO5 strain [smooth] on SCD+MC medium). Thus, a drop in MYO5 expression is correlated with WOR1 expression decay.
Fig 8.
Expression of WOR1 and MYO5 in strains with different sets of MYO5 copies grown in media that were inductive (SCD-MC) or repressive (SCD+MC) for the MET3 promoter. The total-RNA extract was subjected to quantitative RT-PCR to validate the conditional expression of MYO5 in opaque cells (O), in cells from “wrinkled” colonies (W), and in cells from “smooth” colonies (S) and also to evaluate the expression of WOR1 in these types of cells. All gene expression was normalized to actin gene expression (fold increase or decrease). Calibrators are Δmyo5/MYO5-MC and Δmyo5/MYO5+MC for MYO5 expression and Δ/Δ myo5-MC and Δ/Δ myo5+MC for WOR1 expression (-MC, grown in SCD-MC; +MC, grown in SCD+MC). Values are means ± standard deviations. Negative values indicate repression.
Deletion of SLA2 influences the white-opaque transition in the same manner as deletion of MYO5.
In C. albicans, deletion of SLA2 leads to morphology changes similar to those for myo5 deletion, such as actin cytoskeleton disorganization, hyphal formation, and endocytic defects (5, 39), while transcript profiles of the Δ/Δ myo5 and Δ/Δ sla2 strains revealed a common set of genes whose expression was significantly modified (15). We found that the Δ/Δ sla2 mutant grown on SCD plates at 24°C is also unable to switch to the opaque phase; no opaque cell sectors were observed in >1,200 colonies scored.
Formation of polarized projections in myo5 mutants.
Shmooing cells in C. albicans have myosin I at the tips of the mating projections, a situation similar to that in germ tubes. Because white-phase cells lacking both copies of MYO5 are unable to form germ tubes (38), it was of interest to know whether opaque-phase cells lacking MYO5 could still generate shmoos in the same manner as wild-type opaque cells. We used both the Δ/Δ myo5 mutant with ectopic expression of WOR1 and the conditional knockout for MYO5 to investigate whether the presence of Myo5p in cells is necessary for the formation of shmoos, and if so, what their morphology is. Cells of the a/a Δ/Δ myo5 MET3-WOR1 strain growing on SCD-MC plates for 5 days were inoculated overnight (O/N) in SCD-MC or SCD+MC liquid medium at 24°C. Cells derived from “wrinkled” and “smooth” types of colonies of the Δmyo5/MET3-MYO5 strain were grown O/N in SCD+MC liquid medium. Then each type of cell was mixed with an equivalent amount of α/α MYO5/MYO5-GFP or α/α CDC12/CDC12-GFP opaque cells from O/N SCD cultures. The mating mixture was cultivated in SCD-MC or SCD+MC liquid medium for 5 to 7 h to allow the development of shmoos, and the cell mixture was analyzed under a fluorescent phase-contrast microscope. MTLa and MTLα cells could be distinguished through their Cdc12-GFP or Myo5-GFP markers (Fig. 9). We used the mating mixture instead of treating a/a cells with artificial pheromone α, because the former procedure avoids the need for multiple additions of α-pheromone peptide to compensate for pheromone degradation (29), and it is easy to compare the shmoo morphologies of the myo5-defective cells and the wild-type cells in one field of view. As noted, Δmyo5/MET3-MYO5 cells under loss-of-expression conditions form shmoo-like structures if they are derived from “wrinkled” colonies (Fig. 9A) but not if they are derived from smooth colonies (Fig. 9B). It is possible that these polarized projections are formed in cells when the balance of Myo5 and Wor1p is perturbed, because similar structures form in myo5 disruption cells expressing ectopic WOR1 (Fig. 5). It is not clear whether the hypha-like cells from “wrinkled” colonies further generate shmoos, because they are already extensively polarized. However, single cells are able to generate conjugation tubes of normal morphology. The shapes of cells generating these shmoos in many cases were different from the classic shape of wild-type opaque-phase cells (Fig. 9A); they looked roundish and abnormally big.
Fig 9.
