Abstract
Avian pathogenic Escherichia coli (APEC) is associated with extraintestinal infections in poultry causing a variety of diseases collectively known as colibacillosis. The host and bacterial factors influencing and/or responsible for carriage and systemic translocation of APEC inside the host are poorly understood. Identification of such factors could help in the understanding of its pathogenesis and in the subsequent development of control strategies. Recombination-based in vivo expression technology (RIVET) was used to identify APEC genes specifically expressed during infection in chickens. A total of 21 clones with in vivo-induced promoters were isolated from chicken livers and spleens, indicative of systemic infection. DNA sequencing of the cloned fragments revealed that 12 of the genes were conserved E. coli genes (metH, lysA, pntA, purL, serS, ybjE, ycdK [rutC], wcaJ, gspL, sdsR, ylbE, and yjiY), 6 of the genes were phage related/associated, and 3 genes were pathogen specific (tkt1, irp2, and eitD). These genes are involved in various cellular functions, such as metabolism, cell envelope and integrity, transport systems, and virulence. Others were phage related or have yet-unknown functions.
INTRODUCTION
Avian pathogenic Escherichia coli (APEC) is associated with extraintestinal infections in chickens, turkeys, ducks, and other avian species, causing a variety of diseases collectively known as colibacillosis. The infections are responsible for severe economic losses in the poultry industry worldwide (7, 25, 29). A number of virulence factors have been identified as associated with APEC infections (23, 29, 43, 83), some of which have been shown to contribute to pathogenicity. They include autotransporter genes (28, 54), iron uptake systems (13, 14, 36, 75), lipopolysaccharide (LPS) O antigens (66), K1 capsule (66), certain fimbrial adhesins (21, 50), and secretion systems (24). However, the mechanisms underlying pathogenicity are still poorly understood (29). The availability of the complete genome sequence of APEC O1:K1:H7 in combination with experimental infection models in avian hosts provides the basis for comprehensive understanding of APEC pathogenesis (4, 42).
With genome-wide analyses in recent years, several new molecular approaches have provided a better understanding of the molecular mechanisms of pathogenicity of microorganisms (35, 60, 89). Recently, techniques such as selective capture of transcribed sequences (SCOTS), signature-tagged mutagenesis (STM), genomic suppression subtractive hybridization (GSSH), and transcriptome analysis using microarrays were applied to APEC to identify different genes associated with virulence and/or pathogenicity in chickens (27, 55, 56, 81). Although some of these techniques provide evidence that certain genes may contribute to virulence, it is of interest to determine which genes are actually expressed in vivo, as they may be required for APEC to cause infection and could be potential vaccine candidates (16). Identification of differentially active promoters in vivo and, if possible, those for putative and unknown virulence genes would provide more information on pathogenesis.
One of the methods that has been used successfully in bacteria to study the promoters that are specifically active in vivo and not in vitro is in vivo expression technology (IVET) (60, 61). Recombination-based in vivo expression technology (RIVET) is a variant of the original in vivo expression technology that uses a promoterless site-specific recombinase and a pair of recombinase target sequences flanking a selectable marker. When active promoters are fused to the promoterless recombinase, induction of recombinase expression occurs. This subsequently causes recombination of the two target sequences, deleting the intervening marker, and permanently changes the bacterial phenotype that can be detected after gene expression has ceased (10, 74). RIVET has been used in several Gram-negative and Gram-positive bacteria to identify promoters and/or genes that are induced in vivo (5, 9, 11, 12, 33, 38, 58, 80, 86, 91).
In this study, the use of RIVET for the identification of APEC promoters and/or genes induced specifically in vivo during infection in chickens is described, and the findings suggest that there is a range of genes that are induced as a response of the pathogen to the new environment.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. APEC strain CH2 (O78) was isolated from an infected chicken and is virulent in experimentally infected chickens (90). Strains containing plasmids pKD46 (22) and pSC101-BAD-Cre-tet (3) were incubated at 30°C unless otherwise indicated. Those containing plasmid pR6K-rpsL-Neor (34) were incubated at 37°C. Luria-Bertani (LB) medium and SOC medium were prepared as described elsewhere (76). All bacteria were routinely grown in LB broth or agar medium at 37°C unless otherwise indicated. Antibiotics for plasmid and/or recombinant selection were added at the following concentrations: ampicillin (Amp), 100 μg ml−1; kanamycin (Kan), 50 μg ml−1; streptomycin (Str), 100 μg ml−1; and tetracycline (Tet), 5 μg ml−1. A streptomycin-resistant (Strr) derivative of APEC CH2, designated CH2-Strr, was obtained by serial culturing in increasing concentrations of streptomycin, as previously described (88). E. coli strain Top10F′ (Invitrogen, Paisley, Scotland) was used as the cloning strain.
Table 1.
Bacteria and plasmids used in this study
| Strain or plasmid | Relevant feature(s) | Reference |
|---|---|---|
| Strains | ||
| APEC CH2 | O78 serotype isolated from infected chicken | 90 |
| APEC CH2-StrR | Streptomycin-resistant derivative of APEC CH2 | This study |
| APEC CH2-StrR/pKD46 | APEC CH2-Strr derivative containing pKD46 (oriR101 bla PBAD-λ gam bet exo) | This study |
| CH2LoxP | APEC CH2-Strr derivative containing loxP-rpsL-neo-loxP cassette in the fiu gene | This study |
| Plasmids | ||
| pFLAG-MAC | Cytoplasmic expression vector | Sigma |
| pSC101-BAD-Cre-tet | Cre expression vector under the promoter PBAD | 3 |
| pR6K-rspL-neo | R6K gamma ori, rpsL-neo cassette | 34 |
| pHT001 | Derivative of pFLAG-MAC | This study |
| pHT002 | pHT001 containing promoterless cre | This study |
| pHT003 | pHT002 containing E. coli lpp promoter | This study |
DNA manipulation.
DNA manipulations were performed as described elsewhere (76). Electrocompetent cells were prepared using standard procedures unless otherwise indicated (76). Electroporation was carried out as previously described (88). CH2 genomic DNA, plasmid DNA, and DNA fragments were purified using commercial kits purchased from Fermentas (St. Leon-Rot, Germany). DNA restriction and modification enzymes were purchased from Fermentas and used as recommended by the manufacturer. PCR amplifications were performed using AccuPrime Taq High Fidelity polymerase (Invitrogen) or SuperTaq polymerase (SphaeroQ, Leiden, The Netherlands). Oligonucleotides were ordered from Sigma-Aldrich (Bornem, Belgium) (Table 2).
