Abstract
Previous studies have demonstrated the capability of Corynebacterium glutamicum for anaerobic succinate production from glucose under nongrowing conditions. In this work, we have addressed two shortfalls of this process, the formation of significant amounts of by-products and the limitation of the yield by the redox balance. To eliminate acetate formation, a derivative of the type strain ATCC 13032 (strain BOL-1), which lacked all known pathways for acetate and lactate synthesis (Δcat Δpqo Δpta-ackA ΔldhA), was constructed. Chromosomal integration of the pyruvate carboxylase gene pycP458S into BOL-1 resulted in strain BOL-2, which catalyzed fast succinate production from glucose with a yield of 1 mol/mol and showed only little acetate formation. In order to provide additional reducing equivalents derived from the cosubstrate formate, the fdh gene from Mycobacterium vaccae, coding for an NAD+-coupled formate dehydrogenase (FDH), was chromosomally integrated into BOL-2, leading to strain BOL-3. In an anaerobic batch process with strain BOL-3, a 20% higher succinate yield from glucose was obtained in the presence of formate. A temporary metabolic blockage of strain BOL-3 was prevented by plasmid-borne overexpression of the glyceraldehyde 3-phosphate dehydrogenase gene gapA. In an anaerobic fed-batch process with glucose and formate, strain BOL-3/pAN6-gap accumulated 1,134 mM succinate in 53 h with an average succinate production rate of 1.59 mmol per g cells (dry weight) (cdw) per h. The succinate yield of 1.67 mol/mol glucose is one of the highest currently described for anaerobic succinate producers and was accompanied by a very low level of by-products (0.10 mol/mol glucose).
INTRODUCTION
Succinate is a C4-dicarboxylate with interesting prospects for a sustainable chemical industry when produced biotechnologically in large amounts from renewable carbon sources (17). It could serve as a precursor for the production of a great variety of important bulk chemicals, such as tetrahydrofurane (THF), 1,4-butanediol, γ-butyrolactone, or maleic anhydride, which are currently produced petrochemically (33, 61). Therefore, succinate was identified as one of the top 12 building block chemicals from biomass by the U.S. Department of Energy (57).
A number of natural succinate producers which accumulate succinate as a major product of their fermentative catabolism have been described, predominantly rumen bacteria belonging to the family Pasteurellaceae, such as Actinobacillus succinogenes (15), Mannheimia succiniciproducens (30), and Basfia succiniciproducens (48), or Anaerobiospirillum succiniciproducens (44), a member of the Succinivibrionaceae family. Optimization of production conditions and/or manipulation of metabolic pathways led to the development of succinate production processes with high yield (>1 mol/mol glucose) and productivity (14, 31, 34, 35). However, most of the described strains require complex medium and show high by-product accumulation (mainly pyruvate and acetate), increasing the costs associated with production and purification.
Also, Escherichia coli forms succinate as an end product of mixed-acid fermentation. Several metabolic engineering strategies were explored to optimize the anaerobic metabolism of E. coli for increased succinate production (52, 56). One of the most beneficial approaches was based on the design of a metabolic pathway which is functionally similar to that of natural producers from the Pasteurellaceae family, such as A. succinogenes (64). Another approach used a combination of production pathways, such as the reductive tricarboxylic acid (TCA) cycle with the glyoxylate shunt (45) and the introduction of a heterologous carboxylation reaction (53). A further interesting approach for the optimization of succinate production with E. coli combined rational metabolic engineering with directed evolution techniques (23).
Corynebacterium glutamicum is another organism capable of succinate production. It is a Gram-positive soil bacterium with generally regarded as safe (GRAS) status and the most important species for industrial amino acid production, with its major products being l-glutamate (2.1 million tons/year) and l-lysine (1.4 million tons/year). The genome of C. glutamicum is known (19, 27, 59), and numerous genetic tools allowing genetic engineering are available (29). Extensive knowledge on the metabolism and its regulation of this organism has been obtained within the past decades (6, 7). Based on this knowledge, metabolic engineering was applied successfully to develop strains capable of producing a variety of other metabolites besides amino acids, such as putrescine (47), ethanol (21), or isobutanol (3, 50). However, C. glutamicum shows limited anaerobic growth by nitrate respiration to nitrite, which cannot be reduced further, and accumulates (37, 51). When aerobically grown cells of C. glutamicum are incubated under anaerobic conditions in the absence of nitrate, they metabolize glucose to a mixture of l-lactate, acetate, and succinate. This conversion is not coupled to growth (22). Transport of succinate into the medium was shown to involve the product of cg2425, the first succinate exporter to be identified (10, 18).
A C. glutamicum R strain for anaerobic succinate production which lacks the ldhA gene for the NAD+-dependent lactate dehydrogenase and overexpresses the native pyruvate carboxylase gene pyc has been described (38). Thereby, the formation of lactate, which is the dominant product of anaerobic glucose catabolism, was prevented and the provision of oxaloacetate was improved. This strain accumulated succinate with a yield of 1.4 mol/mol glucose and formed acetate as a by-product with a yield of 0.29 mol/mol glucose. The oxidation of pyruvate to acetate provides additional reducing equivalents for conversion of oxaloacetate to succinate in the reductive TCA cycle (38, 58). We recently developed an aerobic succinate producer strain of C. glutamicum, which initially also had the problem of forming large quantities of acetate. However, by deleting the genes encoding the known pathways of acetate formation, i.e., phosphotransacetylase (pta) plus acetate kinase (ackA), pyruvate:menaquinone oxidoreductase (pqo), and acetyl coenzyme A (CoA):CoA transferase (cat), we could drastically reduce the synthesis of this by-product (32).
In this study, two problems of anaerobic succinate formation with C. glutamicum were addressed. First, we tackled the problem of acetate formation by deleting the genes responsible for its formation. Second, we aimed at improving the succinate yield from glucose, which is currently limited by the availability of reducing equivalents for conversion of oxaloacetate to succinate in the reductive TCA cycle. For this purpose, formate was tested as an additional donor of NADH and carbon dioxide. Using the above-mentioned approaches, we obtained a strain which produced 1.13 M succinate in 53 h with a succinate yield of 1.67 mol/mol glucose, a very low by-product level, and a volumetric productivity of 21 mM h−1 in fed-batch fermentation with glucose, formate, and carbon dioxide as carbon sources.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
All bacterial strains and plasmids used or constructed in the course of this work are listed in Table 1. C. glutamicum strains were routinely cultivated at 30°C in brain heart infusion (BHI) medium (Difco Laboratories, Detroit, MI) supplemented with 111 mM glucose or in CGXII medium containing (NH4)2SO4 (20 g/liter), urea (5 g/liter), KH2PO4 (1 g/liter), K2HPO4 (1 g/liter), MgSO4 · 7H2O (0.25 g/liter), CaCl2 (10 mg/liter), FeSO4 · 7H2O (10 mg/liter), MnSO4 · H2O (0.1 mg/liter), ZnSO4 · 7H2O (1 mg/liter), CuSO4 · 5H2O (0.2 mg/liter), NiCl2 · 6H2O (20 μg/liter), biotin (0.4 mg/liter), 3-morpholinopropanesulfonic acid (MOPS) (42 g/liter), and 222 mM glucose as the carbon and energy source. The medium differed from the originally described CGXII medium (28) by a 100-fold-reduced MnSO4 concentration, a doubled biotin concentration, and the lack of protocatechuate. In anaerobic production experiments including formate as an additional substrate, 110 mM sodium formate was added to the CGXII medium during aerobic cultivation of the cells. The pH of the CGXII medium was adjusted to pH 7 with KOH. E. coli DH5α was routinely grown in LB medium at 37°C. If appropriate, kanamycin (25 μg/ml for C. glutamicum or 50 μg/ml for E. coli) and 0.5 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) were added.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| C. glutamicum ATCC 13032 | Wild-type strain, biotin auxotroph | 1 |
| C. glutamicum ΔldhA | ATCC 13032 derivative with an in-frame deletion of the ldhA gene | This work |
| C. glutamicum BOL-1 | ATCC 13032 derivative with in-frame deletions of cat, pqo, pta-ackA, and ldhA | This work |
| C. glutamicum BOL-2 | BOL-1 derivative with chromosomal integration into the Δpta-ackA locus of the pycP458S gene from C. glutamicum DM1727 under the control of the tuf promoter | This work |
| C. glutamicum BOL-3 | BOL-2 derivative with chromosomal integration into the Δpqo locus of the fdh gene from M. vaccae under the control of the tuf promoter from C. glutamicum | This work |
| E. coli DH5α | F− ϕ80dlacΔ(lacZ)M15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK−, mK+) deoR thi-1 phoA supE44 λ−gyrA96 relA1 | Invitrogen |
| Plasmids | ||
| pAN6 | Kanr; C. glutamicum/E. coli shuttle vector for regulated gene expression (Ptac, lacIq, pBL1 oriVC.glutamicum, pUC18 oriVE.coli) derived from pEKEx2 | 9 |
| pAN6-pycP458S | Kanr; pAN6 derivative containing the pycP458S gene from C. glutamicum DM1727 under the control of the tac promoter | 32 |
| pAN6-gap | Kanr; pAN6 derivative containing the gapA gene from C. glutamicum ATCC 13032 under the control of the tac promoter | This work |
| pBBR1MCS2-fdh | Kanr; donor of the fdh gene from M. vaccae | Bringer-Meyer, unpublished |
| pEKEx2 | Kanr; C. glutamicum/E. coli shuttle vector for regulated gene expression (Ptac, lacIq, pBL1 oriVC.glutamicum, pUC18 oriVE.coli) | 8 |
| pEKEx2-fdh | Kanr; pEKEx2 derivative containing the fdh gene from M. vaccae under the control of the tac promoter | This work |
| pK19mobsacB | Kanr; vector for allelic exchange in C. glutamicum (pK18 oriVE.coli sacB lacZα) | 46 |
| pK19mobsacB-Δcat | Kanr; pK19mobsacB derivative containing a 1.1-kb overlap extension PCR product (SalI/XbaI) which covers the flanking regions of the C. glutamicum cat gene | 55 |
| pK19mobsacB-Δpqo | Kanr; pK19mobsacB derivative containing a 1-kb overlap extension PCR product (HindIII/EcoRI) which covers the flanking regions of the C. glutamicum pqo gene | 32 |
| pK19mobsacB-Δpqo::XbaI-BamHI | Kanr; pK19mobsacB derivative containing a 1-kb overlap extension PCR product (HindIII/EcoRI) which covers the flanking regions of the C. glutamicum pqo gene and possesses an XbaI/BamHI cloning site within the 23-bp overlap region | This work |
| pK19mobsacB-Δpqo::Ptuf-fdh | Kanr; pK19mobsacB-Δpqo::XbaI-BamHI derivative containing a 1.4-kb overlap extension PCR product (XbaI/BamHI) of the tuf promoter (178 bp) from C. glutamicum fused to the fdh gene (1,205 bp) from M. vaccae | This work |
| pk19mobsacB-Δpta-ΔackA | Kanr; pK19mobsacB derivative containing a 1.1-kb overlap extension PCR product (HindIII/XbaI) which covers the flanking regions of the C. glutamicum pta-ackA genes | 32 |
| pk19mobsacB-Δpta-ΔackA::Ptuf-pycP458S | Kanr; pK19mobsacB derivative containing a 5-kb double overlap extension PCR product (HindIII/XbaI) which carries the tuf promoter (178 bp) from C. glutamicum ATCC 13032 fused to the pyc gene (3,422 bp) from C. glutamicum DM1727 within the flanking regions of the C. glutamicum pta-ackA genes | This work |
| pK19mobsacB-ΔldhA | Kanr; pK19mobsacB derivative containing a 1.1-kb overlap extension PCR product (HindIII/EcoRI) which covers the flanking regions of the C. glutamicum ldhA gene | This work |
Kanr, kanamycin resistant.