Polarized growth in strains with conditional and true knockouts of the MYO5 gene. Mating mixtures consist of MTLa/MTLa MET3-MYO5/Δmyo5 cells from “wrinkled” or “smooth” colonies and MTLα/MTLα CDC12/CDC12-GFP opaque cells (A and B) or a/a MET3-WOR1 Δ/Δ myo5 and MTLα/MTLα MYO5/MYO5-GFP cells (E and F) (see the explanation in the text). (A) Cells from “wrinkled” colonies are still able to generate shmoos (blue arrows) and stimulate shmooing in α/α opaque cells with the CDC12-GFP marker (white arrows). (B) Cells from “smooth” colonies (black arrow) demonstrate white morphology and are unable to shmoo. Yellow arrows point to opaque MTLα/MTLα CDC12/CDC12-GFP cells that also do not shmoo. (C and D) Phenotypes of pure a/a MET3-WOR1 Δ/Δ myo5 cells in initial cultures in SCD-MC and SCD+MC liquid media, respectively. Blue arrows show shmoo-like formation in cells grown under MET3-inducing conditions (C). (E) Δ/Δ myo5 cells with ectopic expression of WOR1 generate the same shmoo-like formations (blue arrows) in the mating mixture as in the initial culture. White arrows show shmoos in opaque α/α cells with the MYO5-GFP marker. (F) Under WOR1 shutoff conditions. MTLa/MTLa MET3-WOR1 Δ/Δ myo5 cells (blue arrow) convert to white phase, and the opaque α/α MYO5/MYO5-GFP cells (white arrow) do not form shmoos. Fluorescent images are superimposed on differential interference contrast images. Bars, 5 μm (A, E, and F), 4 μm (B), 10 μm (C), and 12.7 μm (D). An inverted microscope was used. Magnification, ×40.
The culture of the Δ/Δ myo5 cells with ectopic WOR1 expression also possesses considerable morphological complexity. Untreated cells form polarized projections that are morphologically similar to shmoos; thus, it is impossible to say whether a given projection was induced by pheromone or formed spontaneously (Fig. 9C and E). However, these Δ/Δ myo5 MET3-WOR1 opaque cells are able to mate, suggesting that a fraction of the polarized projections are functional. As well, these Δ/Δ myo5 MET3-WOR1 mutant cells express pheromone, as evidenced by the fact that cells of opposite mating types are induced to generate shmoos (Fig. 9E). Turning off ectopic WOR1 expression completely abolishes the opaque shape of the cells and blocks the shmoo-like morphology (Fig. 9F).
DISCUSSION
The white-opaque switch is a central component in the mating process of the fungal pathogen C. albicans. Only cells that have switched to the opaque state by activating a feed-forward transcriptional loop controlled by the Wor1p transcriptional regulator are able to mate (6, 54, 55). The regulatory circuit controlling WOR1 expression involves several other transcription factors, including Efg1p, Wor2p, and Czf1p, and is sensitive to environmental conditions, such as CO2 levels, N-acetylglucosamine concentration, and temperature (21, 23, 53, 55). Intriguingly, proper maintenance of this transcriptional circuit also requires components of the cytoskeletal regulation network, including the motor protein Myo5p, the only member of the myosin I protein family in C. albicans, and Sla2p, an actin-binding protein that plays a significant role in growth control and morphogenesis.
In fungi, myosin I is required for proper function of the actin cytoskeleton (8, 13, 27), and in C. albicans this protein is required for processes such as endocytosis, hyphal formation, proper chitin deposition, efficient separation of yeast form cells, and proper distribution of cell membrane lipids (38). In contrast to S. cerevisiae, where the myosin I-encoding genes MYO3 and MYO5 are critical for proper proliferation and, in many strain backgrounds, are essential for viability (17, 19), the proliferation rates of white-form yeast cells of C. albicans are not dramatically affected by the loss of MYO5 (38).