Table 2.
Oligonucleotides used in this study
| Name | Sequence (5′–3′) | Reference |
|---|---|---|
| HT53Fa | CCCGATACCCCAGTTTGGCGTCGGCGACCCAATAACCTTCGGTAAATGCTGGCGTCCCCACCGCGCCGTCataacttcgtatagcatacattatacgaagttatGGCCTGGTGATGATGGCGGGATCG | 88 |
| HT53Ra | CCGAAATTCTCGCGTCCGGTAGCGGATACTACCCGCACGATGACGGTGATTTCTGAACAAGTGATTAAAGataacttcgtataatgtatgctatacgaagttatTCAGAAGAACTCGTCAAGAAGGCG | 88 |
| HT57F | TGGCTGTGAGCAAGAAGGTTCTTGGC | 88 |
| HT57R | ACGCGGATGACACGCTGGTTGT | 88 |
| HT86 F | CTACAAGGACGACGATGACAA | This study |
| HT86 R | TCAGGCTGAAAATCTTCTCTC | This study |
| CRESEQ | CATTTTCCAGGTATGCTCAG | 38 |
Primer used to isolate the rpsL-neo cassette from plasmid pR6K-rpsL-neo. From the 5′ end, 70 bp are homologs of the target region; the lowercase boldface letters are loxP sites, while the italicized letters represent priming sites to the template plasmid pR6K-rpsL-neo.
Construction of the RIVET host strain.
An APEC CH2-Strr strain harboring a chromosomally located loxP-rpsL-neo-loxP cassette was constructed as previously described for the strain APEC 1-Strr (88). Briefly, APEC CH2-Strr strains were made electrocompetent and transformed with plasmid pKD46 encoding the lambda Red recombinase by electroporation. For the integration of the loxP cassette, the lambda Red recombineering technique (22) was used, whereby APEC CH2-Strr containing pKD46 was made electrocompetent and approximately 0.1 μg to 0.3 μg of PCR products (amplified using primers HT53F and HT53R) containing the loxP-rpsL-neo-loxP cassette (LoxP cassette) flanked by regions homologous to the fiu gene was electroporated. The kanamycin-resistant (Kanr) colonies obtained were purified selectively at 37°C and then grown at 43°C to cure the plasmid pKD46. The strain was designated CH2LoxP.
Construction of a vector containing promoterless cre.
The plasmid pFLAG-MAC (5.1 kb; Sigma) was digested by EcoRV to remove the FLAG coding sequence, part of the lacI gene, and the ribosomal binding site (RBS) and recircularized to form plasmid pHT001 (3.7 kb). An EcoRI/XhoI restriction fragment from plasmid pSC101-BAD-Cre-tet containing a promoterless Cre recombinase gene (cre) was cloned into pHT001 digested with EcoRI/XhoI to form pHT002 (4.7 kb) (Fig. 1A). PCR amplification, using primers HT86F and HT86R flanking the cloning site, was performed to verify the proper insertion of the fragment containing the cre gene.
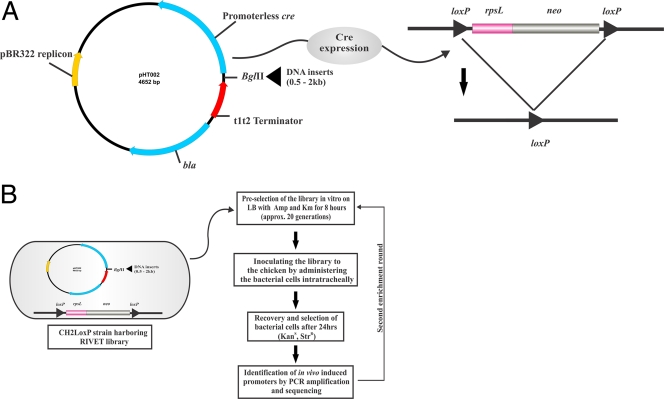
Fig 1.
RIVET strategy for use in APEC to identify promoters that are induced in vivo. (A) An rpsL-neo cassette flanked by two copies of site-specific recombination sequences (loxP) was integrated into the APEC CH2 Strr chromosome. An APEC CH2 promoter trap library was constructed by cloning random DNA fragments in plasmid pHT002 upstream from a promoterless cre gene. If an active promoter is thus cloned, the recombinase Cre is expressed, resulting in a homologous recombination of the loxP sites. This results in the irreversible excision of the rpsL-neo cassette. (B) The APEC CH2 promoter trap library (RIVET library) was transformed into the CH2LoxP strain. After preselection on kanamycin to eliminate bacterial cells containing active promoters under in vitro conditions, the resulting library was administered intratracheally into a chicken. Irreversible changes in the antibiotic resistance phenotype of the CH2LoxP strain allowed selection of resolved bacteria containing in vivo-induced promoters. After isolation of the bacteria, the plasmids contacting in vivo-induced promoters were subjected to a second enrichment round.
Construction of an APEC CH2 RIVET library.
The APEC CH2 RIVET library was made by partial digestion of APEC CH2 genomic DNA with Sau3AI, size fractionating it on a 1% agarose gel, and cloning the fragments (0.5 to 2 kb) into the BglII site of pHT002. The ligation mixture was transformed into E. coli Top10F′ (Invitrogen). The colonies obtained (approximately 60,000) were pooled, and the plasmid DNA was isolated from the cells. Random clones were picked, and the average insert size was determined. The plasmid DNA was transformed into CH2LoxP strains and plated on LB with Amp and Kan. Approximately 28,000 colonies of the APEC RIVET library were collectively resuspended in LB containing 15% glycerol and stored in aliquots at −80°C.
Screening of the APEC CH2 RIVET library for in vivo-induced genes.
Animal studies were approved by the Ethical Committee for Animal Experiments of the Katholieke Universiteit Leuven according to international regulations (project number P119/2009). Approximately 1 × 109 CFU from the APEC RIVET library was administered intratracheally using a feeding needle (malleable metal, silicone tip, sterile, plastic Luer hub, 20 gauge; AgnTho's AB, Lidingo, Sweden) into four 3-week-old broiler chickens (Ross; Belgabroed NV, Merksplas, Belgium), which were housed in a disinfected stable and supplied with water and feed ad libitum. After 24 h, the chickens were sacrificed by cervical dislocation, and their livers and spleens were collected aseptically and homogenized in sterile phosphate-buffered saline (PBS) (1 g of tissue per 1 ml sterile PBS). The dilutions were plated on MacConkey agar with Amp. After incubation at 37°C for 24 h, the colonies obtained were then replica plated on MacConkey agar with Amp and Str and incubated again at 37°C for 24 h. The bacteria that had lost the LoxP cassette due to differential expression of cre regained Str resistance and hence grew in this medium. The loss of the LoxP cassette was further verified by Kan sensitivity (Kans) and colony PCR using primers HT57F and HT57R. The colonies obtained on MacConkey agar with Amp and Str were picked up, and plasmids were isolated and purified for sequencing of the inserts using primers CRESEQ and HT86R, annealing in cre and upstream from the inserted DNA fragment, respectively, on pHT002. The sequences were analyzed using BLASTX (http://www.ncbi.nih.gov) and compared to the sequences available in GenBank.