Recombinant DNA work.
The enzymes for recombinant DNA work were obtained from Roche Diagnostics (Mannheim, Germany) or New England BioLabs (Frankfurt am Main, Germany). All oligonucleotides were synthesized by Eurofins MWG Operon and are listed in Table S1 in the supplemental material. Routine methods like PCR, restriction, or ligation were carried out according to standard protocols (43). All PCR products were generated with KOD Hot Start polymerase (Novagen, Darmstadt, Germany). Plasmids were isolated from E. coli with the QIAprep Spin miniprep kit (Qiagen, Hilden, Germany). E. coli was transformed by the RbCl method (16). Transformation of C. glutamicum was performed as described previously (54). All plasmid constructs described below were controlled by DNA sequencing (LGC Genomics, Berlin, Germany).
Construction of deletion mutants, chromosomal gene replacements, and plasmids.
C. glutamicum mutants with in-frame deletions of ldhA (ΔldhA), pta-ack (Δpta-ack), pqo (Δpqo), and cat (Δcat) as well as the chromosomal integrations of the pycP458S gene from C. glutamicum DM1727 and the fdh gene from Mycobacterium vaccae, both under the control of the tuf promoter from C. glutamicum wild type, were constructed via a two-step homologous recombination procedure as described previously (36) using the suicide vector pK19mobsacB (46). For construction of pK19mobsacB-Δldh, the regions up- and downstream (approximately 500 bp each) of the ldhA gene to be deleted were amplified using two pairs of oligonucleotides designated ΔldhA1-for/ΔldhA2-rev and ΔldhA3-for/ΔldhA4-rev (see Table S1 in the supplemental material), respectively. The resulting DNA fragments served as the template for an overlap extension PCR with oligonucleotides ΔldhA1-for and ΔldhA4-rev, forming a PCR product of about 1 kb which was digested with the restriction enzymes HindIII and EcoRI and cloned into pK19mobsacB cut with the same enzymes. The construction of the plasmids for the in-frame deletions of pta-ack (Δpta-ack) and pqo (Δpqo) was described previously (32). Plasmid pK19mobsacB-Δcat was kindly provided by V. F. Wendisch (University of Bielefeld).
Plasmid pK19mobsacB-Δpta-ΔackA::Ptuf-pycP458S was constructed for replacing the chromosomal pta-ackA genes with the pycP458S gene from C. glutamicum DM1727 under the control of the tuf promoter region from C. glutamicum wild type. For this purpose, the up- and downstream regions (approximately 650 bp each) of the pta-ackA operon and the tuf promoter region (178 bp upstream of the coding sequence of the tuf gene) were amplified from chromosomal DNA of C. glutamicum wild type using the oligonucleotide pairs mΔpta-ack1-for/mΔpta-ack2-rev, mΔpta-ack3-for/mΔpta-ack4-rev, and Δpta-ack-Ptuf-for/GTG-Ptuf-rev, respectively. The pycP458S gene was amplified from plasmid pAN6-pycP458S (32) using the oligonucleotides Ptuf-pyc-for and pyc-Δpta-ack-rev (see Table S1 in the supplemental material). By overlap extension PCR, the pta upstream region and the tuf promoter region were fused with oligonucleotides mΔpta-ack1-for/GTG-Ptuf-rev and the pycP458S gene was fused with the ackA downstream region using oligonucleotides Ptuf-pyc-for/mΔpta-ack4-rev. After purification, the resulting PCR products served again as templates for another overlap extension PCR with the oligonucleotide pair mΔpta-ack1_for/mΔpta-ack4_rev. The resulting PCR product of about 5 kb was digested with the restriction enzymes HindIII and XbaI and cloned into pK19mobsacB cut with the same enzymes. In the resulting plasmid, the pyc gene starts with the native GTG codon.
Plasmid pK19mobsacB-Δpqo::Ptuf-fdh was constructed for replacing the chromosomal pqo gene with the fdh gene from M. vaccae under the control of the tuf promoter. First, the regions up- and downstream (approximately 500 bp each) of the Δpqo deletion region were amplified with the oligonucleotide pairs Δpqo1-for/Δpqo2-XbaI-BamHI-rev and XbaI-BamHI Δpqo3-for/Δpqo4-rev, respectively (see Table S1 in the supplemental material). The two PCR products served as the templates for an overlap extension PCR with oligonucleotide pair Δpqo1-for/Δpqo4-rev. The PCR product of about 1 kb, which carried XbaI and BamHI cloning sites at the fusion site, was digested with HindIII and EcoRI and cloned into pK19mobsacB cut with the same enzymes. The resulting plasmid was named pK19mobsacB-Δpqo::XbaI-BamHI. The DNA fragment covering Ptuf-fdh was constructed by amplifying the tuf promoter region (178 bp upstream of the tuf gene) from chromosomal DNA of C. glutamicum wild type, using the oligonucleotide pair Ptuf-for/Ptuf-rev, and the fdh gene from plasmid pBBR1MCS2-fdh (kindly provided by S. Bringer-Meyer, Forschungszentrum Jülich), using the oligonucleotide pair Ptuf-fdh-for/fdh-rev. The two PCR products served as templates for an overlap extension PCR with oligonucleotides Ptuf-for/fdh-rev. The resulting PCR product of 1.4 kb was digested with XbaI and BamHI and cloned into pK19mobsacB-Δpqo::XbaI-BamHI cut with the same enzymes.
The transfer of the resulting deletion/integration plasmids into C. glutamicum and selection for the first and second recombination events were performed as described previously (36). Kanamycin-sensitive and saccharose-resistant clones were tested by colony PCR analysis with an oligonucleotide pair designated Δ-gene/operon-out-fw and Δ-gene/operon-out-rv. Only in the case of checking the fdh integration was the oligonucleotide pair Δpqo-out2-for/Δpqo-out2-rev used (see Table S1 in the supplemental material). Clones which had the desired in-frame deletion of the gene/operon revealed a 1-kb fragment in which all nucleotides except the first 6 codons (in the case of the Δcat deletion, the first 7 codons) and the last 12 codons were replaced by a 21-bp tag. Clones with the desired chromosomal integration of Ptuf-pycP458S and Ptuf-fdh led to PCR products of 5 kb and 3.5 kb, respectively.
For the construction of the expression plasmid pAN6-gap, the gapA gene was amplified using the oligonucleotide pair gap-for/gap-rev and chromosomal DNA of C. glutamicum wild type. The PCR product (1 kb) was digested with NdeI and NheI and cloned into pAN6 cut with the same enzymes. Plasmid pAN6-pycP458S was described elsewhere (32). The construction of plasmid pEKEx2-fdh was performed by amplifying the fdh gene from plasmid pBBR1MCS2-fdh using the oligonucleotide pair fdh-PstI-RBS-for and fdh-BamHI-rev. Besides the recognition sites for the restriction endonucleases PstI and BamHI, the oligonucleotides also introduced a ribosome binding site (AAGGA) ending 9 nucleotides upstream of the start codon (see Table S1 in the supplemental material). The resulting PCR product (1.2 kb) was digested with PstI and BamHI and cloned into plasmid pEKEx2 cut with the same enzymes.
Production of biomass for anaerobic succinate production.
Five milliliters of BHI medium supplemented with 111 mM glucose was inoculated with a single colony of the desired C. glutamicum strain from a fresh BHI agar plate, and the culture was incubated on a rotary shaker for 16 h at 30°C. Subsequently, the cells were used to inoculate a 500-ml baffled shake flask with 50 ml of CGXII medium containing 222 mM glucose. After approximately 8 h of incubation at 130 rpm and 30°C, the cells were used to inoculate a 5-liter baffled shake flask containing 600 ml CGXII medium to an optical density at 600 nm (OD600) of 1.2. The culture was incubated on a rotary shaker for 16 h at 30°C and 85 rpm. The medium contained 110 mM sodium formate in addition to the 222 mM glucose when cultivating cells for subsequent production experiments with formate as an additional substrate.
Anaerobic succinate production in batch mode.
