Abstract
Contamination of food contact surfaces with pathogens is considered an important vehicle for the indirect transmission of food-borne diseases. Five different cleaning cloths were assessed for the ability to remove viruses from food contact surfaces (stainless steel surface and nonporous solid surface) and to transfer viruses back to these surfaces. Cleaning cloths evaluated include two different cellulose/cotton cloths, one microfiber cloth, one nonwoven cloth, and one cotton terry bar towel. Four viral surrogates (murine norovirus [MNV], feline calicivirus [FCV], bacteriophages PRD1 and MS2) were included. Removal of FCV from stainless steel was significantly greater (P ≤ 0.05) than that from nonporous solid surface, and overall removal of MNV from both surfaces was significantly less (P ≤ 0.05) than that of FCV and PRD1. Additionally, the terry towel removed significantly fewer total viruses (P ≤ 0.05) than the microfiber and one of the cotton/cellulose cloths. The cleaning cloth experiments were repeated with human norovirus. For transfer of viruses from cloth to surface, both cellulose/cotton cloths and microfiber transferred an average of 3.4 and 8.5 total PFU, respectively, to both surfaces, and the amounts transferred were significantly different (P ≤ 0.05) from those for the nonwoven cloth and terry towel (309 and 331 total PFU, respectively). There was no statistically significant difference (P > 0.05) in the amount of virus transfer between surfaces. These data indicate that while the cleaning cloths assessed here can remove viruses from surfaces, some cloths may also transfer a significant amount of viruses back to food contact surfaces.
INTRODUCTION
Each year in the United States and worldwide, human noroviruses (HuNoVs) are the leading cause of nonbacterial gastroenteritis, being associated with 80 to 90% of reported outbreaks (17). Moreover, HuNoVs are the primary cause of food-borne disease outbreaks reported in the United States, causing 5.5 million (58%) illnesses each year (37). Economically, HuNoVs cost approximately $625 per case—equivalent to $3.7 billion each year with respect to food-borne illnesses attributable to HuNoVs (39). Noroviruses may be transferred to people via a direct route (i.e., person to person) or indirectly (i.e., fecal-oral route) by contact with fomite (inanimate) surfaces (6) and ingestion of contaminated food and water (33).
Contaminated fomite surfaces have been well-documented to be a route of HuNoV transmission, especially with respect to outbreaks in enclosed environments such as long-term care facilities, hospitals, cruise ships, camping trips, and military settings (18, 19, 25, 34, 54). One of the critical factors of HuNoV transmission is its ability to persist long term in the environment. Lamhoujeb et al. (2009) reported that HuNoVs can persist on stainless steel and polyvinylchloride (PVC) surfaces for from 1 to more than 7 weeks, depending on the surface, temperature, and relative humidity (24). In addition, once a surface becomes contaminated, virus particles can easily be transferred between inanimate and animate objects (e.g., from contaminated surfaces to hands and vice versa) (3, 20). It is still unclear how many food-borne disease outbreaks are a direct result of transmission of HuNoVs to foodstuffs via contaminated food contact surfaces. However, because of the extremely low infectious dose (as few as 18 infectious virus particles) (49), high number of viruses shed during infection (1011 and 106 genomic copies per gram of stool or vomit, respectively), and ability to persist, fomite surfaces are considered to be a major route in the spread of HuNoV gastroenteritis (27, 36).
Cleaning with chemical disinfectants and sanitizers is considered an important step in preventing the transmission of HuNoVs from contaminated surfaces. The efficacy of various cleaning compounds (alcohols, quaternary ammonium compounds, sodium hypochlorite) against both HuNoVs and HuNoV surrogates (murine norovirus [MNV], feline calicivirus [FCV], MS2 bacteriophage) on different surface materials, primarily stainless steel, melamine, and PVC, has been assessed (3, 16, 31). It is important to note that most studies are based on inactivation of cultivable HuNoV surrogates since there is no cell culture assay to measure the infectivity of HuNoVs. Overall, the studies investigating disinfectants on surfaces agree that the most effective compound against HuNoVs is sodium hypochlorite (NaClO). However, the concentration (5,000 ppm, or 15.6 ml bleach in 1 liter of water) of NaClO determined to be most effective for inactivation of HuNoVs far exceeds what is mandated (i.e., 200 ppm) for sanitizing food contact surfaces in the Food Code published by the U.S. Food and Drug Administration (FDA) (50). In order to use this high concentration of NaClO on food contact surfaces, the area that is disinfected must then be rinsed with clean water and receive a final wipe down with a 200-ppm bleach solution (50). Furthermore, prior to sanitization, the area should also be cleaned with detergent and water in order to remove food residues and maintain the expected efficacy of the sanitizing compound (46). Moreover, the concentration and contact time required for initial inactivation of HuNoV may damage (i.e., oxidize) stainless steel surfaces, the surfaces predominantly used and recommended for food preparation. Because of this, quaternary ammonium and ethyl alcohol compounds are most often used during regular cleaning and sanitizing of food contact surfaces, even though these sanitizers are not very effective against nonenveloped viruses such as HuNoVs and other enteric viruses (e.g., rotavirus, hepatitis A virus, and adenovirus) (16, 45).
Another component involved in the cleaning and sanitizing of food contact surfaces is the cloth used to wipe these surfaces during regular and intermittent cleaning of food preparation areas and tabletops. Because cleaning cloths come in contact with potentially contaminated surfaces, their ability to remove pathogens from surfaces as well as their potential to transfer pathogens to clean surfaces must be evaluated. Tebbutt (1988) (48) evaluated disposable and reusable disinfectant cloths for cleaning Formica surfaces inoculated with fecal bacteria and concluded that while reusable cloths are more convenient and less expensive, these cloths are often not disinfected properly, and thus, use of disposable cloths may reduce the risk of cross contamination. More recent studies have also reported on the effectiveness of select cleaning cloths (e.g., nonwoven fiber, microfiber, and generic kitchen cloths) for removal of bacteria (22, 23); however, none have evaluated cloths for their effectiveness against viral pathogens, and few have evaluated the cloth as a source for potential cross contamination within food service industry environments (48).
