Abstract
We recently proposed a scout model of the microbial life cycle (S. S. Epstein, Nature 457:1083, 2009), the central element of which is the hypothesis that dormant microbial cells wake up into active (so-called scout) cells stochastically, independently of environmental cues. Here, we check the principal prediction of this hypothesis: under growth-permissive conditions, dormant cells initiate growth at random time intervals and exhibit no species-specific lag phase. We show that a range of microorganisms, including environmental species, Escherichia coli, and Mycobacterium smegmatis, indeed wake up in a seemingly stochastic manner and independently of environmental conditions, even in the longest incubations conducted (months to years long). As is implicit in the model, most of the cultures we obtained after long incubations were not inherently slow growers. Of the environmental isolates that required ≥7 months to form visible growth, only 5% needed an equally long incubation upon subculturing, with the majority exhibiting regrowth within 24 to 48 h. This apparent change was not a result of adaptive mutation; rather, most microbial species that appear to be slow growers were in fact fast growers with a delayed initiation of division. Genuine slow growth thus appears to be less significant than previously believed. Random, low-frequency exit from the nongrowing state may be a key element of a general microbial survival strategy, and the phylogenetic breadth of the organisms exhibiting such exit indicates that it represents a general phenomenon. The stochasticity of awakening can also provide a parsimonious explanation to several microbiological observations, including the apparent randomness of latent infections and the existence of viable-but-nonculturable cells (VBNC).
INTRODUCTION
The environment poses formidable challenges for any organism, and microbes are no exception. Dormancy has been considered the principal tool for microbial survival since the dawn of microbiology, as processes are more difficult to corrupt if they are not occurring. The best-known example of dormancy is the microbial spore, but nonsporulating species are believed to have an inactive state as well (39). While the nature of such a state remains essentially unknown, it is abundantly clear that microbial cells of nonsporulating species can enter some form of suspended animation and thus survive a number of unfavorable conditions, from starvation to antibiotic treatment (31, 32, 35). Ultimately the cells must exit dormancy. Recognizing the right moment to do so poses a significant challenge, and presently there is little understanding of how the resumption of activity occurs. A conventional, though rarely spelled out, assumption is that this happens in response to environmental cues, as is the case with the resting seeds of plants and hibernating animals. There is evidence that in at least some species dormant spores and cells can be induced to grow by metabolites of growing kin (33, 34, 41). This, however, leaves the question of what induced the first cell in this growing population. We have recently proposed a model of the microbial life cycle built around an opposite idea, explaining that such exit could be achieved by stochastic awakening into activity, a process leading to active cells we termed scouts (14, 15). In principle, this model is applicable to spore-forming species as well; induction and stochastic germination are not necessarily mutually exclusive. Note that at least one research group reported that the stochastic germination of Bacillus subtilis spores was the likeliest explanation for the empirically observed germination pattern (37).
The proposed model postulates that scout formation is analogous to several well-known cases of epigenetic, noise-driven, stochastic bistability (1, 9, 13, 29, 30). A scout is a cell that exits dormancy as a result of essentially random events that occur at low frequency, such as a stochastic change in the expression or repression of a master regulatory gene. The scout is neither a genetic variant nor a specialized cell. Indeed, the scout may be identical to a typical cell in an actively growing population. The appearance of scouts effectively divides a clonal population into two phenotypes, the dormant and active, which alternate in dominance depending on the environmental conditions.
The term scout comes from the proposed function: the scout is a newly active cell that explores resources that are available at the moment. If adverse conditions persist, the scout dies after it has exhausted its internal resources, but it is followed by a succession of other scouts stochastically formed within the same population. If a scout forms under growth-permissive conditions it will establish a new population, thus achieving the main objective: the multiplication of the population's genome. Once the environmental conditions again become adversarial the revived population will return to dormancy, initiating a new round of the cycle. The model thus proposes that a generalized microbial population achieves survival under hostile conditions, and growth under favorable ones, by cycling between dormancy and activity. One possible method to accomplish this transition to activity is random awakening/germination into scouts. Such random exit from a nongrowing state that is independent from environmental cues may have relevant consequences for microbiology and health sciences (14).
The scout model makes the following prediction: in nutrient medium, dormant cells form colonies at random intervals during extended incubation periods, and exhibit, somewhat counterintuitively, no specific lag phase. In the first of the two companion papers, we test this prediction by observing the awakening pattern of environmental cells, both spore forming and non-spore forming, and of dormant Escherichia coli and Mycobacterium smegmatis. We also discuss implications of this pattern for the nature of slow-growing microorganisms, viable-but-nonculturable cells (VBNCs), and latent infections. The second paper will consider the importance of our findings for the optimization of the microbial discovery process.
MATERIALS AND METHODS
Environmental microorganisms: soil microorganisms. (i) Sample collection and preparation.
Five terrestrial soils were collected at various times during 2009 and 2010. The soils represented both forest and open-field environments from across the United States. Samples of ∼10 g were pulverized with mortar and pestle and air dried at room temperature, followed by heating for 15 min at 55°C to enrich for spore-forming species (27). Cell suspensions were generated by vortexing 1 g of prepared soil in a total volume of 10 ml with sterile deionized water for 30 min.
(ii) Growth conditions.