The cells from the 600-ml culture (see above) were harvested by centrifugation (5,000 × g, 4°C, 20 min), resuspended in 600 ml 0.9% NaCl solution containing 111 mM glucose and 200 mM NaHCO3, and transferred into a 1.4-liter bioreactor (Multifors multifermenter system; Infors, Einsbach, Germany). The cell suspension, which had OD600 values between 18 and 29, was kept at 30°C and stirred at 300 rpm. Where indicated, 220 mM sodium formate was added as an additional substrate. The bioreactor was firmly closed except for an overpressure one-way valve and a sampling tube with a rubber valve. Anaerobic conditions were achieved by rapid consumption of the remaining oxygen by the cells and by preventing aeration in the hermetically sealed bioreactor. The pH was automatically controlled at a value of 6.9 by addition of 3 M KOH.
Anaerobic succinate production in fed-batch mode.
Fed-batch experiments were performed in a manner similar to that of the batch experiments but used cells from the 1.2-liter culture instead of the 0.6-liter culture. The cells were resuspended in 450 ml 0.9% NaCl solution containing 222 mM glucose, 240 mM sodium formate, and 250 mM NaHCO3 and transferred into the 1.4-liter Multifors bioreactor (final OD600 of approximately 50). Feeding included three additional pulses of 222 mM glucose combined with 250 mM NaHCO3 after 7.5, 20, and 39 h and four pulses of sodium formate (2 of 200 mM and 2 of 290 mM) after 7.5, 20, 36, and 53 h of cultivation. The pH was kept at pH 6.9 by automated addition of 4 M KOH. After feeding with NaHCO3, the pH increased to between 7.2 and 7.4 and then returned to pH 6.9 by the acid production of the cells.
Growth parameter determination.
Growth was followed by measuring the optical density at 600 nm (OD600) with an Ultrospec 500 pro spectrophotometer (Amersham Biotech, Freiburg, Germany). The biomass concentration was calculated from OD600 values using an experimentally determined correlation factor of 0.25 g cells (dry weight) (cdw)/liter for an OD600 of 1 (26).
Quantification of glucose and organic acids in the culture supernatant.
Glucose and organic acids in the culture supernatants were quantified using an Agilent 1100 liquid chromatography (LC) system (Agilent Technologies, Waldbronn, Germany) equipped with a 300- by 8-mm organic acid column (polystyrol-divinylbenzol resin; CS Chromatographie Service GmbH, Langerwehe, Germany) and a guard cartridge (40 by 8 mm) filled with the same material. Isocratic elution was performed for 38 min at 40°C with 100 mM sulfuric acid at a flow rate of 0.4 ml/min. Glucose was detected via an Agilent 1100 refractive index detector, and the organic acids were detected via an Agilent 1100 diode array detector at 215 nm. Quantification was performed using calibration curves obtained with external standards.
Measurement of formate dehydrogenase activity in cell extracts.
Five milliliters of BHI medium supplemented with 111 mM glucose was inoculated with a single colony of the desired C. glutamicum strain from a fresh BHI agar plate and incubated on a rotary shaker for 8 h at 30°C. Subsequently, the cells were used to inoculate a 500-ml baffled shake flask containing 50 ml CGXII medium with 222 mM glucose. After approximately 16 h of incubation at 130 rpm and 30°C, the cells were used to inoculate a second 500-ml baffled shake flask with 50 ml of the same medium to an OD600 of 1, and the culture was incubated on a rotary shaker at 30°C and 130 rpm until it reached an OD600 of 7 to 8. Then the cells were harvested by centrifugation, washed in 100 mM potassium phosphate buffer (pH 6.5), and disrupted by sonication. The crude extracts were centrifuged at 10,000 × g for 10 min at 4°C, and the supernatants were used as cell extracts. The formate dehydrogenase activity was determined spectrophotometrically at 340 nm in an assay mixture containing 200 mM sodium formate, 2 mM NAD+, and 100 mM potassium phosphate buffer (pH 7.0) as described previously (2). One unit of enzyme activity (U) was defined as the amount of enzyme catalyzing the reduction of 1 μmol NAD+ per min at 30°C. Protein concentrations were determined by the method of Bradford (5), using bovine serum albumin as a standard.
RESULTS
Minimizing acetate formation during anaerobic succinate production from glucose by C. glutamicum.
As described in the introduction, a C. glutamicum R strain for anaerobic succinate production which lacks the ldhA gene for the NAD+-dependent lactate dehydrogenase and overexpresses the native pyruvate carboxylase gene pyc has been described (38). This strain formed acetate as a major by-product with a yield of 0.29 mol/mol glucose. Although the oxidation of pyruvate to acetate provides additional reducing equivalents for conversion of oxaloacetate to succinate in the reductive TCA cycle (38, 58), it also leads to a loss of carbon and complicates downstream processing. We therefore aimed at the development of a succinate production strain of C. glutamicum with minimized acetate formation.
Starting with the C. glutamicum type strain ATCC 13032, we deleted the genes for all known acetate-forming pathways in C. glutamicum, i.e., the cat gene for acetyl-CoA:CoA transferase (55, 58), the pqo gene for pyruvate:menaquinone oxidoreductase (49), and the pta-ackA genes coding for phosphotransacetylase and acetate kinase (41). To abolish lactate production, the ldhA gene for l-lactate dehydrogenase (22) was deleted. The resulting strain, C. glutamicum Δcat Δpqo Δpta-ack ΔldhA, was named BOL-1 (Fig. 1). As increased C3 carboxylation efficiency was shown to be a key factor for high succinate production (22), strain BOL-1 was transformed with plasmid pAN6-pycP458S (32) for overproduction of the homologous pyruvate carboxylase (40) with a P458S amino acid exchange, which was reported to be beneficial for the reaction (20). Alternatively, the pycP458S gene under the control of the strong constitutive promoter of the tuf gene (coding for the elongation factor Tu) was chromosomally integrated into the deleted pta-ack region of the BOL-1 strain, resulting in strain C. glutamicum Δcat Δpqo Δpta-ack::Ptuf pycP458S ΔldhA (BOL-2). For comparison with C. glutamicum R ΔldhA-pCRA717 (38) and as a reference, C. glutamicum ΔldhA/pAN6-pycP458S was constructed.
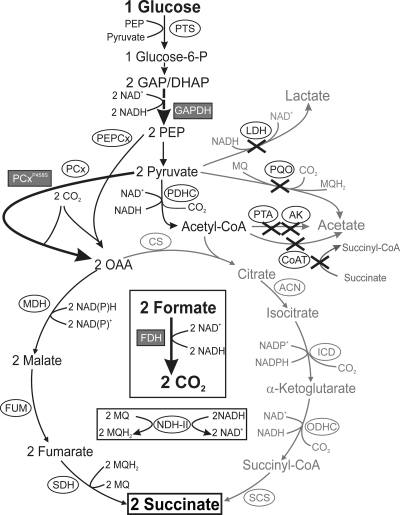
Fig 1.
Schematic representation of the metabolic pathways relevant for succinate production by C. glutamicum under anaerobic conditions. Reactions leading to by-products or which are presumably not relevant are displayed in gray. Enzymes whose genes were deleted in the course of this work are indicated by “X.” Enzymes whose genes were overexpressed are highlighted in gray boxes, and the arrows for the corresponding reactions are thickened. Additional reactions not directly involved in succinate synthesis but important for cofactor supply, cofactor conversion, and CO2 production are displayed in boxes within the TCA cycle. Abbreviations: ACN, aconitase; AK, acetate kinase; CoAT, acetyl-CoA:CoA transferase; CS, citrate synthase; DHAP, dihydroxyacetone phosphate; FDH, formate dehydrogenase (from M. vaccae); FUM, fumarase; GAP, glyceraldehyde 3-phosphate; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; ICD, isocitrate dehydrogenase; LDH, l-lactate dehydrogenase; MDH, malate dehydrogenase; NDH-II, type II NADH dehydrogenase; OAA, oxaloacetate; ODHC, 2-oxoglutarate dehydrogenase complex; PEP, phosphoenolpyruvate; PEPCx, PEP carboxylase; PCx, pyruvate carboxylase; PCxP458S, pyruvate carboxylase with a P458S exchange; PDHC, pyruvate dehydrogenase complex; PTA, phosphotransacetylase; PQO, pyruvate:menaquinone oxidoreductase; SCS, succinyl-CoA synthetase; SDH, succinate dehydrogenase.
Succinate production of strains ΔldhA/pAN6-pycP458S, BOL-1/pAN6-pycP458S, and BOL-2 was tested under anaerobic conditions in 0.9% (wt/vol) NaCl solution containing 111 mM glucose at a constant pH of 6.9 (with 3 M KOH) using a Multifors bioreactor system. To ensure the necessary supply of CO2 for C3 carboxylation, 200 mM NaHCO3 was added. Three independent fermentations with different biomass concentrations, resulting in different kinetics, were performed for each strain. In Fig. 2, one representative fermentation for each of the tested strains is shown. The average specific process parameters and final product titers, including standard deviations, are shown in Table 2.
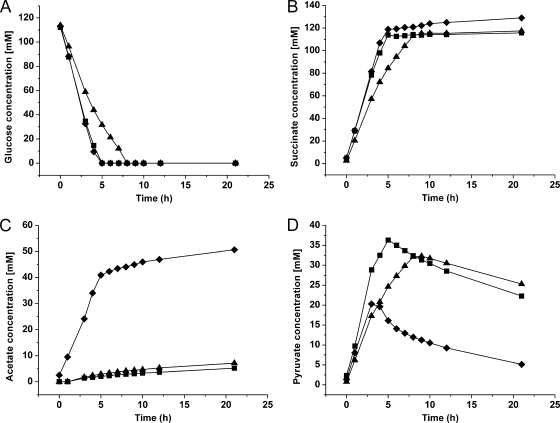
Fig 2.