Therefore, the goal of the present study was to evaluate five different cleaning cloths for their ability to remove HuNoVs, MNV, FCV, and bacteriophages MS2 and PRD1 from a stainless steel surface and a nonporous acrylic-based, solid surface. The potential for each cleaning cloth type to transfer viruses back to the surfaces was also evaluated. To our knowledge, this is the first study to evaluate cleaning cloth efficacy for removal of viruses from food contact surfaces and transfer of viruses from cloth to surface.
MATERIALS AND METHODS
Preparation of virus stocks.
MS2 and PRD1 bacteriophages were prepared as described previously, with modifications for MS2 (1, 15). Briefly, MS2 (ATCC 16696-B1) and PRD1 bacteriophages were generated using the double-agar-layer (DAL) method and Escherichia coli C3000 (ATCC 15597) and Salmonella enterica subspecies enterica serovar Typhimurium LT2 (ATCC 19585) bacterial hosts, respectively. The bacteriophages were then extracted from cell lysates with an equal volume of chloroform (Sigma, St. Louis, MO), sterile filtered, centrifuged at 4,000 × g for 30 min at 4°C, aliquoted, and stored at −80°C. Stock titers were determined by the DAL method. Both MS2 and PRD1 were selected because of their prior use as surrogates for enteric viruses, such as NoV and human adenoviruses, respectively (8, 12). MNV (type 1) and FCV (strain F9) stocks were propagated in monolayers of RAW 264.7 (ATCC TIB-71) and Crandall Reese feline kidney (CrFK) cells, as described previously (14). MNV and FCV stock titers were determined by plaque assay as described by Gibson and Schwab (2011) (14). Both MNV and FCV were chosen because of their prior use as surrogates for the study of HuNoVs (4, 8, 16, 26).
Human norovirus was prepared from a diarrheal stool sample as described previously, with modifications (1). Briefly, a stool sample positive for Norwalk virus GI.1 (denoted substrain 8fIIb and kindly provided by Kellogg Schwab, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD) was diluted in 1× phosphate-buffered saline (PBS) to make a 10% (wt/vol) stool suspension. The stool suspension was vortexed for 5 min, followed by centrifugation at 3,000 × g for 10 min at 4°C to pellet the suspended solids and clarify the sample. The supernatant was removed, placed in a new tube, and stored at 4°C. While the exact concentration of HuNoV particles in the stool sample was unknown, positive amplicons could be detected up to a dilution of 1:10,000 of the prepared stool sample by heat release and real-time reverse transcription-PCR (RT-PCR) (refer to Table 1 for primer/probe information; for PCR conditions, see below).
Table 1.
Primers and probes for detection of human norovirus and hepatitis G virusa
| Microorganism | GenBank accession no. | Primer or probe name | Primer/probeb final concn (nM) | Sequencec | Product size (bp) | Product region | Reference |
|---|---|---|---|---|---|---|---|
| Human norovirus (GI) | M87661 | Cog1-F | 1,000 | 5′-CGYTGGATGCGNTTYCATGA-3′ | 85 | ORF1-ORF2 junction | 21 |
| Cog1-R | 1,000 | 5′-CTTAGACGCCATCATCATTYAC-3′ | |||||
| Ring1A-TP probe | 100 | 5′-FAM-AGATYGCGATCYCCTGTCCA-BHQ-3′ | |||||
| Ring1B-TP probe | 100 | 5′-FAM-AGATCGCGGTCTCCTGTCCA-BHQ-3′ | |||||
| Hepatitis G virus | U44402 | HepG-F | 400 | 5′-CGGCCAAAAGGTGGTGGATG-3′ | 185 | 5′ UTRd | 40 |
| (internal standard) | HepG-R | 400 | 5′-CGACGAGCCTGACGTCGGG-3′ | ||||
| HepG probe | 200 | 5′ AGGTCCCTCTGGCGCTTGTGGCGAG-3′ |
Hepatitis G virus was the RNA internal standard for analysis of inhibition.
The 6-carboxyfluorescein (FAM) quencher is black hole quencher 1 (BHQ-1).
Mixed bases in degenerate primers and probes are as follows: Y = C and T; R = A and G; B = C and T, G; N = A, C, T, and G.
UTR, untranslated region.
Real-time RT-PCR assay for NoV.
For detection of HuNoVs, real-time RT-PCR was completed using a Mastercycler ep realplex4 system (Eppendorf, Hamburg, Germany). Prior to amplification, HuNoV RNA was extracted by heat release at 95°C for 5 min as described previously (41). For the HuNoV-positive control during real-time RT-PCR, HuNoV RNA was extracted from the prepared stool sample using a QIAamp DNA blood minikit and buffer AVL with carrier RNA (Qiagen, Valencia, CA), following the manufacturer's protocols. The positive-control HuNoV RNA was aliquoted and stored at −80°C.
Amplification of HuNoV RNA was performed in 25-μl reaction mixtures containing 12.5 μl of 2× master mix (QuantiTect Probe RT-PCR kit; Qiagen, Valencia, CA), 5 U RNase inhibitor (Promega, Madison, WI), custom primers (Integrated DNA Technologies, Coralville, IA) and dual-labeled TaqMan probes (Biosearch Technologies, Novato, CA) at the final concentrations reported in Table 1, 5 μl of prepared sample, and diethyl pyrocarbonate (DEPC)-treated water for the remaining volume. Real-time RT-PCR amplification was performed under the following conditions: reverse transcription for 30 min at 50°C and denaturation for 15 min at 95°C, followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 60 s.
Real-time RT-PCR assay for inhibition.