Ten-fold serial dilutions were made and plated at various densities in 2% SMS agar (0.125 g/liter casein [MP Biochemicals, San Diego, CA], 0.1 g/liter starch from potato [Sigma, St. Louis, MO], 1 g/liter Casamino Acids [Difco, Franklin Lakes, NJ], 20 g/liter Bacto agar [Difco]). Plates were sealed in Ziploc bags (1.75 mil) and incubated in a humidified chamber at room temperature. Growth was scored under a dissecting microscope (6.5× to 50× magnification range; Zeiss Stemi 2000) weekly for the first month and monthly thereafter for 3 more months. After incubation, 96-well plates containing between 1 and 3 colonies per well were chosen for microbial isolation and identification.
(iii) DNA analysis.
From these plates, all visible colonies were subsampled for subculturing, and 407 were isolated into pure culture as determined by visual observation using a dissecting scope and 16S rRNA gene sequencing. For molecular identification, chromosomal DNA was isolated from approximately 106 cells after 5 min of vigorous agitation in the presence of 50 mg of glass beads (106 nm or smaller) and 100 μl of H2O in a 0.5-ml Eppendorf tube. The PCR-aided amplification of the 16S rRNA gene was carried out using chromosomal DNA, GoTaq Green master mix (Promega, Madison, WI), and universal primers 27F and 782R (Table 1) (2). PCR thermocycler parameters included 30 cycles at 95°C for 30 s, 45°C for 30 s, and 72°C for 105 s. The amplified DNA fragment (∼720 bp) was cleaned up and sequenced at Macrogen (Rockville, MD) using primer 782R and compared by BLAST alignment to the nucleotide collection in GenBank. Sequences were edited using Chromas Lite 2.01 (Technelysium Pty. Ltd., Brisbane, Australia) and clustered into operational taxonomic units (OTUs) based on 99, 97, and 95% sequence similarity cutoff values. This was achieved by first making all possible pairwise sequence alignments using ClustalW at default settings and calculating percent sequence similarities, followed by the clustering of the sequences into OTUs by employing the mean unweighted-pair group method and using average linkages as implemented in the OC clustering program (http://www.compbio.dundee.ac.uk/Software/OC/oc.html). From each OTU, the sequence least different from the other members of the cluster was compared to the NCBI database using the BLAST search function. The top culturable and taxonomically identified hits were used to establish the identity of the OTUs. For soil samples enriched for spore formers, OTUs combining rRNA gene sequences sharing ≥99% identity were considered to belong to the same species, as is commonly accepted for actinobacteria (42, 43).
Table 1.
Sequences of primers used for genetic analysis
| Primer name | Sequence |
|---|---|
| 27F | 5′-AGA GTT TGA TCC TGG CTC AG-3′ |
| 782R | 5′-GAT TAG ATA CCC TGG TAG-3′ |
| 1492R | 5′-GGT TAC CTT GTT ACG ACT T-3′ |
| 907R | 5′-CCG TCA ATT CCT TTA AGT TT-3′ |
| 357F | 5′-CCTACG CGA GGC AGC AG-3′ |
Environmental microorganisms: marine microorganisms. (i) Sample collection and preparation.
Marine cells were obtained from a sample of intertidal sand collected on 31 October 2007 from Massachusetts Bay on the U.S. northeast seashore. The sample was vortexed for 5 cycles of 15 min each, and dislodged cells in the supernatant were counted by epifluorescence (Zeiss Axioskop 50 compound microscope at 1,000× magnification).
(ii) Growth conditions.
Counted cells were mixed with 0.1× lysogeny broth (LB) in natural seawater to a final density of 40 cells/ml. A 12-channel micropipetter was used to distribute aliquots of 50 μl containing (on average) single cells into individual wells of 31 384-well microtiter plates (designated the single-cell format), each containing a row of wells with sterile medium to control for contamination. The plates were sealed with Parafilm, separated into three groups to serve as replicates, placed into humidifying chambers, and incubated at room temperature in the dark for 18 months. Plates were scored for growth under a dissecting scope (Zeiss Stemi 2000 at 6.5× magnification) at monthly intervals. New growth was removed after 5, 6, 12, and 18 months of incubation. No contamination was observed in any of the control wells.
(iii) Genetic analysis.
Microbial biomass was subcultured in the same medium for identification via 16S rRNA gene sequencing. Universal bacterial primers 27F and 1492R were used to perform PCR-aided amplification. In some cases, seminested PCR-aided amplification was performed using 27F and 1492R, followed by 27F and 907R or 357F and 907R (Table 1). Amplicon sequencing was performed commercially, and the resulting sequences were imported into the ARB database (28). Marine species were identified as the smallest clusters of sequences on a phylogenetic tree produced within the ARB database. These were comprised of sequences sharing a ≥97% sequence similarity cutoff value and were considered species as accepted for most bacteria (44). Representative sequences from each cluster were entered into the GenBank database using the BLAST search tool, and the closest cultured and taxonomically identified relative was recorded.
(iv) Subculture.
Out of the total of 496 wells showing growth, 406 were successfully subcultured, and the time interval required for regrowth was noted. Twenty randomly selected isolates were also subcultured in a single-cell format. Cells grown in the original isolation experiment were enumerated as described above, diluted in fresh medium (0.1× LB in seawater), and administered to one 96-well plate per isolate in the single-cell format. A row of wells in each plate was inoculated with sterile medium to control for contamination. The plates were observed, and the growth scored, for at least 1 month.
Escherichia coli.