Representative anaerobic batch fermentation of C. glutamicum ΔldhA/pAN6-pycP458S (♦; biomass concentration, 6.35 g cdw/liter), C. glutamicum BOL-1/pAN6-pycP458S (▲; biomass concentration, 6.00 g cdw/liter), and C. glutamicum BOL-2 (■; biomass concentration, 5.33 g cdw/liter) showing glucose consumption (A), succinate formation (B), acetate formation (C), and pyruvate formation (D). The experiments were performed in a Multifors bioreactor system, and the cells, pregrown aerobically, were resuspended in a solution containing 0.9% (wt/vol) NaCl solution, 111 mM glucose, and 200 mM NaHCO3. The pH was kept constant at 6.9 by addition of 3 M KOH. Note that the different rates cannot be directly compared, as the biomass concentrations differed. The average values of the final process parameters, including standard deviations from three independent fermentations for each strain, are displayed in Table 2.
Table 2.
Comparison of different C. glutamicum strains with respect to relevant parameters for anaerobic succinate production from glucose in a batch fermentationa
| C. glutamicum strain | Yield of succinate/glucose (mol/mol) | Rate (mmol per g cdw per h) of: |
Amt of glucose consumed (mM) | Final titer (mM) of: |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Glucose uptake | Succinate production | Succinate | Acetate | Pyruvate | Malate | Fumarate | α-Ketoglutarate | |||
| ΔldhA/pAN6-pycP458S | 1.16 ± 0.02 | 3.68 ± 0.07 | 3.70 ± 0.17 | 113 ± 3 | 130 ± 4 | 52 ± 2 | 5 ± 0 | 8 ± 2 | 2.2 ± 0.7 | 3 ± 1 |
| BOL-1/pAN6-pycP458S | 1.00 ± 0.03 | 2.29 ± 0.20 | 2.33 ± 0.18 | 115 ± 3 | 116 ± 3 | 5 ± 1 | 24 ± 2 | 6 ± 1 | 3.0 ± 0.3 | 14 ± 0 |
| BOL-2 | 1.03 ± 0.04 | 4.24 ± 0.32 | 4.20 ± 0.26 | 112 ± 4 | 116 ± 2 | 5 ± 1 | 23 ± 2 | 7 ± 2 | 3.3 ± 0.8 | 12 ± 1 |
| BOL-3 | 1.05 ± 0.02 | 4.28 ± 0.39 | 4.37 ± 0.34 | 115 ± 1 | 121 ± 3 | 6 ± 1 | 26 ± 2 | 4 ± 2 | 0.6 ± 0.4 | 13 ± 1 |
| BOL-3/pAN6 | 1.01 ± 0.02 | 3.41 ± 0.22 | 3.38 ± 0.25 | 116 ± 1 | 118 ± 4 | 5 ± 0 | 24 ± 1 | 6 ± 1 | 2.9 ± 0.4 | 15 ± 0 |
| BOL-3/pAN6-gap | 1.07 ± 0.02 | 2.20 ± 0.12 | 2.25 ± 0.08 | 116 ± 3 | 124 ± 3 | 6 ± 0 | 20 ± 1 | 4 ± 0 | 1.1 ± 0.4 | 14 ± 0 |
The cells were pregrown aerobically and subsequently resuspended in 600 ml 0.9% (wt/vol) NaCl solution containing 111 mM glucose and 200 mM NaHCO3 in a bioreactor. The pH of the cell suspension was maintained at pH 6.9 by addition of 3 M KOH. Rates were calculated for the period in which glucose was completely consumed. The final concentrations of products and by-products were determined after 21 ± 1 h. All values are the average data with standard deviations from three independent batch fermentations.
The reference strain C. glutamicum ΔldhA/pAN6-pycP458S utilized glucose with a specific rate of 3.68 ± 0.07 mmol per g cdw per h and produced succinate with a specific rate of 3.70 ± 0.17 mmol per g cdw per h. In total, 130 ± 4 mM succinate was accumulated in the supernatant during batch fermentation with a yield of 1.16 ± 0.02 mol/mol glucose. Simultaneously, 52 ± 2 mM acetate was formed, corresponding to a yield of 0.46 ± 0.01 mol/mol glucose. Pyruvate accumulated during the production process simultaneously to glucose consumption, reaching a maximal level of more than 20 mM. After glucose was depleted, pyruvate was partially consumed, leading to a final concentration of 5 ± 0 mM after 21 h. Besides pyruvate, malate (8 ± 2 mM), fumarate (2.2 ± 0.7 mM), and α-ketoglutarate (3 ± 1 mM) were also detected in the supernatant after 21 h. Whereas malate and fumarate presumably are formed by the reductive branch of the TCA cycle, α-ketoglutarate must be formed by the oxidative branch (Fig. 1).
As expected, strain BOL-1/pAN6-pycP458S showed an approximately 90% decreased final acetate titer (5 ± 1 mM) compared to that of the reference strain C. glutamicum Δldh/pAN6-pycP458S. However, the glucose consumption rate (2.29 ± 0.20 mmol per g cdw per h) and the succinate production rate (2.33 ± 0.18 mmol per g cdw per h) were reduced by 37 to 38% compared to those of the reference strain. The strain BOL-1 reached a succinate titer of 116 ± 3 mM, which corresponded to the maximal theoretical yield (1.00 ± 0.03 mol succinate/mol glucose) that is possible when glucose is metabolized exclusively by glycolysis. The absence of the pyruvate-dissimilating pathways leading to acetate in the BOL-1/pAN6-pycP458S strain led to an increased excretion of pyruvate (final titer, 24 ± 2 mM) and α-ketoglutarate (14 ± 0 mM), whereas the concentrations of malate (6 ± 1 mM) and fumarate (3 ± 0.3 mM) remained nearly unchanged.
The BOL-2 strain showed the same final titers and product yields as strain BOL-1/pAN6-pycP458S; however, the rates of glucose consumption (4.24 ± 0.32 mmol per g cdw per h) and succinate production (4.20 ± 0.26 mmol per g cdw per h) were 80% and 85% higher, respectively. These rates even exceeded those of the reference strain ΔldhA/pAN6-pycP458S by 15% and 14%, respectively.
Increasing the supply of redox equivalents for anaerobic succinate production from glucose by coutilization of formate.
The strain BOL-2 was able to rapidly metabolize glucose to succinate under anaerobic conditions up to its theoretical maximal yield of 1 mol/mol glucose with minimized accumulation of by-products (lactate and acetate). In order to increase the succinate yield, additional reducing equivalents are required. For this purpose, we wanted to introduce the fdh gene from Mycobacterium vaccae (11), which codes for an NAD+-dependent formate dehydrogenase (FDH), into strain BOL-2. Theoretically, the coutilization of glucose and formate could allow an exergonic “homosuccinate fermentation” according to the following reaction:
In a first set of experiments, the fdh gene was overexpressed in strain BOL-2 using the expression plasmid pEKEx2-fdh. However, in the presence of formate, this plasmid led to a complete stop of glucose utilization, although neither formate utilization nor product formation was observed (data not shown). It was supposed that initial oxidation of minor amounts of formate led to an excess of NADH, which inhibited the activity of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and blocked glycolysis. This idea was supported by previous studies, in which it was proposed that high NADH levels can possibly inhibit GAPDH and slow down glycolysis in C. glutamicum during anaerobic alanine production (25). To reduce or eliminate the proposed GAPDH inhibition, two additional changes were introduced into the BOL-2 strain. (i) A single copy of the fdh gene under the control of the strong constitutive promoter of the tuf gene was chromosomally integrated into the Δpqo locus, resulting in strain C. glutamicum Δcat Δpqo::Ptuf fdh Δpta-ack::Ptuf pycP458S ΔldhA (BOL-3). The single, constitutively expressed fdh copy should lead to a decreased FDH level, resulting in reduced FDH activity and, consequently, a lowered NADH production rate. Ultimately, this should allow the NADH reoxidizing reactions to prevent NADH accumulation and GAPDH inhibition. Enzyme activity measurements confirmed that the specific NAD+-dependent FDH activity of strain BOL-3 (0.024 ± 0.004 U/mg protein) was 81% lower than that of strain BOL-2/pEKEx2-fdh (0.130 ± 0.001 U/mg protein). The control strains BOL-2 and BOL-2/pEKEx2 showed no NAD+-dependent FDH activity at all. (ii) Jojima and coworkers previously demonstrated that overexpression of the gapA gene can be used as an efficient approach to improve glucose consumption in C. glutamicum R under anaerobic conditions that lead to high NADH/NAD+ ratios (25). Therefore, we constructed plasmid pAN6-gap, which allows overexpression of the homologous gapA gene from C. glutamicum, and transferred this plasmid into the BOL-3 strain, resulting in BOL-3/pAN6-gap. The strain with empty vector was also constructed as a control (BOL-3/pAN6).
To determine the influence of formate on succinate production, strains BOL-2, BOL-3, BOL-3/pAN6, and BOL-3/pAN6-gap were incubated under anaerobic conditions in 0.9% (wt/vol) NaCl solution containing 111 mM glucose and 200 mM NaHCO3 with and without 220 mM sodium formate. The experiments were performed in a Multifors bioreactor system at pH 6.9, adjusted with 3 M KOH. Three independent fermentations with different biomass concentrations, leading to different kinetics, were performed for each strain. Representative fermentations for each strain are shown in Fig. 3A for the experiments with the glucose/formate mixture and in Fig. 3B for the experiments with glucose as the sole substrate. The average specific process parameters and final product titers, including standard deviations, are listed in Tables 2 and 3 for the experiments with and without formate, respectively.
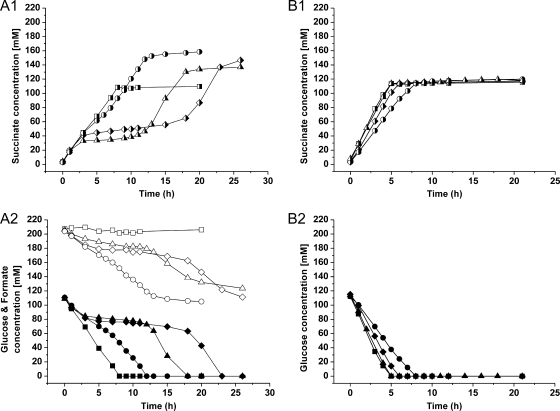
Fig 3.