Because of the role of Myo5p in the formation of the highly polarized hyphae of white-form cells, it was of interest to establish its role in the formation of the dramatically polarized mating projections of opaque cells. In budding opaque cells, as in budding white cells, the GFP-tagged Myo5p was localized in discrete patches to sites of polarized growth, forming a concentration of signal at the sites of bud emergence and bud growth. When the opaque cells were induced to form a mating projection by the addition of purified pheromone or by the addition of cells of the opposite mating type, the Myo5-GFP signal localized to the tip of the growing projection. In these shmooing C. albicans cells, Myo5p displays both a cap and flat patches clustered at the shmoo tip; this pattern was also observed for the unconventional myosin Myo2p in shmooing cells of S. cerevisiae (28).
Interestingly, attempts to directly test the role of MYO5 in mating projection formation were complicated by the inability to generate the mating-competent opaque state in myo5 mutants, a failure attributable to a possible role for Myo5p in the establishment of the stable feed-forward transcriptional circuit. As well, sla2 mutants were unable to switch to the opaque state, suggesting a general involvement in some aspect of actin cytoskeletal regulation in this process. Only by ectopically expressing WOR1 under the control of the MET3 promoter were we able to induce an opaque phenotype in a myo5 deletion background, and this opaque phenotype was not stably maintained after shutoff of the MET3 promoter. This pattern was similar to the result of expressing WOR1 ectopically in a/α white cells: as long as ectopic expression was continued, the opaque-like state was maintained, but when ectopic expression was shut off, the opaque state did not persist (54).
Ectopic expression of WOR1 in a myo5 deletion background produces shmoo-like protrusions in cells even in the absence of a pheromone stimulus. These cells are able to mate, showing that myo5-independent polarized growth is capable of allowing zygote formation, but this could be occurring in a pheromone-independent manner. However, it is clear that in opaque cells, extended polarized growth is possible in the absence of Myo5p; this contrasts with the situation in white cells, where hyphal development is Myo5p dependent (37).
Intriguingly, cells in which MYO5 transcription is controlled from the MET3 promoter also cannot maintain the stable opaque phenotype after suppression of the MET3 promoter. It appears that blocking MYO5 expression disrupts the WOR1 feedback loop that maintains the opaque state, suggesting a possible role for Myo5p in the regulation of gene expression. Myo5p has also been implicated in transcriptional control in white-form cells. Deletion of MYO5 as well as SLA2 generated modified white-cell transcription profiles; genes such as ORF19.5302 and CHR1 were significantly overexpressed in the deletion strains, while CHT2 and CHT3 were underexpressed in the mutants (39). In some cases, the modified expression in white cells was correlated with a modified phenotype, such as chitin deposition (39). In mammalian cells, an unconventional myosin I isoform, termed nuclear myosin I (NMI), is found in the nucleus (40), where it is involved in transcription by RNA polymerase I and II (40, 41). However, it has not been established whether myosin I in yeast functions directly in transcription, and no evidence for nuclear localization was obtained from the Myo5-GFP fusion. Thus, it is currently unclear how Myo5p functions in determining the stability of the Wor1p transcriptional circuit. Sla2p, which has functions similar to those of Myo5p, also has a similar effect on the white-opaque transition. Both genes play important roles in organizing the actin cytoskeleton (5, 13, 16, 38); thus, perhaps Myo5p/Sla2p-dependent transport of messages or proteins is necessary to establish and maintain the circuit. It is intriguing that She4p, which is implicated in message localization in S. cerevisiae, is a Myo5p binding protein (50, 51); it would be informative to test whether the C. albicans homolog is involved in the stability of the Wor1p transcription circuit.
Overall, since the proliferation of yeast-form white cells is essentially unaffected by the loss of Myo5, it is somewhat surprising that myo5-defective yeast form opaque cells created by ectopic expression of Wor1p have a significant proliferation defect. This could suggest that there is some fundamental difference in the involvement of the actin cytoskeleton in the budding processes of white and opaque-form cells. However, further work will be necessary to determine the molecular basis for this intriguing connection between cell type determination and actin-based motor proteins in C. albicans.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Daniel Dignard and Doreen Harcus for help with processing C. albicans and for their technical expertise.
This work was supported by CIHR grants MOP42516 to M.W. and MOP-84231 to B.T.