Real-time RT-PCR inhibition analysis using a hepatitis G virus (HGV) armored RNA standard (Asuragen, Austin, TX) was completed for all samples as described previously (15) using the primers and probes listed in Table 1. Briefly, each 25-μl reaction mixture contained 12.5 μl of 2× master mix (QuantiTect Probe RT-PCR kit; Qiagen), 5 U RNase inhibitor (Applied Biosystems), 400 nM primers (Invitrogen) and 200 nM dual-labeled TaqMan probe (Biosearch Technologies, Novato, CA), 5 μl of prepared sample, 2 μl of a known amount of HGV RNA, and DEPC-treated water for the remaining volume. Real-time RT-PCR amplification was performed under the following conditions: reverse transcription for 30 min at 50°C and denaturation for 15 min at 95°C, followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 60 s. The quantity of HGV measured in the unknown sample was compared to the quantity measured in corresponding HGV-positive controls. Each batch of samples assayed for inhibition included a negative control of HGV master mix containing DEPC-treated water substituted for HGV RNA and at least 3 positive-control reaction mixtures containing only HGV RNA and no sample. A sample was deemed uninhibited if the cycle threshold (CT) of the seeded HGV was less than 1 cycle higher than the mean of the expected CT obtained from the HGV-positive controls. Conversely, a sample was determined to be inhibited if the CT of the seeded HGV was more than 1 cycle higher than the mean of the expected CT obtained from the HGV-positive controls. Complete absence of a CT value for seeded HGV was indicative of total inhibition of the real-time RT-PCR.
Assessment of virus removal from surfaces.
Five cleaning cloths were assessed for removal of viral surrogates and HuNoVs from stainless steel and solid surfaces. Two different blended cellulose/cotton cloths (cellulose, 70%; cotton, 30%), microfiber, nonwoven wipes (viscose, 50%; polyester, 50%), and generic cotton terry bar towels (100% cotton) were assessed. Cloths were selected on the basis of current (e.g., cotton terry bar towels) and/or potential (e.g., microfiber) use by the food service industry and commercial availability. The cloths were cut into 5-cm2 pieces, placed in sterilization pouches (VWR, Radnor, PA), and autoclaved at 121°C and 15 lb/in2 for 15 min. Stainless steel sheets (type 304/14 gauge and type 430/15 gauge; Advance Tabco, Edgewood, NY) and 3- by 3-in. (7.6-cm2) 100% acrylic-based, nonporous solid surface samples (13-mm-thick Wilsonart laminate; Wilsonart International, Inc., Temple, TX) were used for all experiments. Surfaces were sterilized before the beginning of the described study and after each experiment. For stainless steel, the surfaces were initially cleaned with soap and water, wrapped in aluminum foil, and autoclaved at 121°C and 15 lb/in2 for 15 min. After each experiment, the stainless steel surfaces were exposed to UV light for 30 min in a biosafety level 2 (BSL-2) hood, followed by application of a 10% solution of household bleach for 10 min. The stainless steel surfaces were then autoclaved, cleaned again, wrapped in aluminum foil, and autoclaved again. For the acrylic-based solid surface, the same protocol was followed, except the solid surface could not be autoclaved.
One hundred microliters of a prepared virus cocktail containing on average of 5.4 × 105, 1.1 × 106, 1 × 105, and 7.4 × 105 PFU of each viral surrogate (MS2, PRD1, FCV, and MNV, respectively) was inoculated by random spotting onto either a 7.6-cm2 area of solid surface or stainless steel. One hundred microliters of 1× PBS was also inoculated onto one 7.6-cm2 surface as a negative control. For HuNoVs, 100 μl (corresponding to 10,000 RT-PCR units [RT-PCRU]) of prepared, undiluted HuNoV-containing stool was used for inoculation. Inoculated surfaces were allowed to dry completely for 30 to 45 min in a BSL-2 hood.
With sterile forceps, a 5-cm2 piece of sterile cloth was dampened with high-quality, Milli-Q lab water (Millipore, Billerica, MA) and placed on the 7.6-cm2 surface. The surface was wiped by hand 3 times vertically and 3 times horizontally. Gloves were worn throughout the process and sprayed with 70% ethanol between cloths. We attempted to minimize variability between experimental replicates due to differences in the pressure applied to the cloth while wiping by having the same person conduct all of the removal and transfer experiments.
The surfaces and cleaning cloths were then eluted as described below in “Recovery of viruses from cloths and surfaces.” The cleaning cloths were eluted for recovery only of viral surrogates and not of HuNoV. For every cloth type-surface type paired experiment, each cloth was assessed in triplicate with two positive surface controls (i.e., the surface was inoculated but not wiped), one negative surface control (i.e., a surface inoculated with 1× PBS was wiped), and one negative cloth control (i.e., no wiping was done). All eluates and inocula were assayed for MS2, PRD1, MNV, and FCV as described for the stock preparations. Each assay included both positive (known amount of viral surrogate) and negative (1× PBS) controls. Dilutions for viral surrogates were prepared in 1× PBS. Preliminary experiments in which each virus stock was seeded on the other virus host cell line did not generate any cross-reactive plaque formation, nor did the seeding affect the formation of the expected number of PFU for the host virus (data not shown). Real-time RT-PCR was used for analysis of HuNoV. For real-time RT-PCR, eluates were prepared by making 2-fold serial dilutions in DEPC-treated water, and RNA was extracted by heat release for 5 min at 95°C.
Assessment of virus-to-surface transfer.
Initially, 5-cm2 pieces of cleaning cloths were dampened with high-quality, Milli-Q lab water (Millipore). The damp cloths were subsequently seeded in duplicate with 100 μl of prepared virus cocktail containing approximately 105 to 106 PFU of each viral surrogate (MS2, PRD1, MNV, FCV). The virus cocktail was allowed to equilibrate to the cloth for 1 min. Each 7.6-cm2 surface was wiped with one inoculated cloth 3 times vertically and 3 times horizontally. For each cloth type-surface type paired experiment, one positive-control cloth was also inoculated with the viral surrogate cocktail but not used for wiping. The surfaces and cleaning cloths were then eluted as described below in “Recovery of viruses from cloths and surfaces.” All eluates and inocula were assayed for MS2, PRD1, MNV, and FCV as described for stock preparation and titer determination. Transfer experiments were completed using only viral surrogates and did not include assessment of HuNoVs due to the low stock concentrations and real-time RT-PCR limit of detection.