Succinate formation (half-filled symbols), glucose consumption (filled symbols), and formate utilization (open symbols) during a representative anaerobic batch fermentation with a glucose/formate mixture (panels A1 and A2) or with glucose alone (panels B1 and B2) of C. glutamicum BOL-2 (■; biomass concentrations, 5.50 [A] and 5.33 [B] g cdw/liter), BOL-3 (▲; biomass concentrations, 4.70 [A] and 5.10 [B] g cdw/liter), BOL-3/pAN6 (♦; biomass concentrations, 5.28 [A] and 5.33 [B] g cdw/liter), and BOL-3/pAN6-gap (●; biomass concentrations, 5.70 [A] and 5.70 [B] g cdw/liter). The experiments were performed in a Multifors bioreactor system, and the cells, pregrown aerobically, were resuspended in a solution containing 0.9% (wt/vol) NaCl solution, 111 mM glucose, 200 mM NaHCO3, and 220 mM sodium formate. The pH was kept constant at 6.9 by addition of 3 M KOH. Note that the different rates cannot be directly compared, as the biomass concentrations differed. The average values of the final process parameters, including standard deviations from three independent fermentations for each strain, are displayed in Table 3.
Table 3.
Comparison of different C. glutamicum strains with respect to relevant parameters for anaerobic succinate production from glucose and formate in a batch fermentationa
| C. glutamicum strain | Yield of succinate/ glucose (mol/mol) | Rate (mmol per g cdw per h) of: |
Consumption (mM) of: |
Final titer (mM) of: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Succinate production | Glucose uptake | Formate uptake | Formate | Glucose | Succinate | Acetate | Pyruvate | Malate | Fumarate | α-Ketoglutarate | ||
| BOL-2 | 1.01 ± 0.03 | 2.58 ± 0.25 | 2.57 ± 0.22 | ND | ND | 108 ± 2 | 109 ± 5 | 5 ± 0 | 31 ± 2 | 12 ± 0 | 4 ± 0 | 9 ± 1 |
| BOL-3 | 1.26 ± 0.02 | 2.30 ± 0.06 | 1.76 ± 0.17 | 0.98 ± 0.25 | 84 ± 2 | 111 ± 2 | 141 ± 3 | 2 ± 0 | 23 ± 2 | 14 ± 1 | 5 ± 0 | 5 ± 0 |
| BOL-3/pAN6 | 1.28 ± 0.05 | 2.42 ± 0.15 | 1.67 ± 0.09 | 1.01 ± 0.21 | 77 ± 15 | 104 ± 10 | 133 ± 15 | 2 ± 0 | 23 ± 5 | 12 ± 1 | 4 ± 0 | 5 ± 2 |
| BOL-3/pAN6-gap | 1.41 ± 0.02 | 2.14 ± 0.54 | 1.56 ± 0.42 | 1.11 ± 0.12 | 89 ± 13 | 107 ± 4 | 151 ± 7 | 3 ± 0 | 14 ± 3 | 10 ± 2 | 4 ± 1 | 4 ± 1 |
The cells were pregrown aerobically and subsequently resuspended in 600 ml 0.9% (wt/vol) NaCl solution containing 111 mM glucose, 200 mM NaHCO3, and 220 mM sodium formate in a bioreactor. The pH of the cell suspension was maintained at pH 6.9 by addition of 3 M KOH. Rates were calculated for the period in which glucose was completely consumed for strains BOL-2 and BOL-3/pAN6-gap but for only the first 3 h of glucose utilization for strains BOL-3 and BOL-3/pAN6, which showed a temporary metabolic blockage after those 3 h. The final concentrations of products and by-products were determined after 20 ± 1 h for strains BOL-2 and BOL-3/pAN6-gap and after 26 ± 3 h for strains BOL-3 and BOL-3/pAN6. All values are the average data with standard deviations from three independent batch fermentations. ND, not detected.
In the case of the BOL-2 strain, the addition of formate did not influence the succinate yield (1.01 ± 0.03 mol/mol glucose) but reduced the glucose uptake and succinate production rates by 40%. Thus, the presence of 220 mM formate slowed down the overall process, although it was not consumed in detectable amounts. The acetate level produced remained low (5 ± 0 mM), but the final titers of pyruvate (31 ± 2 mM; +34%), malate (12 ± 0 mM; +71%), and fumarate (4 ± 0 mM; +20%) increased, whereas the α-ketoglutarate titer decreased (9 ± 1 mM; −25%).
In contrast to BOL-2, the BOL-3 strain containing the fdh gene from M. vaccae utilized 84 ± 2 mM formate with an uptake rate of 0.98 ± 0.25 mmol per g cdw per h. The succinate yield of the BOL-3 strain on a glucose/formate mixture increased about 20% to 1.26 ± 0.02 mol/mol glucose compared to 1.05 ± 0.02 mol/mol with glucose alone. The substrate uptake and succinate production rates of the BOL-3 strain for the process with the glucose/formate mixture were calculated only for the first 3 h of production, because afterwards, the strain entered a dormancy phase which lasted for several hours (Fig. 3A). In these first 3 h, the glucose uptake (1.76 ± 0.17 mmol per g cdw per h) and succinate production (2.30 ± 0.06 mmol per g cdw per h) rates of BOL-3 were reduced by 59% and 47% compared to the values on glucose alone (4.28 ± 0.39 mmol per g cdw per h and 4.37 ± 0.34 mmol per g cdw per h, respectively). Thus, the deceleration of the process in the presence of formate that was observed for the BOL-2 strain was also detected for the BOL-3 strain. During the temporary block of the BOL-3 strain utilizing the glucose/formate mixture, very low substrate consumption and product excretion were detected. After several hours, the production continued again with nearly the same rate as in the first 3 h. This behavior was observed in all three analyzed fermentations with formate as a cosubstrate. Interestingly, during glucose/formate coutilization, the BOL-3 strain accumulated less acetate (2 ± 0 mM; −60%) and α-ketoglutarate (5 ± 0 mM; −62%) but more intermediates of the reductive branch of the TCA cycle, namely, malate (14 ± 1 mM; +250%) and fumarate (5 ± 0 mM; +733%), than during production on glucose as the sole substrate. The final excreted pyruvate levels were comparable between the two conditions (23 ± 2 mM to 26 ± 2 mM). As expected, BOL-3/pAN6 exhibited characteristics similar to those of the parental strain BOL-3 except for reduced glucose uptake and succinate production rates during fermentation on glucose as the sole substrate.
In contrast to BOL-3 and BOL-3/pAN6, the strain BOL-3/pAN6-gap showed no temporary process inhibition in the presence of formate as a cosubstrate (Fig. 3A). The BOL-3/pAN6-gap strain consumed 89 ± 13 mM formate (1.11 ± 0.12 mmol per g cdw per h) together with 107 ± 4 mM glucose (1.56 ± 0.42 mmol per g cdw per h) and produced 151 ± 7 mM succinate (2.14 ± 0.54 mmol per g cdw per h) with a yield of 1.41 ± 0.02 mol/mol glucose. This was the best yield achieved during batch fermentation and represented an increase of 32% compared to the value on glucose alone (1.07 ± 0.02 mol/mol glucose). Remarkably, the production process of the BOL-3/pAN6-gap strain on glucose as the sole substrate showed the lowest glucose uptake (2.20 ± 0.12 mmol per g cdw per h) and succinate production (2.25 ± 0.08 mmol per g cdw per h) rates of all tested strains. In the presence of formate, the glucose uptake rate of BOL-3/pAN6-gap dropped by 30% compared to that of glucose alone, while the succinate production rate decreased by only 5%. During formate/glucose coutilization, the BOL-3/pAN6-gap strain accumulated less pyruvate (14 ± 1 mM; −30%), α-ketoglutarate (4 ± 1 mM; −72%), and acetate (3 ± 0 mM; −50%) but showed increased excretion of malate (10 ± 2 mM; +150%) and fumarate (4 ± 1 mM; +263%) compared to the by-product profile on glucose as the sole substrate.
Fed-batch succinate production with strain BOL-3/pAN6-gap on a formate/glucose substrate mixture.
To test the performance of strain BOL-3/pAN6-gap throughout a process in which high concentrations of products should accumulate, two independent fed-batch fermentations were carried out with different biomass concentrations, leading to different kinetics. The two experiments showed comparable final succinate yields (deviation, 5%) and titers (deviation, 10%). One representative experiment is shown in Fig. 4 and described in detail here. Initially, the 0.9% NaCl solution contained 222 mM glucose, 240 mM formate, and 250 mM NaHCO3. After 7.5, 20, and 39 h, 222 mM glucose and 250 mM NaHCO3 were added. After 7.5 and 20 h, 200 mM sodium formate was added, and after 36 and 53 h, 290 mM sodium formate was added. The pH was kept at pH 6.9 by automated addition of 4 M KOH. After feeding with NaHCO3, the pH increased to between 7.2 and 7.4 and then returned to pH 6.9 by the acid production of the cells. Throughout the experiment, the concentrations of glucose and organic acids were measured offline by high-performance liquid chromatography (HPLC).
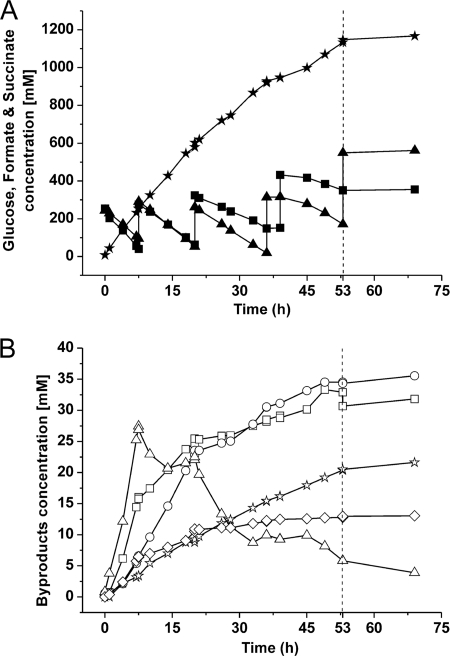
Fig 4.