Footnotes
Published ahead of print 9 March 2012
This article is publication 53122 of the National Research Council of Canada.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Alby K, Bennett RJ. 2009. Stress-induced phenotypic switching in Candida albicans. Mol. Biol. Cell 20:3178–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson J, et al. 1989. Hypha formation in the white-opaque transition of Candida albicans. Infect. Immun. 57:458–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson JM, Soll DR. 1987. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J. Bacteriol. 169:5579–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Askew C, et al. 2009. Transcriptional regulation of carbohydrate metabolism in the human pathogen Candida albicans. PLoS Pathog. 5:e1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Asleson CM, et al. 2001. Candida albicans INT1-induced filamentation in Saccharomyces cerevisiae depends on Sla2p. Mol. Cell. Biol. 21:1272–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bennett RJ, Johnson AD. 2005. Mating in Candida albicans and the search for a sexual cycle. Annu. Rev. Microbiol. 59:233–255 [DOI] [PubMed] [Google Scholar]
- 7. Blamire J, Melnick LM. 1975. The mating reaction in yeast. I. A new mutation involved in the determination of mating-type. Mol. Gen. Genet. 140:243–252 [DOI] [PubMed] [Google Scholar]
- 8. Brown SS. 1997. Myosins in yeast. Curr. Opin. Cell Biol. 9:44–48 [DOI] [PubMed] [Google Scholar]
- 9. Care RS, Trevethick J, Binley KM, Sudbery PE. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34:792–798 [DOI] [PubMed] [Google Scholar]
- 10. Chapa-Y-Lazo B, Lee S, Regan H, Sudbery P. 2011. The mating projections of Saccharomyces cerevisiae and Candida albicans show key characteristics of hyphal growth. Fungal Biol. 115:547–556 [DOI] [PubMed] [Google Scholar]
- 11. Crampin H, et al. 2005. Candida albicans hyphae have a Spitzenkörper that is distinct from the polarisome found in yeast and pseudohyphae. J. Cell Sci. 118:2935–2947 [DOI] [PubMed] [Google Scholar]
- 12. Dignard D, Whiteway M. 2006. SST2, a regulator of G-protein signaling for the Candida albicans mating response pathway. Eukaryot. Cell 5:192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Evangelista M, et al. 2000. A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J. Cell Biol. 148:353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Finley KR, Berman J. 2005. Microtubules in Candida albicans hyphae drive nuclear dynamics and connect cell cycle progression to morphogenesis. Eukaryot. Cell 4:1697–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fischer-Parton S, et al. 2000. Confocal microscopy of FM4-64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. J. Microsc. 198:246–259 [DOI] [PubMed] [Google Scholar]
- 16. Gale CA, et al. 2009. SLA2 mutations cause SWE1-mediated cell cycle phenotypes in Candida albicans and Saccharomyces cerevisiae. Microbiology 155:3847–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geli MI, Riezman H. 1996. Role of type I myosins in receptor-mediated endocytosis in yeast. Science 272:533–535 [DOI] [PubMed] [Google Scholar]
- 18. Gola S, Martin R, Walther A, Dunkler A, Wendland J. 2003. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 20:1339–1347 [DOI] [PubMed] [Google Scholar]
- 19. Goodson HV, Anderson BL, Warrick HM, Pon LA, Spudich JA. 1996. Synthetic lethality screen identifies a novel yeast myosin I gene (MYO5): myosin I proteins are required for polarization of the actin cytoskeleton. J. Cell Biol. 133:1277–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hnisz D, Schwarzmüller T, Kuchler K. 2009. Transcriptional loops meet chromatin: a dual-layer network controls white-opaque switching in Candida albicans. Mol. Microbiol. 74:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang G, Srikantha T, Sahni N, Yi S, Soll DR. 2009. CO2 regulates white-to-opaque switching in Candida albicans. Curr. Biol. 19:330–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang G, et al. 2006. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 103:12813–12818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang G, et al. 2010. N-Acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 6:e1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Janbon G, Sherman F, Rustchenko E. 1998. Monosomy of a specific chromosome determines l-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 95:5150–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kohrer K, Domdey H. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194:398–405 [DOI] [PubMed] [Google Scholar]