E. coli strain K-12 W3110 was kindly provided by K. Lewis (Northeastern University, Boston, MA). Cultures were exposed to antibiotics by following standard protocols for studying persister cells (23, 24). Cells were grown to mid-exponential phase in LB, challenged with either 500 μg/ml of gentamicin or 50 μg/ml of ofloxacin for 3 h, washed twice in fresh LB by centrifugation/resuspension, and diluted to 20 cells/ml. Cells were distributed into 32 384-well microtiter plates in single-cell format. The Parafilm-sealed plates were incubated for 2 to 8 weeks, and visual growth was periodically scored. In addition, a dilution series of the same cells was plated on standard LB petri dishes with 1.5% Bacto agar.
Mycobacterium smegmatis.
M. smegmatis strain mc2155 was kindly provided by K. Lewis (Northeastern University). Cells were grown in Difco Middlebrook 7H9 broth supplemented with bovine serum albumin, dextrose, NaCl, catalase, and 20% Tween 80 for 48 h to mid-stationary phase. This medium is optimal for M. smegmatis growth (4). To induce dormancy, 1-μl loops of the culture were inoculated into 7H9 broth. The vials were capped and incubated at 37°C with slow shaking for 2 weeks until the depletion of oxygen, which was monitored using methylene blue. This protocol, including the size of the cultivation tubes, follows prior recommendations (11, 49), with minor modifications. After dormancy induction, cultures were lightly sonicated and vortexed to disrupt clumps, and cells were counted by epifluorescence after staining with 4′,6-diamidino-2-phenylindole (DAPI), diluted to 20 or 200 cells/ml, and distributed into 10 384-well microtiter plates in single-cell format. Growth was scored visually at 2 and 5 days and weekly thereafter. In a separate experiment, we incubated dormant cells for 4 weeks and subcultured material that appeared early (72 h) as well as late (2 weeks) in the experiment. The subcultures were established as described above with cells diluted in fresh medium to a concentration of 1 and 10 cells per well. Growth was scored visually in the same manner as that described above.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of soil microorganisms have been deposited in the NCBI database under accession numbers JQ419503 to JQ419717. The 16S rRNA gene sequences of marine microorganisms have been deposited in the NCBI database under accession numbers HQ446854 to HQ446856, GQ262723, and JQ660963 to JQ661255.
RESULTS
Awakening of environmental cells.
A large percentage of environmental microorganisms are thought to be nongrowing at any given time (45), and environmental samples may thus serve as sources of dormant cells. We sampled soils and marine sediments, and we used microbial cells from these habitats to examine the patterns of their growth in two sets of cultivation experiments.
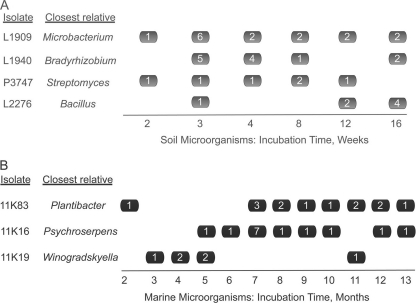
In the first set, we enriched for spore-forming bacteria by preheating dried soil samples prior to inoculation on solid media. The resulting petri dishes were observed during a period of 16 weeks, colonies were counted after 1, 2, 3, 4, 8, 12, and 16 weeks of growth, and colony material was subsampled for subculturing and taxonomic identification via 16S rRNA gene analyses. We successfully subcultured and identified 407 isolates, which fell into 211 OTUs based on 99% 16S rRNA gene sequence identity. Almost three-quarters of these OTUs belonged to the phylum Actinobacteria, in which this (99%) level of identity is equated with species status (42, 43). Most species were met only rarely and are thus not informative of a temporal pattern of their cells' growth resumption. To reconstruct this pattern, we chose species whose representatives were detected >10 times each. Four species satisfied this criterion, two of them spore forming (Bacillus and Streptomyces species; 100 and 99.9% 16S rRNA gene identity, respectively) and two non-spore forming (Bradyrhizobium and Microbacterium species; 99.0 and 99.6% 16S rRNA gene identity, respectively). In each of the four species, the cells formed visible colonies at widely different time intervals and exhibited few signs of a species-specific time lag (Fig. 1A). In some species, there were indications that more cells formed visible growth at a specific time point (e.g., relatives of Streptomyces in week 1 or Microbacterium in week 3), but by and large the growth events were spread evenly across long periods of time.
Fig 1.
Temporal pattern of growth resumption of selected microorganisms from soil (A) and marine (B) samples. Values inside the boxes indicate the number of isolates observed at the given time point. Note that the closest cultivated relatives of the marine isolates are all fast growers forming visible colonies within 24 to 94 h (26, 35, and O. Nedashkovskaya, personal communication).
In the second set of cultivation experiments, we obtained a mix of 11,000 cells randomly taken from a marine sediment sample and placed them into microtiter plates in nutrient medium such that each well received a single cell on average. This enabled observations of the initiation of the growth of individual cells unimpeded by the growth of other cells. During a uniquely long incubation lasting for 1.5 years, we periodically scored visible growth and observed a total of 502 wells with growing biomass. We then successfully subcultured 406 isolates, identified 310 of them via the rRNA approach, and grouped their 16S rRNA gene sequences into 86 OTUs based on 97% sequence identity. As is common for most bacteria, these OTUs are referred to as species (44). We then reconstructed, for species met multiple times, the temporal pattern of their cells forming visible growth. As was the case with soil microorganisms, different cells of the same species formed such growth at dramatically different times, e.g., 13 instances of growth spread over as many months in Plantibacter sp. (98% 16S rRNA gene identity) (Fig. 1B).