Recovery of viruses from cloths and surfaces.
Prior to assessment of cleaning cloths, a method for elution of the viruses from the surfaces as well as from the cleaning cloths was optimized. For elution of viruses from the surfaces, a method described previously by Taku et al. (2002) was used with modifications (47). Briefly, 600 μl of elution buffer (0.05 M glycine [pH 6.5], 0.1% Tween 80, and 0.3 M NaCl) was added to the 7.6-cm2 areas of inoculated surfaces. The elution buffer was allowed to contact the surface for 10 min. Using a sterile cell scraper (Fisher Scientific, Pittsburgh, PA), the surface was scraped to spread the elution buffer and detach adhered viruses. The elution buffer was then collected from the surface using a micropipette. The total volume of buffer containing viruses was recorded in order to calculate recovery efficiency.
For elution of viruses from cleaning cloths, the cloths (5-cm2 pieces) were placed in 50-ml polypropylene tubes containing 20 ml of elution buffer. The tubes containing cloths were processed at room temperature for 30 min with shaking at 150 rpm. After shaking, the elution buffer containing viruses from the cloths was removed from the tube and placed in a new 50-ml polypropylene tube. The total volume of buffer containing viruses was recorded in order to calculate recovery efficiency.
Statistical analyses.
For all experiments, statistical analyses were performed using JMP software (version 9.0; SAS, Cary, NC). Virus PFU values were transformed to logarithmic values to achieve a normal distribution. One-way analysis of variance (ANOVA) and Tukey-Kramer honestly significant difference (HSD) were used when performing two or more mean comparisons within a data set. In all cases, statistical significance was set at an α value of ≤0.05.
RESULTS
Recovery efficiency of elution method.
To assess the efficiency of the elution method, virus recoveries were calculated for both the surfaces and the cleaning cloths for virus removal and transfer experiments, respectively. Values for recovery efficiency were based on the number of PFU reported for the positive controls (surface or cloth) divided by the number of PFU in the inoculum. The optimized elution method described in the Materials and Methods section achieved average overall efficiencies of recovery from the surfaces of >100, 37, 41, and 57% of MS2 (n = 14), PRD1 (n = 14), MNV (n = 9), and FCV (n = 8), respectively. Elution efficiencies from cleaning cloths during transfer experiments for MS2 (n = 38), PRD1 (n = 40), and FCV (n = 10) were >100, >100, and 36%, respectively. Elution efficiency of MNV from cleaning cloth transfer experiments could not be calculated because the values for the positive controls for the assay (i.e., MNV of a known concentration analyzed simultaneously with the samples to ensure the validity of the plaque assay) were significantly lower than expected; thus, the PFU counts obtained from elution of the cleaning cloths were not considered valid and these data were excluded.
Removal of viruses from solid surface and stainless steel.
On average, the five cleaning cloths removed 2.85 log10 and 3.15 log10 units of the viral surrogates on solid surface (Fig. 1A) and stainless steel (Fig. 1B), respectively. Statistically significant differences (P = 0.0031) between surfaces were seen for removal of FCV, with stainless steel surfaces yielding greater removal. In addition, removal of MNV from both surfaces combined was significantly less than removal of FCV and PRD1 (P = 0.0016 and 0.0004, respectively) (Fig. 1). Comparing total virus removal by cleaning cloth type, the terry bar towel removed significantly fewer viruses than the cellulose/cotton blend 1 (P = 0.0064) and the microfiber (P = 0.0016) cloths. Initially, the microfiber was used as a dry cloth, but upon further consideration, a dampened microfiber cloth was used for remaining removal experiments. The dry microfiber removed an average of ≤10 PFU (P ≤ 0.05) (data not shown), while the dampened microfiber cloth performed similarly to the other cloths. Therefore, data for microfiber removal of viruses presented in Fig. 1 include only the log10 amount removed by damp microfiber.
Fig 1.
Virus removal from solid surface (A) and stainless steel (B) by cleaning cloth and virus type. Error bars indicate standard deviations. *, only dry microfiber was included in these data for MNV removal, and thus, the data were excluded from statistical analyses, as dry microfiber was determined to be significantly different from dampened cleaning cloths; a, statistically significant difference (P = 0.0031) in removal of FCV from solid surface and stainless steel; b, statistically significant difference in removal of MNV from both surfaces compared to FCV and PRD1 removal (P = 0.0016 and 0.0004, respectively).
Data collected on removal of HuNoVs by cleaning cloths are presented in Table 2. These data are results from elution of the surface after wiping with the contaminated cloths. Results for cellulose/cotton cloth 2 are shown for only one experiment (performed in triplicate) on each surface, as this cloth became unavailable for use in the remaining replications. During preliminary analyses, total inhibition (i.e., no detection of the HGV RNA standard within the sample matrix, as indicated by lack of a CT value) was detected in samples undiluted and diluted to 2−1 (data not shown); therefore, all samples were analyzed at a 2−2 dilution. Even still, samples diluted to 2−2 exhibited partial (i.e., a shift or increase in the expected CT value for the HGV RNA standard) and/or total inhibition. The impact of inhibitors on the real-time RT-PCR assays makes quantitative interpretation of these data nearly impossible.
Table 2.