Nucleotide sequence accession numbers.
The sequences of the selected 21 unique clones were submitted to GenBank with accession numbers JQ582670, JQ582671, JQ582672, JQ582673, JQ582674, JQ582675, JQ582676, JQ582677, JQ582678, JQ582679, JQ582680, JQ582681, JQ582682, JQ582683, JQ582684, JQ582685, JQ582686, JQ582687, JQ582688, JQ582689, and JQ582690.
RESULTS
Adaptation of RIVET screening for APEC.
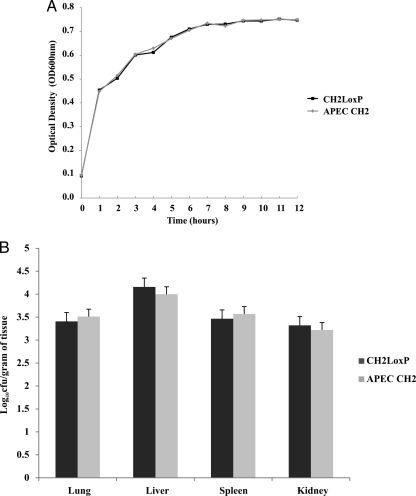
The general scheme for RIVET is shown in Fig. 1A. The Cre-lox system was shown to be functional in APEC strains (88), which suggests that the Cre recombinase is suitable for the construction of a RIVET screening system in this bacterium. The reporter strain CH2LoxP was constructed by integrating a LoxP cassette in the chromosomally located fiu gene, one of the 12 iron receptor genes in APEC encoding an iron-regulated outer membrane protein known as ferric iron uptake protein (19, 68, 69). The LoxP cassette contains a floxed rpsL-neo marker where rpsL is used for positive selection of colonies with an active promoter while neo is used for negative selection. Confirmation of integration was done using primers flanking the fiu gene and locus-specific primers as described previously for the APEC1LoxP strain (88). There were no significant differences between CH2LoxP and APEC CH2 in terms of in vitro growth for 12 h or survival in vivo inside the host for 48 h (Fig. 2A and B). The stability of integration of the LoxP cassette was confirmed in vitro and in vivo. The CH2LoxP strain was still Kanr after growth without antibiotic selection pressure for 96 h with 4 subculturing steps in between, followed by plating on LB-Kan. In vivo, Kanr colonies could be isolated 48 h after infection of the chicken host with CH2LoxP. PCR analysis confirmed that all selected colonies contained an intact LoxP cassette (data not shown). The whole LoxP cassette was also sequenced to confirm the orientation of the loxP sites. Taken together, these experiments indicate that the cassette was stably integrated into CH2LoxP and maintained during infection in chickens and that this did not affect its virulence or growth under standard laboratory conditions.
Fig 2.
Comparison of growth and survival of APEC CH2 and its isogenic mutant CH2LoxP strain. (A) Growth comparison of the two strains in vitro. There were no significant differences in terms of growth in LB medium after growing the strains separately for 12 h. (B) The in vivo survival of the strains was evaluated. The strains were separately administered intratracheally into chickens. After 48 h, the bacteria were isolated from the lungs, livers, spleens, and kidneys of four different chickens. There were no significant differences between the two strains in terms of CFU counts from organs.
In the next step, the promoter trap plasmid pHT002, containing a promoterless cre gene and a pBR322 origin of replication, was created. It also contained a bla gene for Amp resistance, an E. coli terminator to prevent transcriptional readthrough, and a BglII site for the insertion of the APEC CH2 genomic library. As a positive control, a derivative of pHT002 was made by cloning a strong E. coli constitutive promoter from the lpp gene encoding E. coli lipoprotein (40) at the BglII site and designated pHT003. Plasmid pHT002 and its derivative, pHT003, were introduced into the reporter strain CH2LoxP to evaluate their functionality. The strains were plated on LB-Amp and LB agar plates with Amp and Kan and with Amp and Str. After 48 h of growth, none of the pHT002 transformants became Strr while more than 98% of the pHT003 transformants became Strr, suggesting excision of the rpsL marker (data not shown). PCR using the HT57 primer pair was performed on CH2LoxP containing pHT003 from LB agar plates with Amp and confirmed that the LoxP cassette was indeed deleted. These results confirm the suitability of the reporter strain CH2LoxP, together with the promoter trap plasmid pHT002, for the RIVET application.
Construction of the APEC CH2 RIVET library in pHT002.
A genomic library of APEC CH2 was constructed by cloning random genomic DNA fragments of CH2 (0.5 kb to 2 kb) into the BglII site of pHT002. The E. coli Top10F′ strain was used as an intermediate cloning host. A total of approximately 28,000 colonies were obtained in the CH2LoxP strain (RIVET library) after preselecting the strains on LB agar plates with Amp and Kan. This selection excluded in vitro active promoters, which would induce the expression of cre and subsequently cause the deletion of the loxP cassette, making the strain Kans. Insert analysis of the library was performed by PCR using primers CRESEQ and HT86R (Table 2) on 90 randomly picked clones containing pHT002 derivatives, and it revealed the estimated average size of the insert to be approximately 1 kb (data not shown). To evaluate the library redundancy, 27 inserts were sequenced, suggesting that the library was random and not redundant. The genome coverage was estimated using the formula of Clarke and Carbon (18) and found to be higher than 99%.
RIVET screen in the chicken host model.
