Abstract
Advenella mimigardefordensis strain DPN7T was genetically modified to produce poly(3-mercaptopropionic acid) (PMP) homopolymer by exploiting the recently unraveled process of 3,3′-dithiodipropionic acid (DTDP) catabolism. Production was achieved by systematically engineering the metabolism of this strain as follows: (i) deletion of its inherent 3MP dioxygenase-encoding gene (mdo), (ii) introduction of the buk-ptb operon (genes encoding the butyrate kinase, Buk, and the phosphotransbutyrylase, Ptb, from Clostridium acetobutylicum), and (iii) overexpression of its own polyhydroxyalkanoate synthase (phaCAm). These measures yielded the potent PMP production strain A. mimigardefordensis strain SHX22. The deletion of mdo was required for adequate synthesis of PMP due to the resulting accumulation of 3MP during utilization of DTDP. Overexpression of the plasmid-borne buk-ptb operon caused a severe growth repression. This effect was overcome by inserting this operon into the genome. Polyhydroxyalkanoate (PHA) synthases from different origins were compared. The native PHA synthase of A. mimigardefordensis (phaCAm) was obviously the best choice to establish homopolythioester production in this strain. In addition, the cultivation conditions, including an appropriate provision of the carbon source, were further optimized to enhance PMP production. The engineered strain accumulated PMP up to approximately 25% (wt/wt) of the cell dry weight when cultivated in mineral salts medium containing glycerol as the carbon source in addition to DTDP as the sulfur-providing precursor. According to our knowledge, this is the first report of PMP homopolymer production by a metabolically engineered bacterium using DTDP, which is nontoxic, as the precursor substrate.
INTRODUCTION
Polythioesters (PTE) are sulfur-containing polymers, which in comparison to the corresponding polyoxoesters (see Fig. S1 in the supplemental material) exhibit interesting features such as higher melting points, lower solubility, and increased heat stability (20, 21, 32). One further remarkable difference between polyhydroxyalkanoates (PHA) and PTE regards their biodegradability (47). Whereas PHA are degraded into simple organic compounds by many microorganisms, including bacteria and fungi, which are present in all natural environments (27), no organisms or habitats in which degradation of PTE takes place are known (22). PTE are therefore considered nonbiodegradable. The biological persistence of poly(3-mercaptopropionic acid) (PMP) may be a valuable property for certain applications. Considering the actual and prospective development of public opinion and environmental consciousness, industry may search for persistent materials which are biotechnically produced from renewable resources (47).
Chemical synthesis of PTE was first described in 1951 (35, 42). However, due to its difficult preparation, the requirement of toxic reagents, low yields, and high costs, it was never produced on a large scale and commercialized. The first report of PTE production by a biotechnical process attracted much attention (30). Up to now, two strains have been used successfully for PTE production on the laboratory scale.
The first strain which synthesized PTE at all was Ralstonia eutropha H16; it carried out PTE synthesis by utilizing its inherent metabolism (30). Different organic sulfur compounds (OSC) such as 3-mercaptopropionic acid (3MP), 3,3′-thiodipropionic acid (TDP), 3,3′-dithiodipropionic acid (DTDP), 3-mercaptobutyric acid (3MB), and 3-mercaptovaleric acid (3MV) were used as precursors in R. eutropha H16 for PTE production. Depending on the organic sulfur compound used, poly(3HB-co-3MP), poly(3HB-co-3MB), or poly(3HB-co-3MV) was produced. Since the natural poly(3-hydroxybutyrate) (PHB) biosynthesis pathway could not be suppressed (28), production of PTE homopolymer is unlikely to be achieved in R. eutropha H16.
PTE homopolythioesters were first produced by an engineered recombinant Escherichia coli strain which harbors genes encoding enzymes for the nonnatural BPEC pathway (31). The BPEC pathway contains the genes encoding butyrate kinase (Buk) and phosphotransbutyrylase (Ptb) from Clostridium acetobutylicum in addition to the PHA synthase (PhaEC) from Thiocapsa pfennigii (29). Several different PTE homopolymers, such as PMP, poly(3MB), and poly(3MV), were accumulated in the cells of this recombinant strain when the respective precursor substrates were applied. The bioprocess was also optimized at the pilot scale by changing the medium composition (50). Cells with a PMP content of more than 40% (wt/wt [CDW]) were obtained in this optimized process.