Human norovirus removal from solid surface and stainless steel using cleaning cloths
| Cleaning cloth type | Average CT value (SD)a |
|||||
|---|---|---|---|---|---|---|
| Solid surface |
Stainless steel |
|||||
| Rep 1 | Rep 2 | Rep 3b | Rep 1 | Rep 2 | Rep 3b | |
| Cellulose/cotton 1 | 35.31 | Not detected | Not detected | Not detected | 38.55 | Not detected |
| Cellulose/cotton 2 | 37.55 | ND | ND | Not detected | ND | ND |
| Microfiber | 33.90 (1.03) | 32.89 (0.13) | 37.33 (1.95) | 33.06 (0.44) | 33.36 (0.09) | 38.26 |
| Nonwoven | 34.51 (1.88) | 33.67 (0.53) | 39.00 | 33.00 (0.41) | 33.15 (1.24) | 38.28 |
| Cotton terry towel | 33.12 (0.28) | 32.90 (1.16) | 36.13 (1.60) | 37.12 (2.16) | 33.64 (1.27) | 37.84 |
| Positive control | 32.83 | 31.61 | 37.19 | 34.27 | 32.04 | 35.86 |
Values are average CT values from triplicate samples (for each replicate) based on analyses of a 5-μl portion of a 2−2 dilution of eluate recovered from surfaces after wiping with cleaning cloths. The positive control is from a nonwiped, inoculated surface. All samples were partially inhibited (i.e., if the CT of the seeded HGV was more than 1 cycle higher than the mean of the expected CT obtained from the HGV-positive controls) unless otherwise indicated. Rep, replicate; not detected, no virus RNA was detected in the sample; therefore, no CT values are reported; ND, not done. The standard deviation is not reported for those samples with less than 3 CT values; a lack of a CT value indicates that no RNA was amplified and does not indicate that less than 3 samples were assayed.
None of the samples, including the positive controls, were inhibited.
Transfer of viruses to solid surface and stainless steel.
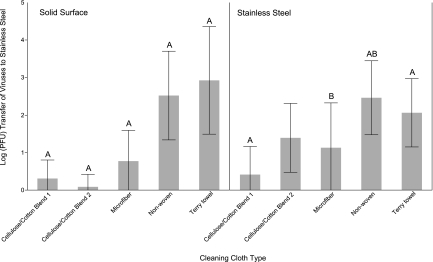
Results of the transfer of total combined virus surrogates (MS2, PRD1, FCV) from the cleaning cloths to stainless steel are provided in Fig. 2. Data for MNV have not been included due to issues occurring with the described plaque assay and a lack of sufficient sample volume remaining to repeat assays. The cellulose/cotton cloths and microfiber transferred an average of 3.4 and 8.5 total virus PFU, respectively, to the solid surface. These three cloths were significantly different (P < 0.0001) from the nonwoven and terry towel cloths for transfer of viruses to solid surface (Fig. 2). For all three viral surrogates (FCV, MS2, PRD1), the nonwoven and terry towel cloths transferred an average of 3.3 × 102 and 8.3 × 102 PFU to the solid surface, respectively. For stainless steel, cellulose/cotton cloth 1 transferred 2.6 PFU, which was significantly different from the amounts for nonwoven and terry towel cloths (P < 0.0001 and P = 0.0009, respectively). In addition, the microfiber cloth transferred to stainless steel significantly less virus (P = 0.0110) than the nonwoven cloth (Fig. 2).
Fig 2.
Total virus (FCV, MS2, PRD1) transfer to solid surface and stainless steel by cleaning cloths. Error bars indicate standard deviations. Letters above the bars represent statistically significant differences (P ≤ 0.05) between cloth types for virus transfer within each surface type.
DISCUSSION
Application of appropriate cleaning tools for the removal of viruses from food contact surfaces—or any surface—is a critical step in preventing the indirect transfer of viruses to persons at food service establishments (6). Therefore, the effectiveness of various cleaning cloths for their removal and subsequent transfer of HuNoVs and HuNoV surrogates on food contact surfaces was studied.
Initially, a method for elution of viruses from surfaces and cleaning cloths was optimized. The method published by Taku et al. (2002) (47) was modified here through the addition of 0.3 M NaCl and 0.1% Tween 80 to the 0.05 M glycine buffer (pH 6.5) that was originally described. The inclusion of NaCl and Tween 80 has been previously described and demonstrated to enhance elution efficiencies from various food and environmental matrices (2, 11, 32, 51). The recovery efficiencies of 37 to >100% for elution of viruses from the surfaces reported here are similar to the recoveries of 32 to 71% for FCV from stainless steel reported by Taku et al. (2002) (47). Our modification of this method was also shown to be effective for recovery of MNV, MS2, PRD1, and HuNoVs from both stainless steel and acrylic, solid surface.
Studies evaluating various cleaning cloths for removal of pathogens within food service environments remain limited, while research in hospital settings (e.g., studies of stainless steel, furniture laminate, and ceramic tile surfaces) has been frequently reported. With respect to hospital settings, microfiber and ultramicrofiber cloths have received the bulk of attention for their potential to improve cleaning efficacy without the need for added detergents (29). Most recently, Smith et al. (2011) (44) evaluated 10 different damp microfiber cloths for removal of pathogens known to cause health care facility-associated infections: Clostridium difficile spores, E. coli, and methicillin-resistant Staphylococcus aureus. The authors reported a mean log10 reduction of 2.21 when single-use damp microfiber cloths were used, while lower reduction levels were reported during repeat use of the cloths to clean a succession of contaminated surfaces. Other studies reported similar or better bacterial reduction values for damp microfiber cloths, depending on the surface and organism (7, 28). Although not directly comparable, the microfiber cloth evaluated in our study had a mean log10 reduction of 3.36 for viruses when used as a damp cloth on both surface types. Therefore, microfiber cloths seem to be effective in the removal of both viruses and bacteria from nonporous surfaces. With respect to food service environments, very few evaluations of cleaning cloths for removal of microbial contamination have been reported, and of those, only insufficiently characterized kitchen fiber cloths, generic cloths, disposable paper wipes, or nonwoven fabric sheets were used (22, 42, 48). Regardless, all of these studies evaluate efficacy against bacterial pathogens and not viruses, and thus, comparison and discussion of virus removal from food contact surfaces by cleaning cloths are not possible.