The RIVET screening approach is depicted in Fig. 1B. The library was grown for 8 h in the presence of Amp and Kan to counterselect against clones displaying cre expression in vitro. The cultures grown were used for intratracheal administration to chickens. To identify genes important for systemic infection, approximately 4,000 clones were recovered from livers and spleens after plating on MacConkey agar with Amp. Replica plating on MacConkey agar with Amp and Str resulted in 139 Strr clones (approximately 3.5%). To verify the primary RIVET results, plasmids representing these 139 clones were isolated, purified, and reintroduced individually by electroporation into the original CH2LoxP strain (Kanr Strs). The resulting colonies were Ampr and Kanr, confirming the absence of cre expression in these clones when grown in vitro. The clones were pooled and subjected to a second round of infection in chickens and screening, as explained above, to identify only promoters that showed reproducible induction in different chickens. Out of 1,320 colonies obtained after plating on MacConkey agar with Amp, 630 colonies were Ampr and Strr (approximately 48%). These clones were subjected to plasmid isolation, purification, and sequencing of the insert in the plasmids. After performing a BLASTX search, the screening resulted in 21 unique clones, of which 12 genes were E. coli conserved genes, 6 were bacteriophage associated, and 3 (tkt1, irp2, and eitD) were pathogen-specific genes. The number of clones isolated per gene is shown in Table 3. Various cellular functions, such as metabolism, adaptation, stress response, cell membrane and integrity, transport systems, virulence, and phage-related or unknown functions, were assigned to the identified genes (Table 3) based on the highest percent identity/similarity with the sequences available in GenBank.
Table 3.
Functions and features of the APEC CH2 genes induced during infection in chickens
| Gene function of in vivo-induced clone | No. of clones isolated | Orf/gene | Similar gene(s) in GenBank |
||
|---|---|---|---|---|---|
| Accession no. | % Identity | Putative gene function and/or feature | |||
| Metabolism | 23 | metH | ACD10039.1 | 93 | 5-Methyltetrahydrofolate-homocycteine methyltransferase |
| 39 | lysA | YP_854078.1 | 100 | Diaminopimelate decarboxylase | |
| 37 | pntA | EFQ02994.1 | 100 | NAD(P) transhydrogenase alpha subunit | |
| 23 | purL | EGW90584.1 | 99 | Phosphoribosylformylglycinamidine synthase | |
| 24 | serS | EGK39589.1 | 100 | Seryl-tRNA synthase | |
| 47 | tkt1 | YP_859417.1 | 100 | Transketolase | |
| 25 | ybjE | YP_851966.1 | 99 | l-Lysine exporter | |
| 25 | rutC (ycdK) | YP_540087.1 | 99 | Pyrimidine catabolism | |
| Cell envelope and integrity | 29 | wcaJ | EHN96105.1 | 97 | Putative colanic acid biosynthesis UDP-glucose lipid carrier |
| Transport systems | 30 | gspL | EFU48119.1 | 99 | Putative type II secretion protein GspL |
| 26 | sdsR | EFW75368.1 | 97 | Putative membrane fusion protein of efflux pump | |
| Virulence | 28 | irp2 | EEH86983.1 | 98 | Yersiniabactin biosynthesis protein |
| 22 | eitD | YP_444144.1 | 93 | Putative iron transport system, membrane component | |
| Phage related | 27 | EGE65170.1 | 99 | Putative minor capsid protein GPC | |
| 26 | EHN80486.1 | 99 | Putative tail component of prophage | ||
| 26 | EGU96028.1 | 100 | Putative replication protein | ||
| 33 | YP_853767.1 | 90 | Bacteriophage V tail/DNA circulation protein | ||
| 31 | ZP_08376451.1 | 99 | Host specificity protein | ||
| 29 | ZP_03357336.1 | 93 | Putative bacteriophage tail protein | ||
| Unknown function | 29 | yjiY | EGB64734.1 | 99 | Putative carbon starvation family protein |
| 30 | ylbE | ACI87286.1 | 99 | Conserved hypothetical protein | |
DISCUSSION
Bacterial pathogens such as APEC encounter unique environments inside the host during infection, so the pathogen has to respond by the coordinated expression of regulatory, metabolic, and virulence genes (30, 64). As it has been suggested that genes induced during infection can play a critical role in virulence (64, 70), in vivo experimental models that allow the recognition of bacterial genes that are expressed during infection are highly advantageous (53). Recently, studies using SCOTS and STM have been used to identify APEC host-induced genes with the goal of finding candidate colonization and disease-promoting factors (27, 55). In this study, RIVET using the Cre/lox system was developed and used for the first time to investigate APEC genes that are host induced in vivo during infection in chickens. The major advantages of RIVET over other strategies include the detection of transient gene induction in a small number of cells. RIVET also allows identification of niche-regulated genes expressed at different levels (11, 70). The disadvantages of SCOTS, such as the instability of bacterial mRNA for the construction of cDNA libraries, the low abundance of mRNA from transiently expressed genes, and the difficulty in isolation of sufficient high-quality mRNA from small populations of bacteria in vivo, do not apply to IVET and RIVET (59). In the STM approach, only limited numbers of mutants can be screened per animal model, and moreover, mutants that are slow growing or nonviable, that contain mutations in genes encoding redundant functions, or that can be complemented in a mixed population may be missed during screening. These disadvantages do not apply to RIVET (59).
In this study, four genes were identified as being involved in amino acid metabolism. They are metH, which encodes cobalamin-dependent methionine synthase (6); serS, which encodes seryl-tRNA synthase (20); lysA, which encodes diaminopimelate decarboxylase (85); and ybjE, which encodes an l-lysine exporter (67). serS and lysA were previously found to be induced in vivo in other pathogens, demonstrating the effectiveness of the system used in this study (31, 52). Moreover, lysA was found to be important for the virulence of Staphylococcus aureus in a murine model of bacteremia (63). Other in vivo approaches have shown that genes involved in amino acid metabolism in Vibrio cholerae (11), E. coli (48), and Helicobacter pylori (12, 47) were induced during mouse infection. The nucleic acid biosynthesis gene purL, which encoded 5′-phosphoribosylformylglycinamide amidotransferase, was identified. The enzyme catalyzes the conversion of 5′-phosphoribosylformylglycinamide to formylglycinamidine in the presence of glutamine and ATP for de novo purine nucleotide biosynthesis (77). The induction of the genes related to amino acid metabolism and nucleic acid biosynthesis could indicate that the requirement for these molecules is high for the survival of the bacteria in the hostile environment inside the chicken host. This was also the case for uropathogenic E. coli (UPEC), which required gluconeogenesis and the tricarboxylic acid (TCA) cycle during urinary tract infection in mice, as the peptides and amino acids were shown to be the primary carbon sources (2). The pntA gene encoding the pyridine nucleotide transhydrogenase alpha subunit, which plays a major role in the production of NADPH (17, 79), was also induced in our APEC strain, indicating the importance of the production of reducing power in the cell for energy metabolism. Previously, the pntB gene encoding the pyridine nucleotide transhydrogenase beta subunit was identified as being induced in E. coli strain i484 in vivo during mouse infection (48). In addition, the ycdK (now called rutC) gene was identified as being induced in vivo. This gene has been shown to be part of a system involved in pyrimidine catabolism and is under the control of the nitrogen starvation regulator ntrC. A rutC insertion mutant lost the ability to utilize pyrimidine nucleosides and bases as the sole source of nitrogen at room temperature (57). The in vivo induction of this gene by APEC is probably due to a nitrogen-limited environment inside the avian host and the metabolism of nucleobases, as was shown in the case of UPEC (1).