Representative anaerobic fed-batch fermentation with C. glutamicum strain BOL-3/pAN6-gap showing succinate production (black stars) during coutilization of glucose (black squares) and formate (black triangles) (A) and formation of the by-products pyruvate (open triangles), acetate (open stars), α-ketoglutarate (open circles), malate (open squares), and fumarate (open diamonds) (B). The dashed line shown after 53 h of incubation marks the end of anaerobic succinate production activity. The experiment was performed in a Multifors bioreactor system, and the cells, pregrown aerobically, were resuspended in 450 ml of a solution containing 0.9% (wt/vol) NaCl, 220 mM glucose, 250 mM NaHCO3, and 220 mM sodium formate. The cell suspension had an initial OD600 of about 50. After 7.5, 20, and 39 h, 222 mM glucose and 250 mM NaHCO3 were added. After 7.5 and 20 h, 200 mM sodium formate was added, and after 36 and 53 h, 290 mM sodium formate was added. The pH was kept at pH 6.9 by automated addition of 4 M KOH. After feeding with NaHCO3, the pH increased to between 7.2 and 7.4 and then returned to pH 6.9 by the acid production of the cells. Two independent fermentations were performed, with both showing comparable results with respect to product yield (5% difference) and titer (10% difference).
As shown in Fig. 4, the succinate concentration increased continuously for 53 h, after which the metabolic activity of the cells ceased, although neither glucose nor formate had been consumed. The stop of metabolism correlated with the fourth addition of sodium formate, indicating that the formate titer of about 500 mM might have been responsible for inhibition of metabolism. Within the first 53 h, succinate was produced to a final titer of 1,134 mM (134 g liter−1). In this time, 679 mM glucose (122 g liter−1) and 772 mM formate (36 g liter−1) had been consumed (Fig. 4A). This resulted in the highest succinate yield described for C. glutamicum of 1.67 mol/mol glucose (1.1 g succinate/g glucose), an average specific succinate production rate of 1.59 mmol per g cdw per h, and a mean volumetric productivity of 21 mM h−1. The average specific glucose uptake rate (0.95 mmol per g cdw per h) observed for strain BOL-3/pAN6-gap during fed-batch fermentation was 40% lower than that for the batch experiment (1.56 ± 0.42 mmol per g cdw per h) and was slightly lower than the average formate uptake rate (1.08 mmol per g cdw per h) in the fed-batch process, which was reduced by only 3% compared to that of the batch experiment. Both the glucose uptake rate and the formate uptake rate decreased after each pulse, but the former decreased more strongly than the latter.
The kinetics of formation of the by-products malate, fumarate, and α-ketoglutarate correlated roughly with the glucose consumption rate, being faster in the initial stages of the fermentation (Fig. 4B). In contrast, acetate was excreted with an approximately constant rate during the whole production process. Pyruvate represented a temporary by-product, as it accumulated up to 27 mM during metabolism of the glucose initially present and then was consumed. In total, the fed-batch process with the BOL-3/pAN6-gap strain resulted in 107 mM by-products (corresponding to 417 mM C) after 53 h (35 mM α-ketoglutarate, 33 mM malate, 20 mM acetate, 13 mM fumarate, and 6 mM pyruvate), which represented only 10% of the total glucose carbon consumed (4,074 mM C).
To judge the efficiency of supplying additional NADH by formate oxidation, the redox balance of the fed-batch process was calculated. As NADH can be converted to MQH2 by NADH dehydrogenase and as malate dehydrogenase can use both NADPH and NADH (4, 12), all three reduced cofactors were calculated as one NADH pool. The calculation was based on the metabolic reactions of strain BOL-3/pAN6-gap, as shown on Fig. 1. Oxidation of glucose to pyruvate via glycolysis delivered 1,358 mM NADH, oxidation of pyruvate to acetyl-CoA or acetate delivered 55 mM, oxidation of isocitrate to α-ketoglutarate delivered 35 mM, and formate oxidation to CO2 delivered 772 mM. Thus, in sum, 2,220 mM NADH was generated. For the conversion of oxaloacetate to succinate via malate and fumarate, 2,314 mM NADH was needed. Thus, the redox balance was close to 96%.
Formate oxidation yields not only NADH but also CO2 for succinate production. In total, 1,862 mM CO2 was available in the fed-batch process: 1,000 mM from NaHCO3, 772 mM from formate oxidation, 55 mM from pyruvate oxidation to acetyl-CoA/acetate, and 35 mM from oxidation of isocitrate to α-ketoglutarate. On the other hand, 1,215 mM CO2 was required for carboxylation of pyruvate (or phosphoenolpyruvate) to oxaloacetate. Thus, at least 10% of the carbon dioxide fixed in succinate originated from formate oxidation. With respect to the entire carbon balance, 5,846 mM C was provided (679 mM glucose, 772 mM formate, 1,000 mM NaHCO3) and 4,953 mM carbon was recovered in products (1,134 mM succinate, 35 mM α-ketoglutarate, 33 mM malate, 20 mM acetate, 13 mM fumarate, 6 mM pyruvate). The gap of 15% is almost certainly due to remaining carbon dioxide, which was not measured.
DISCUSSION
In this work, metabolic engineering was used to develop C. glutamicum strains (based on the ATCC 13032 wild-type strain) for anaerobic succinate production from glucose or glucose/formate mixtures. The specific aims were to reduce by-product formation and to increase the succinate yield. For comparison with the previously described succinate producer C. glutamicum R ΔldhA-pCRA717 (38), C. glutamicum ΔldhA/pAN6-pycP458S was created. By trend, these two strains behaved similarly, but the ATCC 13032-based strain formed 17% less succinate and 59% more by-products than the R-based strain. Besides the different strain backgrounds, the use of the wild-type versus the mutated pyc gene, the use of different plasmids and promoters for pyc expression, and different production conditions could be responsible for the differing results. C. glutamicum ΔldhA/pAN6-pycP458S produced 0.46 mol acetate/mol glucose. To eliminate this major by-product, the genes for the three acetate-forming pathways known in C. glutamicum (cat, pqo, ackA-pta) were deleted, resulting in strain BOL-1. For enhancement of pyruvate carboxylation, either plasmid pAN6-pycP458S was used or pycP458S was chromosomally integrated under the control of the strong constitutive tuf promoter (strain BOL-2). Both strains formed 90% less acetate than C. glutamicum ΔldhA/pAN6-pycP458S. It is still unclear which reactions and enzymes are responsible for the residual acetate production. As discussed previously (32), it might be formed by the hydrolysis and/or conversion of acetyl-CoA to acetate by a number of putative acetyltransferases or hydrolases. As expected, the succinate yield of strains BOL-1/pAN6-pycP458S and BOL-2 was decreased to 1.0 mol/mol glucose compared to strain ΔldhA/pAN6-pycP458S (1.16 mol/mol), since the inhibition of acetate formation from pyruvate also reduced the formation of reducing equivalents (by the pyruvate dehydrogenase complex and/or by pyruvate:menaquinone oxidoreductase), as discussed previously (58).
As the succinate yield of strain BOL-2 was limited by the redox balance, the M. vaccae fdh gene encoding an NAD+-dependent formate dehydrogenase was chromosomally integrated under the control of the tuf promoter, allowing the use of formate as an additional donor of reducing equivalents and, simultaneously, of CO2. All strains carrying the fdh gene (BOL-3, BOL-3/pAN6, and BOL-3/pAN6-gap) showed a significant increase in the succinate yield in the presence of formate (1.3 to 1.4 mol/mol) compared to that in its absence (1.0 to 1.1 mol/mol). In contrast, strain BOL-2 was unable to utilize formate and showed no yield differences on glucose/formate mixtures compared to that with glucose as the sole carbon source. The presence of 220 mM formate reduced the glucose uptake rate and, consequently, also the succinate production rates. As this effect was also observed for strain BOL-2, the inhibition apparently was caused by the mere presence of formate and not by its metabolism. An inhibitory effect of formate on the anaerobic metabolism of resting cells was also described for ethanologenic E. coli LY01 (60). Ethanol production in phosphate-buffered saline at pH 6.8 and supplemented with 217 mM (10 g/liter) formate, conditions very similar to those used in our work, was decreased by 25% compared to that of a reference culture without formate. A possible explanation for the inhibitory effect is that formic acid (pKa, 3.77), but not its anion, can diffuse across the cytoplasmic membrane and thereby dissipates ion gradients and, dependent on the pH gradient, increases the internal anion concentration (42, 60).
In the presence of formate, strain BOL-3 and its derivatives formed fewer oxidized by-products, such as acetate and α-ketoglutarate, and more reduced by-products, such as malate and fumarate, in the supernatant. This was a hint for an increased availability of NADH, which redirected metabolism toward the synthesis of more reduced products. Moreover, it also suggested that the reduction of fumarate to succinate by succinate dehydrogenase (SDH) (Fig. 1) might become a rate-limiting step under these conditions. Thus, an increase of SDH activity could be a starting point for further strain improvement.
The succinate production process of strains BOL-3 and BOL-3/pAN6 on the substrate mixture of glucose plus formate was interrupted by a several-hours-long dormancy phase. This effect was prevented by overexpression of the gapA gene. This supported the assumption that the dormancy phase was caused by an inhibition of GAPDH activity by high NADH levels, as reported for C. glutamicum strain R (25, 39). However, whereas gapA overexpression stimulated glucose consumption by an anaerobic l-alanine producer based on strain R (25), a quite contrary effect was observed for the BOL-3 strain, for which overexpression of gapA reduced the glucose consumption rate. The reason for this difference is not yet clear, but the reduced glucose consumption might improve the balance between NADH generation and reoxidation.