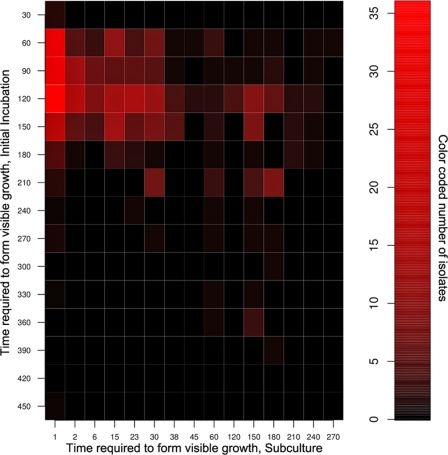
In the original incubation, no visible growth appeared during the first 3 weeks, and more than half of all isolates were not detected until after 120 days of incubation. To check if the time of appearance of an isolate in the first cultivation experiment was reflective of this isolate's inherent growth rate, we subcultured all 502 original marine isolates. Remarkably, upon the subculturing of all isolates obtained here, almost half of them formed visible biomass in less than 48 h (Fig. 2). We then checked if mutational change was involved in such a dramatic drop in the amount of time required for their growth to become visible. The likeliest suspects of mutational change are the isolates that appeared to grow very slowly at first but expanded very quickly upon subculturing. We randomly chose 20 marine isolates from all 502 subcultured isolates. From those we identified four that, in subculture, showed the largest increase in the apparent rate of growth, and again we subcultured the originally grown biomass, now in the single-cell format. Should the mutational change explain their fast growth during subculturing, the subcultured cells should all be fast growers. This is not what we observed, as single cells of such isolates again showed individuality in the timing of growth initiation, differing in that respect by at least an order of magnitude from each other (Fig. 3).
Fig 2.
Time (in days) required to form visible growth during initial isolation of marine microorganisms (y axis) and during subculturing (x axis). Note that the overwhelming majority of isolates observed between 45 and 175 days of the initial incubation regrew within 24 h (red squares in the upper left corner of the heat map).
Fig 3.
Individual cells of four selected isolates show significant differences in subculture with respect to time of growth initiation. Cells were subcultured in the single-cell format, allowing for the observation of when each cell formed visible growth. Here and in all single-cell format experiments, the percentage of wells showing growth is proportional to the number of cells forming visible biomass but is not the percent recovery, because some wells in microtiter plates were unoccupied due to chance.
Awakening of dormant cells in model microbial species.
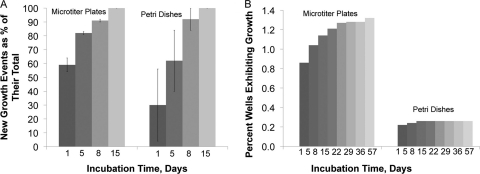
We were interested to know if the phenomenon of random awakening could be observed in standard laboratory strains, such as E. coli. We obtained nongrowing cells of E. coli by killing growing cells with a high dose of bactericidal antibiotics, and we distributed the remaining surviving cells in fresh medium in single-cell format. We observed that individual cells formed growth at different times spread throughout the duration of the experiments (Fig. 4A and B).
Fig 4.
Awakening kinetics of dormant E. coli. (A) After antibiotic challenge, individual surviving cells of E. coli do not initiate growth simultaneously but start growing at different time points spread throughout 2 weeks of incubation. Gradual awakening is apparent in single-cell experiments employing microtiter plates and in conventionally plated petri dishes. Recovery at the end of 2 weeks is designated total recovery and is assigned a value of 100%; earlier data points are presented as fractions of this value, with standard deviations indicated. The results represent data from three independent experiments, two of which employed gentamicin and one ofloxacin, all showing similar kinetics of growth initiation. (B) In a single growth experiment involving cells remaining after gentamicin treatment, we extended the incubation period to 2 months and continued to observe new growth. Note a much lower level of cell recovery in petri dishes versus single-cell experiments in microtiter plates. Note also that gradual awakening is not apparent in petri dishes, likely due to overgrowth by colonies developing early in incubation. Incidentally, this may be one reason why the phenomenon was not observed in earlier studies.
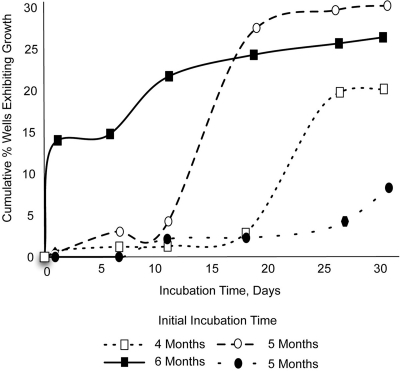
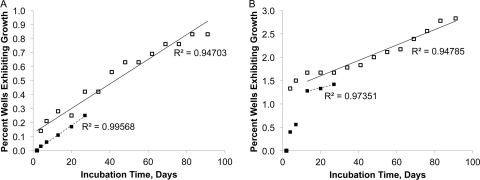
We also checked if the phenomenon could be observed in M. smegmatis. Dormant cells were obtained by following the Wayne and Sohaskey model (49) and were incubated in microtiter plates in single-cell format. Growth events were spread evenly over the incubation time of up to 90 days, and a cumulative curve showed no sign of saturation (Fig. 5A and B).
Fig 5.