The tkt1 homolog represents a gene encoding a transketolase enzyme (42). Generally, transketolases catalyze the reversible transfer of a ketol group in the pentose phosphate pathway (45, 46), responsible for production of essential cell constituents, such as amino acids, NADPH, and several sugar phosphate intermediates (39, 93). The pathway is also believed to play a protective role during oxidative stress by the production of reducing power via NADPH (49, 72). The gene is found in one of the genomic islands in extraintestinal E. coli (ExPEC) and shares 68% and 69% identity with the tktA and tktB genes, respectively, which are also present in ExPEC and E. coli K-12 (42, 93). A recent study demonstrated that the TktA protein is involved in antibiotic and oxidative stress through interaction with MarR, the repressor of the operon marRAB, conferring resistance to multiple antibiotic and oxidative stresses on E. coli (26). This, together with the production of reducing power for energy metabolism, could be the reason why a tkt1 homolog was induced in vivo in the chicken host for survival and colonization of APEC. In previous studies, it was shown that the tktB gene was upregulated when APEC was grown in chicken serum (56). Moreover, APEC could not survive inside the chicken when the tktA gene was knocked out, showing the importance of transketolase enzymes in APEC (55).
In addition, another gene that is involved in stress response after encountering the host environment was identified. The gene, known as sdsR (yjcR), is found in the operon sdsRQP encoding a putative multidrug efflux pump involved in sensitivity control against sulfur drugs (82). The gene is under the control of the LeuO protein, a global regulator controlling a number of genes, including stress response genes and virulence-related genes. Due to stresses, such as upon entry into stationary phase, increased expression of LeuO was observed in several E. coli strains, which also activated the operon sdsRQP (82).
The only gene identified as being associated with the cell envelope and integrity is wcaJ. The gene encodes a putative UDP glucose lipid carrier transferase in the biosynthesis of colanic acid (CA). CA is one of the exopolysaccharides (EPS) that are produced by E. coli, forming a thick mucoid matrix on the cell surface (84). It was demonstrated that a CA-deficient mutant of E. coli O157:H7 was less tolerant of acid, heat, osmotic, and oxidative stresses (15, 62). Also, an APEC wcaE mutant was unable to survive inside chickens after infection (55). As wcaJ was identified in this APEC strain as in vivo induced, this suggests that CA is produced to confer protection against host environmental stresses, such as the body temperature of chickens (42°C) and the acid environment in macrophages, where it was demonstrated that APEC O1:K1:H7 could survive intracellularly (73). Moreover, using an STM approach, an APEC wcaE mutant was shown to be attenuated in vivo (55), further confirming the importance of CA for APEC survival in vivo.
The gspL gene encodes one of the proteins of the putative type II secretion system (T2SS) (GspL-like protein). In the sequenced APEC O1 chromosome, it is located in pathogenicity island I (PAI IAPEC O1) (42). The T2SS, also known as the general secretory pathway (GSP), is one of the systems used by most Gram-negative bacteria to transport folded proteins, including toxins, proteases, cellulases, and lipases, from the periplasm to the extracellular medium through the outer membrane channel (44). Induction of this gene indicates that the operon coding for the T2SS could be active and induced in the chicken host. APEC might use the T2SS to secrete proteins responsible for pathogenesis or its fitness inside the chicken host. In UPEC, it was actually shown by mutant analysis that the T2SS is important for persistence in the urinary tract, particularly in renal tissues (51). Also, it was shown that this system is responsible for the translocation of a variety of proteins that mediate pathogenic effects, including the pore-forming toxin aerolysin of Aeromonas hydrophila (37), the cholera toxin of V. cholerae (78), and the heat-labile (LT) toxin of enterotoxigenic E. coli (ETEC) (87). Recently, transcriptome analysis of APEC O1 indicated that the T2SS was significantly downregulated when the bacteria were grown in chicken serum compared to growth in LB. This contrasting finding could be due to the fact that the analysis was limited to serum (56) or due to the heterogeneity of the APEC strains used in the different studies. Furthermore, it might be due to the fact that gspL is only transiently induced in chickens. Our RIVET screen could have detected the induction of gspL in vivo, as it can detect even transient gene induction leaving irreversible phenotypic change to the bacteria inside the host.
The irp2 gene encodes high-molecular-weight protein 2 (HMWP2), which is involved in yersiniabactin (siderophore) biosynthesis and is located in the Yersinia high-pathogenicity island (HPI) in APEC O1 (32, 42). The HPI was shown to be involved in the virulence of Yersinia pestis, Yersinia enterocolitica, and Yersinia pseudotuberculosis and APEC O1 (56, 71). The available free iron in host tissue is, significantly, in low concentrations, forcing APEC to acquire iron from the host. The induction of this gene in vivo indicates that iron acquisition systems are required for APEC survival in the host. It was also demonstrated in a recent study on APEC O1 that genes involved in iron uptake were among the significantly upregulated genes (56). It was hypothesized that the increased virulence of HPI-positive Enterobacteriaceae, including APEC, is caused by yersiniabactin, which not only provides iron to the bacterium, which is normally bound to iron-binding proteins produced by hosts, but also prevents reactive oxygen species (ROS) production by innate immune cells, thus promoting the survival of the bacteria in the host (71). Another in vivo-induced gene in connection with iron sequestration systems found in this study is eitD. This gene is located in the eitABCD operon encoding putative iron transport systems identified first on a ColV plasmid of an APEC O2 strain (43), then on a ColBM plasmid of an APEC O1 strain (41), and lately on pAPEC-2 of APEC_7122 (65).