Fed-batch fermentations with strain BOL-3/pAN6-gap delivered the highest reported succinate yield from glucose that was achieved with metabolically engineered C. glutamicum strains, namely, 1.67 mol/mol glucose, which represented 84% of the theoretically possible yield under the conditions used. Of course, it has to be taken into account that part of the carbon stems from carbon dioxide, as maximally 1.5 mol succinate can be formed from 1 mol glucose. The high succinate yield was accompanied by very low by-product formation. In sum, only 417 mM C was found in by-products compared to 4,536 mM C in succinate (1,134 mM). Compared to the batch fermentation, the succinate yield of the fed-batch process was increased about 18%. This might be due to the stepwise decrease of the glucose uptake rate compared to the formate uptake rate, allowing an improved balance of the NADH-forming and NADH-consuming reactions. The reduction of the glucose uptake rate was possibly due to increasing formate concentrations, as indicated by the third formate pulse, after which glucose utilization stopped for 4 h but then continued, and by the fourth pulse, after which the formate concentration exceeded 500 mM, and a complete inhibition of glucose metabolism was observed. This behavior suggests that the negative influence of formate was concentration dependent and that in an industrial process, the formate concentration should be kept below 300 mM.
The efficiency of the fed-batch process was also documented by the redox balance, which was close to 96%, i.e., only 4% of the NADH required for reduction of oxaloacetate to succinate could not be ascribed to the known metabolic pathways (Fig. 1). It showed that the entire amount of NADH produced by formate oxidation was used for succinate production. Furthermore, part of the CO2 formed by formate oxidation was incorporated into succinate. However, further experiments are required to determine how efficient the replacement of NaHCO3 by CO2 derived from formate oxidation could become. Altogether, the results confirmed that both products of formate oxidation were utilized in the described process.
A compilation of some of the most efficient microbial strains for anaerobic succinate production from glucose is shown in Table 4. This summary shows that the new process for anaerobic succinate production from glucose/formate mixtures with the C. glutamicum BOL-3/pAN6-gap strain does not only compete favorably with the producer strain based on C. glutamicum R regarding yield, product titer, volumetric productivity, and by-product formation, it also exceeds the maximal parameters of the most important natural succinate producers A. succinogenes ATCC 55618 FZ 53 and A. succiniciproducens as well as those of the metabolically engineered strains M. succiniciproducens LPK7, E. coli KJ134, and E. coli SBS550MG/pHL314. The compared parameters for the C. glutamicum BOL-3/pAN6-gap strain are based only on the production phase of the process, but even if the substrate and the time invested in the generation of the biomass are taken into account (values displayed in parentheses), the process results are still very competitive. The succinate yield for the newly developed process was calculated based on the utilized glucose. Consumption of formate and/or bicarbonate as a CO2 donor was not taken into account, similar to the processes of the previous studies shown in Table 4. Of course, formate and bicarbonate have to be considered cost factors in an industrial implementation. As formate serves as a donor not only of reducing equivalents but also of carbon dioxide, its use allows both an increased succinate yield and a reduced bicarbonate demand and thus offers advantages compared to processes using bicarbonate only.
Table 4.
Data of some of the most efficient processes described for anaerobic succinate production from glucosea
| Succinate producer | Succinate yield (mol/mol) | Total succinate produced (mM) | Volumetric productivity (mM h−1) | Total by-product concn (mM [mM carbon]) | By-product yield (mol C by-product/mol C glucose) | Process description | Reference |
|---|---|---|---|---|---|---|---|
| C. glutamicum BOL-3/pAN6-gap | 1.67 (1.36b) | 1,134 | 21 (16c) | 107 (417) | 0.10 | Two stages—aerobic growth in minimal medium, anaerobic fed-batch production in saline solution | This study |
| C. glutamicum R ΔldhA-pCRA717 | 1.40 (0.85b) | 1,240 | 27 (21d) | 270 (540) | 0.10 | Two stages—aerobic growth in complex medium, anaerobic fed-batch production in minimal medium | 38 |
| E. coli SBS550MG/pHL314 | 1.60 (1.60) | 350 | 4 (3e) | ∼50 (50) | 0.04 | Two stages—aerobic growth in complex medium, anaerobic fed-batch production in complex medium | 45 |
| E. coli KJ134 | 1.53 | 606 | 6 | 72 (192) | 0.08 | One stage—anaerobic batch process in minimal medium | 24 |
| A. succiniciproducens ATCC 53488 | 1.37 | 180 | 9 | 109 (205) | 0.26 | One stage—anaerobic batch process in complex medium on large scale (80 liters) | 13 |
| A. succinogenes ATCC 55618 FZ 53 | 1.26 | 897 | 12 | 368 (773) | 0.18 | One stage—anaerobic batch process in complex medium | 14 |
| M. succiniciproducens LPK7 | 1.16 | 444 | 15 | 241 (800) | 0.35 | One stage—anaerobic fed-batch process in complex medium | 31 |
For two-stage processes, approximate values calculated for the entire process are displayed in parentheses.
Succinate yield calculated for glucose consumed initially for biomass production and subsequently for succinate production. The factor f (11.52 mmol glucose per g cdw), representing the amount of glucose consumed per gram of biomass formed, was calculated from previous growth studies with wild-type C. glutamicum ATCC 13032 in a bioreactor (32).
Total volumetric productivity calculated for the overall process time (69 h) = biomass cultivation time (16 h) + production time (53 h).
Total volumetric productivity calculated for the overall process time (59 h) = biomass cultivation time (13 h) + production time (46 h).
Total volumetric productivity calculated for the overall process time (113 h) = biomass cultivation time (17 h) + production time (96 h).
A general benefit of the new process in comparison to many previously described ones is the use of simplified production medium (saline solution), which enables lower production and purification costs. A further decrease of the purification cost could be achieved with a production process performed at lower pH. Processes in the pH range of 5.8 to 6.5 were already described for some of the listed species, e.g., A. succinogenes (14) and A. succiniciproducens (13), but never tested for anaerobic substrate conversion with C. glutamicum, thus presenting an interesting opportunity for process optimization.
Another means for improvement of the system presented here for anaerobic succinate production is further strain optimization. The recent identification of the succinate exporter SucE in C. glutamicum (10, 18) offers the interesting opportunity to test the effect of increased sucE expression on the succinate production of the reported strains. As the reaction catalyzed by GAPDH was shown to be critical, its optimization may improve the substrate consumption rates and thus further optimize the current producers. Additional improvements of the entire process, e.g., by (i) optimized cultivation conditions for biomass production, (ii) production in cultivation medium without biomass transfer, (iii) maintenance of optimal substrate concentrations, especially of formate because of its toxicity at high concentrations (62), (iv) in situ product recovery (63), and (v) continued strain improvement may further increase the overall efficiency of the process.
Supplementary Material
ACKNOWLEDGMENTS
Financial support (grant 220-095-08D to M.B.) by the Federal Ministry of Food, Agriculture, and Consumer Protection (BMELV) within the ERA-IB project BioProChemBB is gratefully acknowledged.
We thank Volker F. Wendisch (University of Bielefeld) for providing plasmid pK19mobsacB-Δcat and Stephanie Bringer-Meyer (Forschungszentrum Jülich) for providing plasmid pBBR1MCS2-fdh.