Awakening kinetics of dormant M. smegmatis. Four independent long-term experiments show that dormant cells of M. smegmatis initiate growth at apparently random time points, with cumulative growth curves exhibiting no sign of leveling off even after 3 months of incubation. Each well was inoculated with, on average, either 1 (A) or 10 (B) dormant cells of M. smegmatis.
We recognized that, in principle, this might be a result of genetic variability in the tested population of M. smegmatis. We checked this possibility in a separate experiment, whereby we subcultured cells that initially required 3 to 4 weeks to grow. Without exception, regrowth was observed within 48 to 72 h (data not shown). This indicates that cells whose growth became visible at different time points (Fig. 5) were not inherently different in their growth rates.
DISCUSSION
The main focal point of this research was on how dormant cells wake into activity. As we noted in the introduction, it has long been suspected that microbes survive unfavorable conditions in a state of suspended animation (31, 32, 35). The nature of and awakening from dormancy have received significant attention in spore-forming species, but much less attention has been given to their non-spore-forming counterparts. A number of factors can induce the germination of spores. One recent finding suggests an environmentally relevant mechanism: a growing population of Bacillus subtilis can induce the germination of its spores via peptidoglycan fragments (41). Among non-spore formers, a similar induction of dormant cells by their growing kin is known in mycobacteria (33) and in an environmental isolate (34). Note that a growing population is a prerequisite, begging the question of what woke up the first cell. Here, we checked the possibility that such awakening is random.
Challenges in reconstructing kinetics of microbial awakening.
Interpreting our results meets a significant challenge due to the undefined physiological state of the cells used in our experiments. Ideally, a study of the awakening kinetics of dormant cells would start from a population consisting exclusively of cells that are dormant but viable, with no dead/injured cells mixed in. However, this is not possible, and experiments of this kind universally start from physiologically heterogeneous cell mixes. Preferably, one would want to know the proportion of different physiological states in the mix, but this is difficult as well, as reliance on standard measures (live/dead counts and respiratory dyes) may be misleading. For example, in our and other researchers' experience (17), DAPI (total) cell counts are often significantly larger than the sum of live/dead counts. More significantly, live/dead kits appear to work optimally under conditions of minimal stress (8), an exact opposite of conditions leading to dormancy. Also, the presence of cells in intermediate states of staining further confuses the picture when working with the binary live/dead kit (5). Finally, the basic premise of using such kits (i.e., live cells are active) may not accurately reflect the situation in a bacterial culture (5). Therefore, we are lacking validated and calibrated tools distinguishing between (in same cases overlapping) categories of cells that are dead, dormant, viable, active, injured beyond repair, etc. In the end, it is unclear how to distinguish between a dormant cell that we do not know how to induce to grow and an intact but dead cell.
Under the circumstances, one available strategy is to enrich populations with dormant cells as much as possible, preferably by the use of multiple methods and species, then experiment with the resulting mixes and deduce from the results how and if dormant cells resumed growth. This is the strategy we adopted, obtaining cell mixes enriched for dormant cells in three different ways: naturally enriched environmental populations, cells that survived a high dose of antibiotic (E. coli), and cells obtained via the Wayne and Sohaskey model (M. smegmatis).
Supporting evidence for stochastic awakening.
Our expectation was that few dormant populations would be comprised exclusively of inactive cells, and most would contain at least some cells ready to divide or spores on the verge of germination. If so, such cells/spores would be expected to produce a peak in the number of growth events early in the incubation, and the aspects of randomness in the awakening of dormant cells/spores would be more visible after that initial peak. This is precisely what we observed. It indicates that, in each individual population, many cells/spores were viable and capable of growth, but most did not respond to appropriate nutrient conditions similarly. Instead, they initiated growth at different times and in an apparently random fashion.
This pattern, exhibited by both spore-forming and non-spore-forming bacteria and from both soil and marine environments, may have several explanations. Considering Plantibacter species as an example, it is possible that its 13 isolated populations represented different strains with radically different inherent growth rates. The same level of intraspecific heterogeneity may hold for all species shown in Fig. 1. We checked this hypothesis by subculturing all isolates in an attempt to reproduce their growth rate heterogeneities but did not observe any; indeed, the overwhelming majority of them proved to be equally fast growing (Fig. 2). This indicates that either the majority of isolates share an inherent ability to grow fast or that an adaptive mutational process lifted the initial growth restrictions and led to quick proliferation. Indeed, if the appropriate mutations occurred in different cells at different times, the initial awakening and regrowth in subculture patterns would be in line with the ones observed empirically (Fig. 1B and 2). We view this possibility as unlikely, given that our marine isolates would have to mutate at a rate 8 to 9 orders of magnitude above the accepted range of spontaneous mutation (12). Nonetheless, we tested this possibility by exploring if all cells in subculture were equally fast growing, as would be expected in the case of mutants. Figure 3 shows that they were not, arguing against the idea of genetic change. This leaves us with the following explanation of the empirical observations: in our cultivation experiments, most cells and spores in the inocula were viable but dormant, and they remained so for months (and longer) in the otherwise growth-permissive nutrient medium. They thus showed no response to appropriate nutrients per se. Instead, they initiated growth in a nutrient-independent, intrinsically driven, and apparently stochastic fashion, in accordance with the scout model.