- 26. Lan CY, et al. 2002. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 99:14907–14912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lechler T, Shevchenko A, Li R. 2000. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J. Cell Biol. 148:363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lillie SH, Brown SS. 1994. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. J. Cell Biol. 125:825–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lockhart SR, Zhao R, Daniels KJ, Soll DR. 2003. Alpha-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot. Cell 2:847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Longtine MS, Bi E. 2003. Regulation of septin organization and function in yeast. Trends Cell Biol. 13:403–409 [DOI] [PubMed] [Google Scholar]
- 31. Madden K, Snyder M. 1998. Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol. 52:687–744 [DOI] [PubMed] [Google Scholar]
- 32. Magee BB, Magee PT. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science 289:310–313 [DOI] [PubMed] [Google Scholar]
- 33. Mermall V, Post PL, Mooseker MS. 1998. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science 279:527–533 [DOI] [PubMed] [Google Scholar]
- 34. Middelhoven WJ, Broekhuizen B, van Eijk J. 1976. Detection, with the dye phloxine B, of yeast mutants unable to utilize nitrogenous substances as the sole nitrogen source. J. Bacteriol. 128:851–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morrow B, Anderson J, Wilson J, Soll DR. 1989. Bidirectional stimulation of the white-opaque transition of Candida albicans by ultraviolet irradiation. J. Gen. Microbiol. 135:1201–1208 [DOI] [PubMed] [Google Scholar]
- 36. Nagai S. 1963. Diagnostic color differentiation plates for hereditary respiration deficiency in yeast. J. Bacteriol. 86:299–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oberholzer U, Iouk TL, Thomas DY, Whiteway M. 2004. Functional characterization of myosin I tail regions in Candida albicans. Eukaryot. Cell 3:1272–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oberholzer U, Marcil A, Leberer E, Thomas DY, Whiteway M. 2002. Myosin I is required for hypha formation in Candida albicans. Eukaryot. Cell 1:213–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oberholzer U, Nantel A, Berman J, Whiteway M. 2006. Transcript profiles of Candida albicans cortical actin patch mutants reflect their cellular defects: contribution of the Hog1p and Mkc1p signaling pathways. Eukaryot. Cell 5:1252–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pestic-Dragovich L, et al. 2000. A myosin I isoform in the nucleus. Science 290:337–341 [DOI] [PubMed] [Google Scholar]
- 41. Philimonenko AA, et al. 2004. Dynamics of DNA replication: an ultrastructural study. J. Struct. Biol. 148:279–289 [DOI] [PubMed] [Google Scholar]
- 42. Pruyne D, Bretscher A. 2000. Polarization of cell growth in yeast. J. Cell Sci. 113(Pt 4):571–585 [DOI] [PubMed] [Google Scholar]
- 43. Pruyne D, Bretscher A. 2000. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113(Pt 3):365–375 [DOI] [PubMed] [Google Scholar]
- 44. Rose MD, Winston F, Hieter P. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 45. Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 46. Srikantha T, et al. 2006. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot. Cell 5:1674–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Srikantha T, Soll DR. 1993. A white-specific gene in the white-opaque switching system of Candida albicans. Gene 131:53–60 [DOI] [PubMed] [Google Scholar]
- 48. Sudbery PE. 2001. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol. Microbiol. 41:19–31 [DOI] [PubMed] [Google Scholar]
- 49. Tkacz JS, MacKay VL. 1979. Sexual conjugation in yeast. Cell surface changes in response to the action of mating hormones. J. Cell Biol. 80:326–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Toi H, et al. 2003. She4p/Dim1p interacts with the motor domain of unconventional myosins in the budding yeast, Saccharomyces cerevisiae. Mol. Biol. Cell 14:2237–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wesche S, Arnold M, Jansen RP. 2003. The UCS domain protein She4p binds to myosin motor domains and is essential for class I and class V myosin function. Curr. Biol. 13:715–724 [DOI] [PubMed] [Google Scholar]
- 52. Wilson RB, Davis D, Mitchell AP. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhao R, et al. 2005. Unique aspects of gene expression during Candida albicans mating and possible G1 dependency. Eukaryot. Cell 4:1175–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zordan RE, Galgoczy DJ, Johnson AD. 2006. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc. Natl. Acad. Sci. U. S. A. 103:12807–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. 2007. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 5:e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.