Six phage-related genes were also identified in this study as in vivo induced (Table 3). APEC is believed to carry prophages in its genome (42), which under stress conditions, such as H2O2 and the reactive oxygen species generated and released by leukocytes, may trigger the induction of these prophages inside the host (92). Previously, a study utilizing a SCOTS approach demonstrated that APEC bacteriophage-related genes were expressed in vivo (27). Also, in another study with Streptococcus mitis, it was demonstrated that adhesion of the pathogen to the platelets was phage mediated, where genes for two surface proteins responsible for adhesion (PblA and PblB) resembled phage capsid and tail fiber genes and were surrounded by other phage-like gene sequences (8). This indicates that bacteriophage-related genes coding for virulence properties, such as toxins and adhesins, may also be induced during APEC infection, although it is not yet determined whether these phage-encoded genes are associated with the virulence of the APEC CH2 strain.
Finally, genes with unknown functions were also identified in this study. Two genes, ylbE, encoding a conserved hypothetical protein, and yjiY, which is a paralog of cstA, encoding a peptide tranporter induced during carbon starvation (8, 83), were induced. The fact that these genes are induced in vivo is an interesting observation in the further analysis of their functions.
In conclusion, the fact that we could identify host-induced genes that were previously identified in other studies validates the RIVET approach. However, there appears to be little overlap between the APEC genes identified in the STM study (55) and by SCOTS (27) and those identified by this RIVET screen. This comparison further demonstrates that STM, SCOTS, and RIVET are complementary, and not redundant, functional-genomics screens. This study provides a highlight on the APEC genes that are induced inside the chicken host during infection and that are likely to be involved in the survival and successful colonization of the pathogen in the stressful host environment. To our knowledge, this is the first report to use RIVET in this pathogen. These genes may be required for APEC pathogenesis and could be potential vaccine candidates (16). However, further experiments, such as mutational analysis, should confirm the requirement for these genes by the pathogen during infection in the chicken. Other techniques, such as quantitative real-time PCR or microarray analyses, will be required to confirm whether these conserved E. coli genes are actually expressed at high levels in vivo in infected tissues or during growth in vitro.
ACKNOWLEDGMENTS
We thank F. Stewart (Technische Universität Dresden, Dresden, Germany) for the provision of plasmids pR6K-rpsL-Neo and pSC101-BAD-Cre-tet and T. D. Ho for the provision of plasmid pKD46. Rob Lavigne and Ellen Ons are acknowledged for their valuable discussions. We thank Tom Luyten and Marcel Samian for their technical assistance.
H.N.T. is the recipient of IRO scholarships from Katholieke Universiteit Leuven, Leuven, Belgium.
Footnotes
Published ahead of print 17 February 2012
REFERENCES
- 1. Alteri CJ, Mobley HL. 2012. Escherichia coli physiology and metabolism dictates adaptation to diverse host microenvironments. Curr. Opin. Microbiol. 15:3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alteri CJ, Smith SN, Mobley HL. 2009. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 5:e1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anastassiadis K, et al. 2009. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis. Model Mech. 2:508–515 [DOI] [PubMed] [Google Scholar]
- 4. Antão E, et al. 2009. The chicken as a natural model for extraintestinal infections caused by avian pathogenic Escherichia coli (APEC). Microb. Pathog. 45:361–369 [DOI] [PubMed] [Google Scholar]
- 5. Bachmann H, Kleerebezem M, van Hylckama Vlieg J. 2008. High-throughput identification and validation of in situ-expressed genes of Lactococcus lactis. Appl. Environ. Microbiol. 74:4727–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Banerjee RV, Johnston NL, Sobeski JK, Datta P, Matthews RG. 1989. Cloning and sequence analysis of the Escherichia coli metH gene encoding cobalamin-dependent methionine synthase and isolation of a tryptic fragment containing the cobalamin-binding domain. J. Biol. Chem. 264:13888–13895 [PubMed] [Google Scholar]
- 7. Barnes HJ, Vaillancourt J-P, Gross WB. 2003. Colibacillosis, p 631–652 In Saif YM, et al. (ed), Diseases of poultry, 11th ed Iowa State Press, Ames, IA [Google Scholar]
- 8. Bensing BA, Rubens CE, Sullam PM. 2001. Genetic loci of Streptococcus mitis that mediate binding to human platelets. Infect. Immun. 69:1373–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bron P, Grangette C, Mercenier A, de Vos W, Kleerebezem M. 2004. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 186:5721–5729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Camilli A, Beattie D, Mekalanos J. 1994. Use of genetic recombination as a reporter of gene expression. Proc. Natl. Acad. Sci. U. S. A. 91:2634–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Camilli A, Mekalanos J. 1995. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol. Microbiol. 18:671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Castillo A, Woodruff A, Connolly L, Sause W, Ottemann K. 2008. Recombination-based in vivo expression technology identifies Helicobacter pylori genes important for host colonization. Infect. Immun. 76:5632–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Caza M, Lépine F, Dozois CM. 2011. Secretion, but not overall synthesis, of catecholate siderophores contributes to virulence of extraintestinal pathogenic Escherichia coli. Mol. Microbiol. 80:266–282 [DOI] [PubMed] [Google Scholar]
- 14. Caza M, Lépine F, Milot S, Dozois C. 2008. Specific roles of the iroBCDEN genes in virulence of an avian pathogenic Escherichia coli O78 strain and in production of salmochelins. Infect. Immun. 76:3539–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen J, Lee SM, Mao Y. 2004. Protective effect of exopolysaccharide colanic acid of Escherichia coli O157:H7 to osmotic and oxidative stress. Int. J. Food Microbiol. 93:281–286 [DOI] [PubMed] [Google Scholar]
- 16. Chiang S, Mekalanos J, Holden D. 1999. In vivo genetic analysis of bacterial virulence. Annu. Rev. Microbiol. 53:129–154 [DOI] [PubMed] [Google Scholar]
- 17. Clarke DM, Loo TW, Gillam S, Bragg PD. 1986. Nucleotide sequence of the pntA and pntB genes encoding the pyridine nucleotide transhydrogenase of Escherichia coli. Eur. J. Biochem. 158:647–653 [DOI] [PubMed] [Google Scholar]
- 18. Clarke L, Carbon J. 1976. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell 9:91–99 [DOI] [PubMed] [Google Scholar]
- 19. Curtis N, et al. 1988. Iron-regulated outer membrane proteins of Escherichia coli K-12 and mechanism of action of catechol-substituted cephalosporins. Antimicrob. Agents Chemother. 32:1879–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cusack S, Berthet-Colominas C, Härtlein M, Nassar N, Leberman R. 1990. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature 347:249–255 [DOI] [PubMed] [Google Scholar]