This has interesting implications for the notion of slow-growing species. Microorganisms are often categorized into copiotrophs and oligotrophs (16, 38). The difference in growth rate between these categories is conventionally viewed as an ecological one: the first group is well positioned to opportunistically, and quickly, respond to a nutrient patch, whereas the second one is more competitive, with cells rarely dividing under conditions of continuously low concentrations of substrates. Many natural habitats are typically poor in nutrients, prompting the idea that some, perhaps many, environmental microorganisms are obligate oligotrophs. Apart from being incapable of growth at substrate concentrations above a few mg C/liter (16, 20, 21, 38, 48), these species would also be characterized by an inherently low growth rate. Some of the first long-term cultivation experiments showed that new colonies do indeed appear in petri dishes even after months of incubation (18). Further, the kinetics of colony formation on petri dishes suggested that environmental microorganisms fall into several groups, one of them being slow growers (19). Our observations argue against the prevalence of inherent slow growth as a strategy of all but perhaps a minority of environmental microorganisms. The majority of our isolates regrew within hours or days, and of the isolates that required ≥7 months of incubation during their initial isolation, less than 5% also required ≥7 months to grow in subculture (Fig. 2). In the experiments reported earlier, some microbial cultures might have appeared as bona fide slow growers only superficially, as they did in our experiments. The late appearance of such slow growers might not have been due to the inherently low growth rate but rather to their late awakening, followed by fast growth, as is implicit in the scout model.
These observations appear to be rather general, as they are equally applicable to marine and soil species and both spore and non-spore formers. The molecular mechanism of a process of such applicability is of interest, but it might be difficult to resolve using environmental isolates for which we lack genetic tools. To facilitate future studies of the molecular underpinning of dormancy and growth resumption, we checked if standard model species, such as E. coli, showed similarity to environmental isolates in growth behavior. Our experiments indicate that dormant cells of this species indeed maintain inactivity for at least weeks and months, and they appear to exit the state stochastically in the absence of obvious inducers (Fig. 4). The continuous resumption of E. coli growth after antibiotic challenge was recently reported by several research groups (3, 22, 40). We note, however, that the time scale of these experiments was universally short (minutes to hours), leaving the possibility that the observed wake-up kinetics were due to slight physiological differences among the survivors. Such differences could include, but are not limited to, heterogeneities in the degree of cell damage and the repair needed (10) and are ordinarily used to explain why cells from an apparently homogenous mix, such as a logarithmically growing and shaken culture, would not form colonies at precisely the same time and of precisely the same size. This explanation becomes less likely if colonies keep appearing as the time scale of incubation increases. The time scale of our experiments (weeks to months) virtually eliminates this explanation, instead pointing to a random awakening of surviving dormant cells.
The pattern of scout awakening is reminiscent of the spontaneous reactivation of latent infections (46), and we examined the growth resumption of M. smegmatis, an established model for Mycobacterium tuberculosis (47). This species provided the best illustration so far of how dormant cells resume activity in a time-independent manner (Fig. 5). Note that M. smegmatis is a fast-growing organism, and it is remarkable that, of the cells that ultimately grew, so many waited to initiate growth for several months even though they were continuously exposed to growth-permissive conditions. We checked if genetic change was involved by examining growth kinetics of the progeny, especially since microorganisms are known to mutate with higher frequency under stress (6), such as the conditions of starvation and oxygen deprivation the cells encounter in the Wayne and Sohaskey model. Subculturing isolates that initially took weeks to grow showed that they were not inherently different with respect to their growth rate. Therefore, if cells accumulated mutations during the resting stage, these did not affect their rate of division. As was the case with the environmental isolates, this leaves us with the following interpretation of the empirical observations: dormant cells of M. smegmatis were similar in their growth characteristics and woke up independently of the environmental cues or mutational change. The pattern in which growth events accumulated during long-term incubation is particularly informative (Fig. 5A and B). The best regression proved linear, providing statistical evidence for the randomness of cell awakening in M. smegmatis.
Mechanism of awakening and implications of the scout model.
The regulatory mechanism of scout formation is not known, but it may be similar to the reported cases of noise-driven, stochastic gene expression (9, 29, 30). It is, in principle, possible that the stochastic expression or repression of a master regulatory gene would determine whether the cell remains dormant or wakes into activity and what the relative proportions of these two phenotypes in the otherwise isogenic population would be. This would make the dormant/scout state similar to several well-known and researched examples of microbial bistability (13). While this remains hypothetical, our data provide strong support for the phenomenon itself: nongrowing cells of taxonomically diverse microorganisms can resume growth at widely different intervals and in a seemingly random pattern. The simplest explanation is that their exit from dormancy is not triggered by external factors and is stochastic. Similar patterns of awakening exhibited by phylogenetically unrelated species explored here further point to the possibly general nature of the phenomenon. This may be due to the simplicity of the strategy and its potential benefits for microbial survival. Indeed, if cells in a clonal population of just 10,000 cells exit dormancy at about the rate we observed in M. smegmatis (≈0.01%/day) (Fig. 5A), and under adverse conditions each survives for 10 days, then such a population (i) has 10 active cells at any given time continuously scouting the environment and checking if the conditions are appropriate for growth, (ii) is capable of quick growth response to such conditions at any moment, and (iii) can afford scouting for several decades before going extinct. Interestingly, purely theoretical considerations and mathematical modeling suggest that stochastic switching in behavior can be advantageous over changes due to sensing, at least in infrequently changing environments (25).
A survival strategy based on the stochastic awakening of dormant cells/spores may have interesting implications. For example, in health sciences this strategy may explain why relapses of latent infections have random patterns. It could be implicated in episodes of, e.g., reactivated tuberculosis, which may be due to randomly awakening scouts whose growth, for one reason or another, was not eliminated by the immune system. This is important, because once the mechanism of postulated awakening is resolved, one could envision devising a method of artificially wakening all dormant cells into scouts, followed by treatment with the available antimycobacterial drugs, which today mainly target actively growing cells.