Our study also demonstrates differences in virus removal between surfaces. In the current study, there was significantly greater removal of FCV from the stainless steel surface than from the solid surface (mean log10 reductions, 3.5 and 2.8, respectively), but this difference was not seen for MNV, MS2, and PRD1. The reasons for the difference in removal of FCV are not known but may be due to the variable affinities of FCV adsorption to solid surface and stainless steel. Electrostatic interactions, van der Waals forces, and hydrophobic effects are assumed to play a role in the interactions between virus particles and surfaces (13). Additional factors affecting adsorption include the different intrinsic characteristics, such as the isoelectric point, of the virus. In this experiment, the viral surrogates studied (FCV, MS2, MNV, and PRD1) may have slightly different net surface charges, as demonstrated by the different isoelectric points (4.9, 3.9, unknown [but thought to be similar to that of NoV at 5.0], and 4.2, respectively) (38, 52). Even with these differences, the elution buffer with a pH of 6.5 containing 0.3 M NaCl and 0.1% Tween 80 described here was formulated to inhibit electrostatic and hydrophobic interactions, thus disrupting adsorption to the surface. More likely, the difference in FCV removal is related to the variable efficiency of the plaque assay (i.e., random differences in virus adsorption and/or infectivity of the cells in the plaque assay). Significant differences between cleaning cloths were also reported for removal of both FCV and MNV from stainless steel but not from solid surface. A potential reason for these differences could be attributed to application of inconsistent pressure to the surface when wiping with the cleaning cloths; however, if this were the case, then one would expect all virus types to be affected, since a cocktail of all viruses was used for the inoculum.
Aside from evaluating the removal of HuNoV surrogates by cleaning cloths, we also evaluated HuNoV removal using real-time RT-PCR. These data, however, did not provide a clear picture of the cloths' effectiveness for removal of HuNoVs due to issues with inhibition of the assays. Here we used a clarified stool sample containing HuNoV GI.1 for the inoculum in order to minimize processing of the sample. Unfortunately, stool samples—even if clarified—contain numerous inhibitors (e.g., phenolic compounds, glycogen, fats, cellulose, bacterial debris, and heavy metals) that can impact the results of RT-PCR assays (30). Steps beyond simple clarification that may have helped to alleviate inhibitors in the stool sample include further processing with organic solvents such as Freon or Vertrel XF and purification using cesium chloride or sucrose gradients (10, 43). Initially, we did compare simple clarification by centrifugation with extraction of HuNoVs from the stool sample by Vertrel XF, and there was a slight loss of virus signal with minimal alleviation of inhibition (data not shown). Another possibility to address inhibition would have been to extract RNA from the eluates collected from the surfaces during evaluation of the cloths. Even though inhibition would have been less of an issue, the extraction efficiency for each sample would need to be assessed in order to provide the amount of certainty necessary to accurately quantify these data. In the end, we did not feel that this was warranted for the scope of the present study, though future research should be done to further evaluate HuNoV removal using cleaning cloths through the utilization of cesium chloride-purified HuNoVs and/or HuNoV-like particles (53).
The most obvious and potentially most critical difference in the cloths was demonstrated in the transfer experiments, where the cotton/cellulose and microfiber cloths transferred significantly less—approximately 2 orders of magnitude less—virus than the nonwoven cloths and cotton terry bar towels. Within the food service industry, cotton terry bar towels are most often used for cleaning, as they can be laundered and reused. The data presented in our study indicate that damp cotton terry bar towels transfer an average of 832 and 115 PFU of HuNoV surrogates (FCV, MS2, PRD1) back to solid surface and stainless steel, respectively. Similar levels of transfer were also demonstrated in our study when using a damp nonwoven cloth. Given the low infectious dose of HuNoVs and resistance to environmental degradation, this level of transfer for HuNoV surrogates should be considered when selecting the appropriate cloth for cleaning and for disinfection before reuse. The difference in the cloth transfer levels may be due to differences in fiber density, though this was not investigated in our study. For instance, the cellulose/cotton cloths have a more sponge-like construction, whereas the cotton terry bar towels have a more open construction with loose fabric loops to which the viruses may not adsorb as readily. The majority of data existing on the transfer of viruses to and from porous (e.g., food and hands) and nonporous (e.g., stainless steel and glass) surfaces primarily focus on transfer from hand to surface (3, 5, 20), surface to hand (35), hand to food (5), and surface to food (9); thus, this study represents the first report of the transfer of viruses from contaminated cloths to nonporous surfaces.
There are a few limitations in the design of our study. First, we used a 5- to 6-log10-unit virus inoculum for the removal and transfer experiments. This amount of virus is likely much greater than would be found on contaminated surfaces, not including surfaces contaminated by an episode of vomiting or other symptoms of acute gastrointestinal illness; even so, Julian et al. (2010) (20) concluded that inoculum size did not significantly influence the amount of virus transferred. Second, we elected to wipe inoculated surfaces 3 times vertically and 3 times horizontally without conducting an experimental evaluation of whether different wiping methods would be more appropriate for assessing the efficacy of cleaning cloths or the ability for the cleaning cloths to transfer viruses to surfaces. Previous studies have reported a variety of wiping methods, ranging from simple to complex, for assessing removal of bacteria (22, 23, 28, 48). If future studies for evaluation of cleaning cloths are developed, we feel a standard method would be beneficial for comparison of data across studies. Last, we did not evaluate the cloths in combination with a sanitizing agent. According to the FDA Food Code (§4-501.114), cleaning and wiping cloths should be stored in an approved sanitizing solution for reuse during an undefined period of time. However, we know that the most common sanitizing compounds (i.e., quaternary ammonium) are ineffective against HuNoVs and other viral pathogens; therefore, significantly increased efficacy is not likely. Furthermore, as this is the first study of its kind, we felt it to be important to first establish removal/transfer due to the cloth alone without compounding variables.
Overall, the findings of this study indicate that cleaning cloths composed of certain materials may be a valuable interim cleaning tool in the food preparation environment when time does not permit the use of a sanitizing agent. This study also demonstrated that some cleaning cloths may more readily transfer viruses back to the food contact surface if used after cleaning a contaminated area and present a potential risk to public health. Although effective at virus removal by themselves, future research should involve evaluation of cloths identified to be the most effective all around (i.e., cellulose/cotton blend cloths and microfiber) in combination with sanitizers and other cleaning regimens.
ACKNOWLEDGMENTS
We especially thank Kim Buldoc and Jesse Kay for their assistance in the lab. We also thank S. R. Milillo for her critical review of the manuscript. We acknowledge John Lample for providing the cleaning cloths used for evaluation.