- 21. Dai J, et al. 2010. Suppression subtractive hybridization identifies an autotransporter adhesin gene of E. coli IMT5155 specifically associated with avian pathogenic Escherichia coli (APEC). BMC Microbiol. 10:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Datsenko K, Wanner B. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Delicato E, de Brito B, Gaziri L, Vidotto M. 2003. Virulence-associated genes in Escherichia coli isolates from poultry with colibacillosis. Vet. Microbiol. 94:97–103 [DOI] [PubMed] [Google Scholar]
- 24. de Pace F, et al. 2010. The type VI secretion system plays a role in type 1 fimbria expression and pathogenesis of an avian pathogenic Escherichia coli strain. Infect. Immun. 78:4990–4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dho-Moulin M, Fairbrother M. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30:299–316 [PubMed] [Google Scholar]
- 26. Domain F, Bina XR, Levy SB. 2007. Transketolase A, an enzyme in central metabolism, derepresses the marRAB multiple antibiotic resistance operon of Escherichia coli by interaction with MarR. Mol. Microbiol. 66:383–394 [DOI] [PubMed] [Google Scholar]
- 27. Dozois C, Daigle F, Curtiss R. 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. U. S. A. 100:247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dozois C, et al. 2000. Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect. Immun. 68:4145–4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dziva F, Stevens M. 2008. Colibacillosis in poultry: unravelling the molecular basis of virulence of avian pathogenic Escherichia coli in their natural hosts. Avian Pathol. 37:355–366 [DOI] [PubMed] [Google Scholar]
- 30. Finlay BB, Falkow S. 1989. Common themes in microbial pathogenicity. Microbiol. Rev. 53:210–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Van Immerseel F. 2008. Salmonella enterica serovar Enteritidis genes induced during oviduct colonization and egg contamination in laying hens. Appl. Environ. Microbiol. 74:6616–6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guilvout I, Mercereau-Puijalon O, Bonnefoy S, Pugsley AP, Carniel E. 1993. High-molecular-weight protein 2 of Yersinia enterocolitica is homologous to AngR of Vibrio anguillarum and belongs to a family of proteins involved in nonribosomal peptide synthesis. J. Bacteriol. 175:5488–5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hanin A, et al. 2010. Screening of in vivo activated genes in Enterococcus faecalis during insect and mouse infections and growth in urine. PLoS One 5:e11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heermann R, Zeppenfeld T, Jung K. 2008. Simple generation of site-directed point mutations in the Escherichia coli chromosome using Red(R)/ET(R) recombination. Microb. Cell Fact. 7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hensel M, et al. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400–403 [DOI] [PubMed] [Google Scholar]
- 36. Holden KM, Browning GF, Noormohammadi AH, Markham PF, Marenda MS. 2012. TonB is essential for virulence in avian pathogenic Escherichia coli. Comp. Immunol. Microbiol. Infect. Dis. 35:129–138 [DOI] [PubMed] [Google Scholar]
- 37. Howard SP, Buckley JT. 1985. Protein export by a gram-negative bacterium: production of aerolysin by Aeromonas hydrophila. J. Bacteriol. 161:1118–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang Y, Leming C, Suyemoto M, Altier C. 2007. Genome-wide screen of Salmonella genes expressed during infection in pigs, using in vivo expression technology. Appl. Environ. Microbiol. 73:7522–7530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Iida A, Teshiba S, Mizobuchi K. 1993. Identification and characterization of the tktB gene encoding a second transketolase in Escherichia coli K-12. J. Bacteriol. 175:5375–5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Inouye S, Inouye M. 1985. Up-promoter mutations in the lpp gene of Escherichia coli. Nucleic Acids Res. 13:3101–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johnson T, Johnson S, Nolan L. 2006. Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids. J. Bacteriol. 188:5975–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Johnson T, et al. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 189:3228–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Johnson T, Siek K, Johnson S, Nolan L. 2006. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J. Bacteriol. 188:745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Johnson TL, Abendroth J, Hol WG, Sandkvist M. 2006. Type II secretion: from structure to function. FEMS Microbiol. Lett. 255:175–186 [DOI] [PubMed] [Google Scholar]
- 45. Josephson BL, Fraenkel DG. 1974. Sugar metabolism in transketolase mutants of Escherichia coli. J. Bacteriol. 118:1082–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Josephson BL, Fraenkel DG. 1969. Transketolase mutants of Escherichia coli. J. Bacteriol. 100:1289–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kavermann H, et al. 2003. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 197:813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Khan M, Isaacson R. 2002. Identification of Escherichia coli genes that are specifically expressed in a murine model of septicemic infection. Infect. Immun. 70:3404–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kletzien RF, Harris PK, Foellmi LA. 1994. Glucose-6-phosphate dehydrogenase: a “housekeeping” enzyme subject to tissue-specific regulation by hormones, nutrients, and oxidant stress. FASEB J. 8:174–181 [DOI] [PubMed] [Google Scholar]
- 50. Kobayashi RK, Gaziri LC, Vidotto MC. 2010. Functional activities of the Tsh protein from avian pathogenic Escherichia coli (APEC) strains. J. Vet. Sci. 11:315–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kulkarni R, et al. 2009. Roles of putative type II secretion and type IV pilus systems in the virulence of uropathogenic Escherichia coli. PLoS One 4:e4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lai YC, Peng HL, Chang HY. 2001. Identification of genes induced in vivo during Klebsiella pneumoniae CG43 infection. Infect. Immun. 69:7140–7145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee K, et al. 2007. IVET-based identification of virulence factors in Vibrio vulnificus MO6-24/O. J. Microbiol. Biotechnol. 17:234–243 [PubMed] [Google Scholar]
- 54. Li G, et al. 2010. AatA is a novel autotransporter and virulence factor of avian pathogenic Escherichia coli. Infect. Immun. 78:898–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li G, Laturnus C, Ewers C, Wieler LH. 2005. Identification of genes required for avian Escherichia coli septicemia by signature-tagged mutagenesis. Infect. Immun. 73:2818–2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li G, et al. 2011. Transcriptome analysis of avian pathogenic Escherichia coli O1 in chicken serum reveals adaptive responses to systemic infection. Infect. Immun. 79:1951–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Loh KD, et al. 2006. A previously undescribed pathway for pyrimidine catabolism. Proc. Natl. Acad. Sci. U. S. A. 103:5114–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lowe A, Beattie D, Deresiewicz R. 1998. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol. Microbiol. 27:967–976 [DOI] [PubMed] [Google Scholar]