We further note an implication of our findings for the concept of viable-but-nonculturable cells (VBNC), a hypothetical state of a microbial cell in which it does not grow on a medium typically sufficient to cultivate the species in question (50). VBNCs are thought to be alive and, in principle, capable of growth (and thus, in the case of pathogens, of reinfection), and their existence has significant empirical support (for a recent review, see reference 36). However, the idea has been repeatedly challenged by others (7), who suggested that VBNCs do not exist and are cells that are dead or injured beyond the possibility of repair. The main rationale for this alternative interpretation is that it proved difficult to produce unequivocal evidence for the revival of VBNCs. The principal challenge was that in all populations that were claimed to be in the VBNC state, there always was a small but relentless fraction of cultivable cells. As a consequence, what appeared to be a revival of VBNC could also be interpreted as the regrowth of the few cultivable cells, confusing the picture. Our observations may explain the nature of such cells, and they indicate that attempts to eliminate them are futile. We hypothesize that VBNCs are simply dormant cells that do not respond to environmental cues, and the cultivable fraction contains scouts continuously (and randomly) generated by such dormant cells. If so, these cultivable cells, rather than confusing the picture, might have been the very evidence of the revival researchers tried to prove, and this finding may contribute to resolving the long-standing arguments in VBNC research.
Conclusions.
Observations made on the revival of dormant environmental cells and spores from soil and marine habitats, as well as the standard model species E. coli and M. smegmatis, indicate that such revival is a result of low-frequency, stochastic events that occur during a long period of time and independently of environmental cues, with implications for general microbiology and health sciences.
ACKNOWLEDGMENTS
We thank J. Clardy, R. Kolter, E. Rubin (Harvard University), D. Dubnau (Public Health Research Institute), B. Price (University of California), and K. Lewis (Northeastern University) for the discussion of the ideas expressed in this paper and P. Muller (Northeastern University) for preparing Fig. 4.
This research was supported by NIH grants 1RC1DE020707-01 and R21 DE018026-01A1, NSF grant DEB-0816840, and DOE grants DE-FG02-07ER64507 and DE-FG02-04ER63782 to S.S.E. and 1R43AI091224 to A.S.
Footnotes
Published ahead of print 24 February 2012
This article is contribution 281 of the Marine Science Center, Northeastern University, Nahant, Massachusetts, USA.
REFERENCES
- 1. Avery SV. 2006. Microbial cell individuality and the underlying sources of heterogeneity. Nat. Rev. Microbiol. 4:577–587 [DOI] [PubMed] [Google Scholar]
- 2. Baker GC, Smith JJ, Cowan DA. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541–555 [DOI] [PubMed] [Google Scholar]
- 3. Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625 [DOI] [PubMed] [Google Scholar]
- 4. Belanger AE, Hatfull GF. 1999. Exponential-phase glycogen recycling is essential for growth of Mycobacterium smegmatis. J. Bacteriol. 181:6670–6678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berney M, Hammes F, Bosshard F, Weilenmann HU, Egli T. 2007. Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight kit in combination with flow cytometry. Appl. Environ. Microbiol. 73:3283–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bjedov I, et al. 2003. Stress-induced mutagenesis in bacteria. Science 300:1404–1409 [DOI] [PubMed] [Google Scholar]
- 7. Bogosian G, Bourneuf EV. 2001. A matter of bacterial life and death. EMBO Rep. 2:770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boulos L, Prevost M, Barbeau B, Coallier J, Desjardins R. 1999. LIVE/DEAD BacLight: application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J. Microbiol. Methods 37:77–86 [DOI] [PubMed] [Google Scholar]
- 9. Choi PK, Cai L, Frieda K, Xie XS. 2008. A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science 322:442–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Desnues B, et al. 2003. Differential oxidative damage and expression of stress defense regulons in culturable and non-culturable Escherichia coli cells. EMBO Rep. 4:400–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dick T, Lee BH, Murugasu-Oei B. 1998. Oxygen depletion induced dormancy in Mycobacterium smegmatis. FEMS Microbiol. Lett. 163:159–164 [DOI] [PubMed] [Google Scholar]
- 12. Drake JW. 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. U. S. A. 88:7160–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dubnau D, Losick R. 2006. Bistability in bacteria. Mol. Microbiol. 61:564–572 [DOI] [PubMed] [Google Scholar]
- 14. Epstein SS. 2009. General model of microbial uncultivability, p 131–150 In Epstein SS. (ed) Uncultivated microorganisms. Spinger, Heidelberg, Germany [Google Scholar]
- 15. Epstein SS. 2009. Microbial awakenings. Nature 457:1083. [DOI] [PubMed] [Google Scholar]
- 16. Fry JC. 1990. Oligotrophs. In Edwards C. (ed), Microbiology of extreme environments. Open University Press, Milton Keynes, United Kingdom [Google Scholar]
- 17. Haglund AL, Lantz P, Tornblom E, Tranvik L. 2003. Depth distribution of active bacteria and bacterial activity in lake sediment. FEMS Microbiol. Ecol. 46:31–38 [DOI] [PubMed] [Google Scholar]