Footnotes
Published ahead of print 2 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abe S, Takayama K, Kinoshita S. 1967. Taxonomical studies on glutamic acid producing bacteria. J. Gen. Appl. Microbiol. 13:279–301 [Google Scholar]
- 2. Bäumchen C, Bringer-Meyer S. 2007. Expression of glf Z.m. increases D-mannitol formation in whole cell biotransformation with resting cells of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 76:545–552 [DOI] [PubMed] [Google Scholar]
- 3. Blombach B, et al. 2011. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl. Environ. Microbiol. 77:3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bott M, Niebisch A. 2003. The respiratory chain of Corynebacterium glutamicum. J. Biotechnol. 104:129–153 [DOI] [PubMed] [Google Scholar]
- 5. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 6. Burkovski A. (ed). 2008. Corynebacteria: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 7. Eggeling L, Bott M. 2005. Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, FL [Google Scholar]
- 8. Eikmanns BJ, Kleinertz E, Liebl W, Sahm H. 1991. A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene 102:93–98 [DOI] [PubMed] [Google Scholar]
- 9. Frunzke J, Engels V, Hasenbein S, Gätgens C, Bott M. 2008. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol. Microbiol. 67:305–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fukui K, et al. 2011. Identification of succinate exporter in Corynebacterium glutamicum and its physiological roles under anaerobic conditions. J. Biotechnol. 154:25–34 [DOI] [PubMed] [Google Scholar]
- 11. Galkin A, Kulakova L, Tishkov V, Esaki N, Soda K. 1995. Cloning of formate dehydrogenase gene from a methanol-utilizing bacterium Mycobacterium vaccae N10. Appl. Microbiol. Biotechnol. 44:479–483 [DOI] [PubMed] [Google Scholar]
- 12. Genda T, Nakamatsu T, Ozak H. 2003. Purification and characterization of malate dehydrogenase from Corynebacterium glutamicum. J. Biosci. Bioeng. 95:562–566 [PubMed] [Google Scholar]
- 13. Glassner DA, Datta R. 1992. Process for the production and purification of succinic acid. US patent 5143834
- 14. Guettler MV, Jain MK, Rumler D. 1996. Method for making succinic acid, bacterial variants for use in the process, and methods for obtaining variants. US patent 5,573,931
- 15. Guettler MV, Rumler D, Jain MK. 1999. Actinobacillus succinogenes sp. nov., a novel succinic-acid-producing strain from the bovine rumen. Int. J. Syst. Bacteriol. 49:207–216 [DOI] [PubMed] [Google Scholar]
- 16. Hanahan D. 1985. Techniques for transformation of E. coli, p 109–135 In Glover DM. (ed), DNA cloning, vol 1 IRL Press, Oxford, United Kingdom [Google Scholar]
- 17. Hermann BG, Patel M. 2007. Today's and tomorrow's bio-based bulk chemicals from white biotechnology: a techno-economic analysis. Appl. Biochem. Biotechnol. 136:361–388 [DOI] [PubMed] [Google Scholar]
- 18. Huhn S, Jolkver E, Krämer R, Marin K. 2011. Identification of the membrane protein SucE and its role in succinate transport in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 89:327–335 [DOI] [PubMed] [Google Scholar]
- 19. Ikeda M, Nakagawa S. 2003. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 62:99–109 [DOI] [PubMed] [Google Scholar]
- 20. Ikeda M, Ohnishi J, Hayashi M, Mitsuhashi S. 2006. A genome-based approach to create a minimally mutated Corynebacterium glutamicum strain for efficient l-lysine production. J. Ind. Microbiol. Biotechnol. 33:610–615 [DOI] [PubMed] [Google Scholar]
- 21. Inui M, Kawaguchi H, Murakami S, Vertès AA, Yukawa H. 2004. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J. Mol. Microbiol. Biotechnol. 8:243–254 [DOI] [PubMed] [Google Scholar]
- 22. Inui M, et al. 2004. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J. Mol. Microbiol. Biotechnol. 7:182–196 [DOI] [PubMed] [Google Scholar]
- 23. Jantama K, et al. 2008. Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol. Bioeng. 99:1140–1153 [DOI] [PubMed] [Google Scholar]
- 24. Jantama K, et al. 2008. Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol. Bioeng. 101:881–893 [DOI] [PubMed] [Google Scholar]
- 25. Jojima T, Fujii M, Mori E, Inui M, Yukawa H. 2010. Engineering of sugar metabolism of Corynebacterium glutamicum for production of amino acid l-alanine under oxygen deprivation. Appl. Microbiol. Biotechnol. 87:159–165 [DOI] [PubMed] [Google Scholar]
- 26. Kabus A, Niebisch A, Bott M. 2007. Role of cytochrome bd oxidase from Corynebacterium glutamicum in growth and lysine production. Appl. Environ. Microbiol. 73:861–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kalinowski J, et al. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5–25 [DOI] [PubMed] [Google Scholar]
- 28. Keilhauer C, Eggeling L, Sahm H. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175:5595–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kirchner O, Tauch A. 2003. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 104:287–299 [DOI] [PubMed] [Google Scholar]
- 30. Lee PC, Lee SY, Hong SH, Chang HN. 2002. Isolation and characterization of a new succinic acid-producing bacterium, Mannheimia succiniciproducens MBEL55E, from bovine rumen. Appl. Microbiol. Biotechnol. 58:663–668 [DOI] [PubMed] [Google Scholar]
- 31. Lee SJ, Song H, Lee SY. 2006. Genome-based metabolic engineering of Mannheimia succiniciproducens for succinic acid production. Appl. Environ. Microbiol. 72:1939–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Litsanov B, Kabus A, Brocker M, Bott M. 2012. Efficient aerobic succinate production from glucose in minimal medium with Corynebacterium glutamicum. Microb. Biotechnol. 5:116–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McKinlay JB, Vieille C, Zeikus JG. 2007. Prospects for a bio-based succinate industry. Appl. Microbiol. Biotechnol. 76:727–740 [DOI] [PubMed] [Google Scholar]
- 34. Meynial-Salles I, Dorotyn S, Soucaille P. 2008. A new process for the continuous production of succinic acid from glucose at high yield, titer, and productivity. Biotechnol. Bioeng. 99:129–135 [DOI] [PubMed] [Google Scholar]
- 35. Nghiem NP, Davison BH, Suttle BE, Richardson GR. 1997. Production of succinic acid by Anaerobiospirillum succiniciproducens. Appl. Biochem. Biotechnol. 63–65:565–576 [DOI] [PubMed] [Google Scholar]
- 36. Niebisch A, Bott M. 2001. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 175:282–294 [DOI] [PubMed] [Google Scholar]
- 37. Nishimura T, Vertès AA, Shinoda Y, Inui M, Yukawa H. 2007. Anaerobic growth of Corynebacterium glutamicum using nitrate as a terminal electron acceptor. Appl. Microbiol. Biotechnol. 75:889–897 [DOI] [PubMed] [Google Scholar]
- 38. Okino S, et al. 2008. An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl. Microbiol. Biotechnol. 81:459–464 [DOI] [PubMed] [Google Scholar]
- 39. Omumasaba CA, Okai N, Inui M, Yukawa H. 2004. Corynebacterium glutamicum glyceraldehyde-3-phosphate dehydrogenase isoforms with opposite, ATP-dependent regulation. J. Mol. Microbiol. Biotechnol. 8:91–103 [DOI] [PubMed] [Google Scholar]
- 40. Peters-Wendisch PG, et al. 1998. Pyruvate carboxylase from Corynebacterium glutamicum: characterization, expression and inactivation of the pyc gene. Microbiology 144:915–927 [DOI] [PubMed] [Google Scholar]
- 41. Reinscheid DJ, et al. 1999. Cloning, sequence analysis, expression and inactivation of the Corynebacterium glutamicum pta-ack operon encoding phosphotransacetylase and acetate kinase. Microbiology 145:503–513 [DOI] [PubMed] [Google Scholar]
- 42. Rusell JB. 1992. Another explanation for the toxicity of fermentation acids at low pH: anion accumulation versus uncoupling. J. Appl. Bacteriol. 73:363–370 [Google Scholar]
- 43. Sambrook J, MacCallum P, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 44. Samuelov NS, Lamed R, Lowe S, Zeikus JG. 1991. Influence of CO2-HCO3− levels and pH on growth, succinate production, and enzyme activities of Anaerobiospirillum succiniciproducens. Appl. Environ. Microbiol. 57:3013–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sanchez AM, Bennett GN, San KY. 2005. Novel pathway engineering design of the anaerobic central metabolic pathway in Escherichia coli to increase succinate yield and productivity. Metab. Eng. 7:229–239 [DOI] [PubMed] [Google Scholar]
- 46. Schäfer A, et al. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73 [DOI] [PubMed] [Google Scholar]
- 47. Schneider J, Wendisch VF. 2010. Putrescine production by engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 88:859–868 [DOI] [PubMed] [Google Scholar]
- 48. Scholten E, Dagele D. 2008. Succinic acid production by a newly isolated bacterium. Biotechnol. Lett. 30:2143–2146 [DOI] [PubMed] [Google Scholar]
- 49. Schreiner ME, Riedel C, Holatko J, Patek M, Eikmanns BJ. 2006. Pyruvate:quinone oxidoreductase in Corynebacterium glutamicum: molecular analysis of the pqo gene, significance of the enzyme, and phylogenetic aspects. J. Bacteriol. 188:1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smith KM, Cho KM, Liao JC. 2010. Engineering Corynebacterium glutamicum for isobutanol production. Appl. Microbiol. Biotechnol. 87:1045–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Takeno S, et al. 2007. Anaerobic growth and potential for amino acid production by nitrate respiration in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 75:1173–1182 [DOI] [PubMed] [Google Scholar]
- 52. Thakker C, Martinez I, San KY, Bennett GN. 2012. Succinate production in Escherichia coli. Biotechnol. J. 7:213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thakker C, Zhu J, San KY, Bennett GN. 2011. Heterologous pyc gene expression under various natural and engineered promoters in Escherichia coli for improved succinate production. J. Biotechnol. 155:236–243 [DOI] [PubMed] [Google Scholar]
- 54. van der Rest ME, Lange C, Molenaar D. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52:541–545 [DOI] [PubMed] [Google Scholar]
- 55. Veit A, et al. 2009. Pathway identification combining metabolic flux and functional genomics analyses: acetate and propionate activation by Corynebacterium glutamicum. J. Biotechnol. 140:75–83 [DOI] [PubMed] [Google Scholar]
- 56. Wendisch VF, Bott M, Eikmanns BJ. 2006. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr. Opin. Microbiol. 9:268–274 [DOI] [PubMed] [Google Scholar]
- 57. Werpy T, Peterson G. 2004. Top value added chemicals from biomass. Volume 1. Results of screening for potential candidates from sugars and synthesis gas. US Department of Energy, Oak Ridge, TN: http://www.ascension-publishing.com/BIZ/HD49.pdf [Google Scholar]
- 58. Yasuda K, et al. 2007. Analyses of the acetate-producing pathways in Corynebacterium glutamicum under oxygen-deprived conditions. Appl. Microbiol. Biotechnol. 77:853–860 [DOI] [PubMed] [Google Scholar]
- 59. Yukawa H, et al. 2007. Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 153:1042–1058 [DOI] [PubMed] [Google Scholar]
- 60. Zaldivar J, Ingram LO. 1999. Effect of organic acids on the growth and fermentation of ethanologenic Escherichia coli LY01. Biotechnol. Bioeng. 66:203–210 [DOI] [PubMed] [Google Scholar]
- 61. Zeikus JG, Jain MK, Elankovan P. 1999. Biotechnology of succinic acid production and markets for derived industrial products. Appl. Environ. Microbiol. 51:545–552 [Google Scholar]
- 62. Zelić B, et al. 2003. Fed-batch process for pyruvate production by recombinant Escherichia coli YYC202 strain. Eng. Life Sci. 3:299–305 [Google Scholar]
- 63. Zelic B, Gostovic S, Vuorilehto K, Vasić-Rački D, Takors R. 2004. Process strategies to enhance pyruvate production with recombinant Escherichia coli: from repetitive fed-batch to in situ product recovery with fully integrated electrodialysis. Biotechnol. Bioeng. 85:638–646 [DOI] [PubMed] [Google Scholar]
- 64. Zhang X, Jantama K, Shanmugam KT, Ingram LO. 2009. Reengineering Escherichia coli for succinate production in mineral salts medium. Appl. Environ. Microbiol. 75:7807–7813 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.