- 59. Mahan M, Heithoff D, Sinsheimer R, Low D. 2000. Assessment of bacterial pathogenesis by analysis of gene expression in the host. Annu. Rev. Genet. 34:139–164 [DOI] [PubMed] [Google Scholar]
- 60. Mahan M, Slauch J, Mekalanos J. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686–688 [DOI] [PubMed] [Google Scholar]
- 61. Mahan M, et al. 1995. Antibiotic-based selection for bacterial genes that are specifically induced during infection of a host. Proc. Natl. Acad. Sci. U. S. A. 92:669–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mao Y, Doyle MP, Chen J. 2001. Insertion mutagenesis of wca reduces acid and heat tolerance of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:3811–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mei JM, Nourbakhsh F, Ford CW, Holden DW. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399–407 [DOI] [PubMed] [Google Scholar]
- 64. Mekalanos J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mellata M, Ameiss K, Mo H, Curtiss R. 2010. Characterization of the contribution to virulence of three large plasmids of avian pathogenic Escherichia coli chi7122 (O78:K80:H9). Infect. Immun. 78:1528–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mellata M, et al. 2003. Role of avian pathogenic Escherichia coli virulence factors in bacterial interaction with chicken heterophils and macrophages. Infect. Immun. 71:494–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nagai Y, Ito H, Yasueda H. 2010. Amino acid production: L-lysine, p 1–10 In Flickinger MC. (ed), Encyclopedia of industrial biotechnology: bioprocess, bioseparation, and cell technology, vol 7 John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 68. Newman D, Shapiro J. 1999. Differential fiu-lacZ fusion regulation linked to Escherichia coli colony development. Mol. Microbiol. 33:18–32 [DOI] [PubMed] [Google Scholar]
- 69. Ons E, Bleyen N, Tuntufye H, Vandemaele F, Goddeeris B. 2007. High prevalence iron receptor genes of avian pathogenic Escherichia coli. Avian Pathol. 36:411–414 [DOI] [PubMed] [Google Scholar]
- 70. Osorio C, et al. 2005. Second-generation recombination-based in vivo expression technology for large-scale screening for Vibrio cholerae genes induced during infection of the mouse small intestine. Infect. Immun. 73:972–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Paauw A, Leverstein-van Hall MA, van Kessel KP, Verhoef J, Fluit AC. 2009. Yersiniabactin reduces the respiratory oxidative stress response of innate immune cells. PLoS One 4:e8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pandolfi PP, et al. 1995. Targeted disruption of the housekeeping gene encoding glucose 6-phosphate dehydrogenase (G6PD): G6PD is dispensable for pentose synthesis but essential for defense against oxidative stress. EMBO J. 14:5209–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pourbakhsh SA, et al. 1997. Dynamics of Escherichia coil infection in experimentally inoculated chickens. Avian Dis. 41:221–233 [PubMed] [Google Scholar]
- 74. Rediers H, Rainey P, Vanderleyden J, De Mot R. 2005. Unraveling the secret lives of bacteria: use of in vivo expression technology and differential fluorescence induction promoter traps as tools for exploring niche-specific gene expression. Microbiol. Mol. Biol. Rev. 69:217–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sabri M, et al. 2008. Contribution of the SitABCD, MntH, and FeoB metal transporters to the virulence of avian pathogenic Escherichia coli O78 strain chi7122. Infect. Immun. 76:601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 77. Sampei G, Mizobuchi K. 1989. The organization of the purL gene encoding 5′-phosphoribosylformylglycinamide amidotransferase of Escherichia coli. J. Biol. Chem. 264:21230–21238 [PubMed] [Google Scholar]
- 78. Sandkvist M, Morales V, Bagdasarian M. 1993. A protein required for secretion of cholera toxin through the outer membrane of Vibrio cholerae. Gene 123:81–86 [DOI] [PubMed] [Google Scholar]
- 79. Sauer U, Canonaco F, Heri S, Perrenoud A, Fischer E. 2004. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 279:6613–6619 [DOI] [PubMed] [Google Scholar]
- 80. Saviola B, Woolwine S, Bishai W. 2003. Isolation of acid-inducible genes of Mycobacterium tuberculosis with the use of recombinase-based in vivo expression technology. Infect. Immun. 71:1379–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schouler C, Koffmann F, Amory C, Leroy-Sétrin S, Moulin-Schouleur M. 2004. Genomic subtraction for the identification of putative new virulence factors of an avian pathogenic Escherichia coli strain of O2 serogroup. Microbiology 150:2973–2984 [DOI] [PubMed] [Google Scholar]
- 82. Shimada T, Yamamoto K, Ishihama A. 2009. Involvement of the leucine response transcription factor LeuO in regulation of the genes for sulfa drug efflux. J. Bacteriol. 191:4562–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Skyberg J, et al. 2006. Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E. coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect. Immun. 74:6287–6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Stevenson G, Andrianopoulos K, Hobbs M, Reeves P. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Stragier P, Danos O, Patte JC. 1983. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. II. Nucleotide sequence of the lysA gene and its regulatory region. J. Mol. Biol. 168:321–331 [DOI] [PubMed] [Google Scholar]
- 86. Tamir-Ariel D, Navon N, Burdman S. 2007. Identification of genes in Xanthomonas campestris pv. vesicatoria induced during its interaction with tomato. J. Bacteriol. 189:6359–6371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tauschek M, Gorrell RJ, Strugnell RA, Robins-Browne RM. 2002. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:7066–7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tuntufye HN, Goddeeris BM. 2011. Use of lambda Red-mediated recombineering and Cre/lox for generation of markerless chromosomal deletions in avian pathogenic Escherichia coli. FEMS Microbiol. Lett. 325:140–147 [DOI] [PubMed] [Google Scholar]
- 89. Valdivia R, Falkow S. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007–2011 [DOI] [PubMed] [Google Scholar]
- 90. Vandemaele F, et al. 2005. Immunization with the binding domain of FimH, the adhesin of type 1 fimbriae, does not protect chickens against avian pathogenic Escherichia coli. Avian Pathol. 34:264–272 [DOI] [PubMed] [Google Scholar]
- 91. Veal-Carr W, Stibitz S. 2005. Demonstration of differential virulence gene promoter activation in vivo in Bordetella pertussis using RIVET. Mol. Microbiol. 55:788–798 [DOI] [PubMed] [Google Scholar]
- 92. Wagner PL, Waldor MK. 2002. Bacteriophage control of bacterial virulence. Infect. Immun. 70:3985–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhao G, Winkler ME. 1994. An Escherichia coli K-12 tktA tktB mutant deficient in transketolase activity requires pyridoxine (vitamin B6) as well as the aromatic amino acids and vitamins for growth. J. Bacteriol. 176:6134–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]