- 18. Hattori T. 1976. Plate count of bacteria in soil on a diluted broth as a culture medium. Rep. Inst. Agr. Res. Tohoku Univ. 27:23–30 [Google Scholar]
- 19. Hattori T, et al. 1997. Advances in soil microbial ecology and the biodiversity. Antonie Van Leeuwenhoek 72:21–28 [DOI] [PubMed] [Google Scholar]
- 20. Horowitz A, Krichevsky M, Atlas RM. 1983. Characteristics and diversity of subarctic marine oligotrophic, steno-heterotrophic, and euryheterotrophic bacterial populations. Can. J. Microbiol. 29:527–535 [Google Scholar]
- 21. Ishida Y, Kadota H. 1981. Growth patterns and substrate requirements of naturally occurring obligate oligotrophs. Microb. Ecol. 7:123–130 [DOI] [PubMed] [Google Scholar]
- 22. Joers A, Kaldalu N, Tenson T. 2010. The frequency of persisters in Escherichia coli reflects the kinetics of awakening from dormancy. J. Bacteriol. 192:3379–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13–18 [DOI] [PubMed] [Google Scholar]
- 24. Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kussell E, Leibler S. 2005. Phenotypic diversity, population growth, and information in fluctuating environments. Science 309:2075–2078 [DOI] [PubMed] [Google Scholar]
- 26. Kwon KK, Lee SJ, Park JH, Ahn TY, Lee HK. 2006. Psychroserpens mesophilus sp. nov., a mesophilic marine bacterium belonging to the family Flavobacteriaceae isolated from a young biofilm. Int. J. Syst. Evol. Microbiol. 56:1055–1058 [DOI] [PubMed] [Google Scholar]
- 27. Labeda DP, Shearer MC. 1990. Isolation of actinomycetes for biotechnological applications, p 1–19 In Labeda DP. (ed), Isolation of biotechnological organisms from nature. McGraw-Hill Publishing Co., New York, NY [Google Scholar]
- 28. Ludwig W, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maamar H, Dubnau D. 2005. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 56:615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maamar H, Raj A, Dubnau D. 2007. Noise in gene expression determines cell fate in Bacillus subtilis. Science 317:526–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morita RY. 1982. Starvation-survival of heterotrophs in the marine environment. Adv. Microb. Ecol. 6:117–198 [Google Scholar]
- 32. Morita RY. 1988. Bioavailability of energy and its relationship to growth and starvation survival in nature. Can. J. Microbiol. 34:436–441 [Google Scholar]
- 33. Mukamolova GV, Kaprelyants AS, Young DI, Young M, Kell DB. 1998. A bacterial cytokine. Proc. Natl. Acad. Sci. U. S. A. 95:8916–8921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nichols D, et al. 2008. Short peptide induces an “uncultivable” microorganism to grow in vitro. Appl. Environ. Microbiol. 74:4889–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nookaraju A, Chandrama UP, Park SW. 2010. In vitro tuberization of potato as influenced by plant growth promoting rhizobacteria. K J. Life Environ. Sci. 32:27–34 [Google Scholar]
- 36. Oliver JD. 2010. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 34:415–425 [DOI] [PubMed] [Google Scholar]
- 37. Paidhungat M, Setlow P. 2000. Role of Ger proteins in nutrient and non-nutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182:2513–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Poindexter JS. 1981. Oligotrophy: feast and famine existence. Adv. Microb. Ecol. 5:63–89 [Google Scholar]
- 39. Postgate JR. 1976. Death in macrobes and microbes. Symp. Soc. Gen. Microbiol. 26:1–18 [Google Scholar]
- 40. Roostalu J, Joers A, Luidalepp H, Kaldalu N, Tenson T. 2008. Cell division in Escherichia coli cultures monitored at single cell resolution. BMC Microbiol. 8:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shah IM, Laaberki MH, Popham DL, Dworkin J. 2008. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135:486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stach JE, Bull AT. 2005. Estimating and comparing the diversity of marine actinobacteria. Anotonie Van Leeuwenhoek 87:3–9 [DOI] [PubMed] [Google Scholar]
- 43. Stach JE, et al. 2003. Statistical approaches for estimating actinobacterial diversity in marine sediments. Appl. Environ. Microbiol. 69:6189–6200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stackebrandt E, Goebel BM. 1994. Taxonomic note: a place for DNA:DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteria. Int. J. Syst. Bacteriol. 44:846–849 [Google Scholar]
- 45. Stevenson LH. 1978. A case for bacterial dormancy in aquatic systems. Microb. Ecol. 4:127–133 [DOI] [PubMed] [Google Scholar]
- 46. Stewart GR, Robertson BD, Young DB. 2003. Tuberculosis: a problem with persistence. Nat. Rev. Microbiol. 1:97–105 [DOI] [PubMed] [Google Scholar]
- 47. Tyagi JC, Sharma D. 2002. Mycobacterium smegmatis and tuberculosis. Trends Microbiol. 10:68–69 [DOI] [PubMed] [Google Scholar]
- 48. Van Gemerden H, Kuenen GJ. 1984. Strategies for growth and evolution of microorganisms in oligotrophic habitats, p 25–44 In Hobbie JEL, Williams PJ. (ed), Heterotrophic activity in the sea. Plenum Press, New York, NY [Google Scholar]
- 49. Wayne LG, Sohaskey CD. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139–163 [DOI] [PubMed] [Google Scholar]
- 50. Xu HS, et al. 1982. Survival and viability of non-culturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb. Ecol. 8:313–323 [DOI] [PubMed] [Google Scholar]