This research was funded by a USDA NIFSI grant (award no. 10507316) to P. G. Crandall and S. C. Ricke.
Footnotes
Published ahead of print 10 February 2012
REFERENCES
- 1. Bae J, Schwab K. 2008. Evaluation of murine norovirus, feline calicivirus, poliovirus, and MS2 as surrogates for human norovirus in a model of viral persistence in surface water and groundwater. Appl. Environ. Microbiol. 74:477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baert L, Uyttendaele M, Debevere J. 2008. Evaluation of viral extraction methods on a broad range of ready-to-eat foods with conventional and real-time RT-PCR for norovirus GII detection. Int. J. Food Microbiol. 123:101–108 [DOI] [PubMed] [Google Scholar]
- 3. Barker J, Vipond IB, Bloomfield SF. 2004. Effects of cleaning and disinfection in reducing the spread of norovirus contamination via environmental surfaces. J. Hosp. Infect. 58:42–49 [DOI] [PubMed] [Google Scholar]
- 4. Belliot G, Lavaux A, Souihel D, Agnello D, Pothier P. 2008. Use of murine norovirus as a surrogate to evaluate resistance of human norovirus to disinfectants. Appl. Environ. Microbiol. 74:3315–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bidawid S, Malik N, Adegbunrin O, Sattar SA, Farber JM. 2004. Norovirus cross-contamination during food handling and interruption of virus transfer by hand antisepsis: experiments with feline calicivirus as a surrogate. J. Food Prot. 67:103–109 [DOI] [PubMed] [Google Scholar]
- 6. Boone SA, Gerba CP. 2007. Significance of fomites in the spread of respiratory and enteric viral disease. Appl. Environ. Microbiol. 73:1687–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diab-Elschahawi M, et al. 2010. Evaluation of the decontamination efficacy of new and reprocessed microfiber cleaning cloth compared with other commonly used cleaning cloths in the hospital. Am. J. Infect. Control 38:289–292 [DOI] [PubMed] [Google Scholar]
- 8. D'Souza D, Su X. 2010. Efficacy of chemical treatments against murine norovirus, feline calicivirus, and MS2 bacteriophage. Foodborne Pathog. Dis. 7:319–326 [DOI] [PubMed] [Google Scholar]
- 9. D'Souza DH, et al. 2006. Persistence of caliciviruses on environmental surfaces and their transfer to food. Int. J. Food Microbiol. 108:84–91 [DOI] [PubMed] [Google Scholar]
- 10. Duizer E, et al. 2004. Laboratory efforts to cultivate noroviruses. J. Gen. Virol. 85:79–87 [DOI] [PubMed] [Google Scholar]
- 11. Farrah SR, Shah DO, Ingram LO. 1981. Effects of chaotropic and antichaotropic agents on elution of poliovirus adsorbed on membrane filters. Proc. Natl. Acad. Sci. U. S. A. 78:1229–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ferguson C, et al. 2007. Field scale quantification of microbial transport from bovine faeces under simulated rainfall events. J. Water Health 5:83–95 [DOI] [PubMed] [Google Scholar]
- 13. Gerba C. 1984. Applied and theoretical aspects of virus adsorption to surfaces. Adv. Appl. Microbiol. 30:133–168 [DOI] [PubMed] [Google Scholar]
- 14. Gibson KE, Schwab KJ. 2011. Thermal inactivation of human norovirus surrogates. Food Environ. Virol. 3:74–77 [Google Scholar]
- 15. Gibson KE, Schwab KJ. 2011. Tangential-flow ultrafiltration with integrated inhibition detection for recovery of surrogates and human pathogens from large-volume source water and finished drinking water. Appl. Environ. Microbiol. 77:385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Girard M, Ngazoa S, Mattison K, Jean J. 2010. Attachment of noroviruses to stainless steel and their inactivation, using household disinfectants. J. Food Prot. 73:400–404 [DOI] [PubMed] [Google Scholar]
- 17. Glass RI, Parashar UD, Estes MK. 2009. Norovirus gastroenteritis. N. Engl. J. Med. 361:1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Isakbaeva ET, et al. 2005. Norovirus transmission on cruise ship. Emerg. Infect. Dis. 11:154–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jones EL, Kramer A, Gaither M, Gerba CP. 2007. Role of fomite contamination during an outbreak of norovirus on houseboats. Int. J. Environ. Health Res. 17:123–131 [DOI] [PubMed] [Google Scholar]
- 20. Julian TR, Leckie JO, Boehm AB. 2010. Virus transfer between fingerpads and fomites. J. Appl. Microbiol. 109:1868–1874 [DOI] [PubMed] [Google Scholar]
- 21. Kageyama T, et al. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lalla F, Dingle P. 2004. The efficacy of cleaning products on food industry surfaces. J. Environ. Health 67:17–22 [PubMed] [Google Scholar]
- 23. Lalla F, Dingle P, Cheong C. 2005. The antibacterial action of cloths and sanitizers and the use of environmental alternatives in food industries. J. Environ. Health 68:31–35 [PubMed] [Google Scholar]
- 24. Lamhoujeb S, Fliss I, Ngazoa SE, Jean J. 2009. Molecular study of the persistence of infectious human norovirus on food-contact surfaces. Food Environ. Virol. 1:51–56 [Google Scholar]
- 25. Malek M, et al. 2009. Outbreak of norovirus infection among river rafters associated with packaged delicatessen meat, Grand Canyon, 2005. Clin. Infect. Dis. 48:31–37 [DOI] [PubMed] [Google Scholar]
- 26. Mattison K, et al. 2007. Survival of calicivirus in foods and on surfaces: experiments with feline calicivirus as a surrogate for norovirus. J. Food Prot. 70:500–503 [DOI] [PubMed] [Google Scholar]
- 27. Mokhtari A, Jaykus LA. 2009. Quantitative exposure model for the transmission of norovirus in retail food preparation. Int. J. Food Microbiol. 133:38–47 [DOI] [PubMed] [Google Scholar]
- 28. Moore G, Griffith C. 2006. A laboratory evaluation of the decontamination properties of microfibre cloths. J. Hosp. Infect. 64:379–385 [DOI] [PubMed] [Google Scholar]
- 29. Nilsen SK, Dahl I, Jorgensen O, Schneider T. 2002. Micro-fibre and ultra-micro-fibre cloths, their physical characteristics, cleaning effect, abrasion on surfaces, friction, and wear resistance. Build Environ. 37:1373–1378 [Google Scholar]
- 30. Oikarinen S, et al. 2009. PCR inhibition in stool samples in relation to age of infants. J. Clin. Virol. 44:211–214 [DOI] [PubMed] [Google Scholar]
- 31. Park GW, Sobsey MD. 2011. Simultaneous comparison of murine norovirus, feline calicivirus, coliphage MS2, and GII.4 norovirus to evaluate the efficacy of sodium hypochlorite against human norovirus on a fecally soiled stainless steel surface. Foodborne Pathog. Dis. 8:1005–1010 [DOI] [PubMed] [Google Scholar]
- 32. Park H, Kim M, Ko G. 2010. Evaluation of various methods for recovering human norovirus and murine norovirus from vegetables and ham. J. Food Prot. 73:1651–1657 [DOI] [PubMed] [Google Scholar]
- 33. Patel MM, Hall AJ, Vinjé J, Parashar UD. 2009. Noroviruses: a comprehensive review. J. Clin. Virol. 44:1–8 [DOI] [PubMed] [Google Scholar]
- 34. Riddle MS, et al. 2006. Epidemic infectious gastrointestinal illness aboard U.S. Navy ships deployed to the Middle East during peacetime operations—2000-2001. BMC Gastroenterol. 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rusin P, Maxwell S, Gerba CP. 2002. Comparative surface-to-hand and fingertip-to-mouth transfer efficiency of gram-positive bacteria, gram-negative bacteria, and phage. J. Appl. Microbiol. 93:585–592 [DOI] [PubMed] [Google Scholar]
- 36. Rzezutka A, Cook N. 2004. Survival of human enteric viruses in the environment and food. FEMS Microbiol. Rev. 28:441–453 [DOI] [PubMed] [Google Scholar]
- 37. Scallan E, et al. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schaldach CM, Bourcier WL, Shaw HF, Viani BE, Wilson WD. 2006. The influence of ionic strength on the interaction of viruses with charged surfaces under environmental conditions. J. Colloid Interface Sci. 294:1–10 [DOI] [PubMed] [Google Scholar]
- 39. Scharff RL. 2012. The economic burden from health losses due to foodborne illness in the United States. J. Food Prot. 75:123–131 [DOI] [PubMed] [Google Scholar]
- 40. Schlueter V, et al. 1996. Reverse transcription-PCR detection of hepatitis G virus. J. Clin. Microbiol. 34:2660–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schwab KJ, Estes MK, Neill FH, Atmar RL. 1997. Use of heat release and an internal RNA standard control in reverse transcription-PCR detection of Norwalk virus from stool samples. J. Clin. Microbiol. 35:511–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Scott E, Bloomfield SF. 1990. The survival and transfer of microbial contamination via cloths, hands and utensils. J. Appl. Bacteriol. 68:271–278 [DOI] [PubMed] [Google Scholar]
- 43. Shieh Y, Wait D, Tai L, Sobsey MD. 1995. Methods to remove inhibitors in sewage and other fecal wastes for enterovirus detection by the polymerase chain reaction. J. Virol. Methods 54:51–66 [DOI] [PubMed] [Google Scholar]
- 44. Smith D, Gillanders S, Holah J, Gush C. 2011. Assessing the efficacy of different microfibre cloths at removing surface microorganisms associated with healthcare-associated infections. J. Hosp. Infect. 78:182–186 [DOI] [PubMed] [Google Scholar]
- 45. Solomon EB, Fino V, Wei J, Kniel KE. 2009. Comparative susceptibilities of hepatitis A virus, feline calicivirus, bacteriophage MS2 and bacteriophage phiX-174 to inactivation by quaternary ammonium and oxidative disinfectants. Int. J. Antimicrob. Agents 33:288–289 [DOI] [PubMed] [Google Scholar]
- 46. Takahashi H, Ohuchi A, Miya S, Izawa Y, Kimura B. 2011. Effect of food residues on norovirus survival on stainless steel surfaces. PLoS One 6:e21951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Taku A, et al. 2002. Concentration and detection of caliciviruses from food contact surfaces. J. Food Prot. 65:999–1004 [DOI] [PubMed] [Google Scholar]
- 48. Tebbutt GM. 1988. Laboratory evaluation of disposable and reusable disinfectant cloths for cleaning food contact surfaces. Epidemiol. Infect. 101:367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Teunis PF, et al. 2008. Norwalk virus: how infectious is it? J. Med. Virol. 80:1468–1476 [DOI] [PubMed] [Google Scholar]
- 50. U.S. Department of Health and Human Services 2009. Food code. U.S. Department of Health and Human Services, Washington, DC: http://www.fda.gov/Food/FoodSafety/RetailFoodProtection/FoodCode/FoodCode2009/default.htm 3 January 2012 [Google Scholar]
- 51. Vega E, Garland J, Pillai SD. 2008. Electrostatic forces control nonspecific virus attachment to lettuce. J. Food Prot. 71:522–529 [DOI] [PubMed] [Google Scholar]
- 52. Vega E, Smith J, Garland J, Matos A, Pillai SD. 2005. Variability of virus attachment patterns to butterhead lettuce. J. Food Prot. 68:2112–2117 [DOI] [PubMed] [Google Scholar]
- 53. White LJ, et al. 1996. Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J. Virol. 70:6589–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu HM, et al. 2005. A norovirus outbreak at a long-term-care facility: the role of environmental surface contamination. Infect. Control Hosp. Epidemiol. 26:802–810 [DOI] [PubMed] [Google Scholar]