Because it is still not possible to produce PTE de novo from sulfate and simple carbon sources, which are structurally not related to the constituent mercaptoalkanoic acids, the choice of an appropriate precursor remained a key factor in the process of PTE production. So far, only 3MP, 3MB, and 3MV could be used for the more valuable PTE homopolymer production in recombinant E. coli strains containing the BPEC pathway. However, all three precursors possessing sulfhydryl groups are unstable, expensive reactive, malodorous, or commercially unavailable, and are toxic to cells. Growth of R. eutropha is for example already severely inhibited by only 1 g/liter of 3-mercaptopropionic acid in the medium (33). In contrast, the disulfide DTDP is more stable, cheaper, chemically inert, and less toxic. Unfortunately, E. coli cannot utilize DTDP for growth or PTE biosynthesis. Otherwise, E. coli and R. eutropha grow unsuppressed with other carbon sources even in the presence of 10 g/liter of DTDP in the medium (33). For these reasons, DTDP is considered to be an ideal alternative substrate for PMP homopolymer production. The establishment of PMP production based on these nontoxic and more stable PTE precursors is promising for large-scale applications.
Consequently, Advenella mimigardefordensis DPN7T, which has the capacity to utilize DTDP as the sole carbon source, was previously isolated from mature compost in a waste management facility (15, 54). Meanwhile, this bacterium serves our laboratory as a model organism to study the metabolism of DTDP and related organic sulfur compounds. Transposon mutagenesis was applied to unravel its DTDP degradation pathway and the relevant genes involved (53). DTDP is first cleaved into two molecules of 3MP by a dihydrolipoamide dehydrogenase (LpdA) (55), and the 3MP is further catalyzed to 3-sulfinopropionic acid (3SP) by a 3MP dioxygenase (Mdo) (4). 3SP is then covalently linked to coenzyme A (CoA) to form 3SP-CoA by a succinyl-CoA synthetase (SucCD) (45). This intermediate is then most probably converted to propionyl-CoA by an acyl-CoA dehydrogenase (CaiA) (M. Schürmann, A. Deters, J. H. Wübbeler, and A. Steinbüchel, unpublished data), and further metabolized via the methylcitric acid cycle (53) (Fig. 1A). In this study, a new recombinant homo-PMP production pathway was engineered, optimized, and applied in A. mimigardefordensis by using recently acquired knowledge about DTDP catabolism.
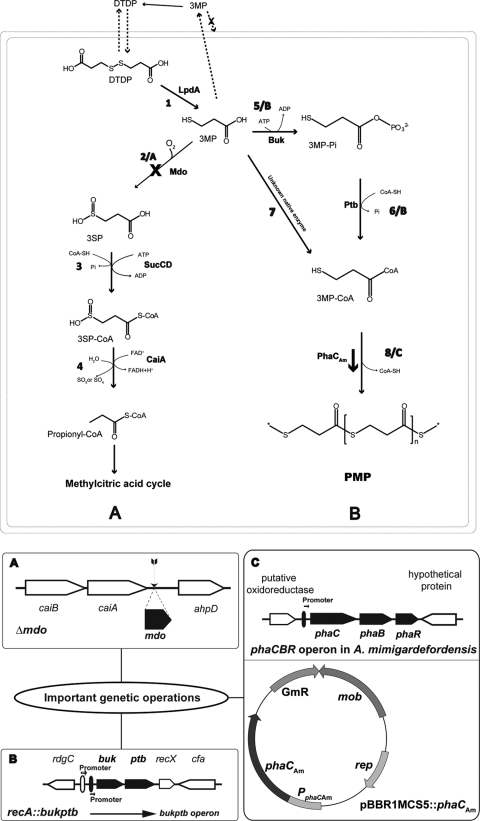
Fig 1.
Predicted DTDP degradation and PMP production pathway. (A) Inherent DTDP degradation pathway of A. mimigardefordensis. (B) DTDP-to-PMP conversion pathway in the engineered strain. The double box indicates the A. mimigardefordensis cell. The deletion of mdo is indicated with a bold X. Dashed arrows across the periplast indicate transmembrane transport of relevant molecules. The bold arrow indicates the overexpression of phaCAm in plasmid pBBR1MCS5:: phaCAm. All reactions catalyzed by enzymes are numbered. Bold A, B, and C indicate three important manipulations for improved PMP production. The detailed genetic operations of these three steps are given at the bottom. The buk-ptb operon originates from C. acetobutylicum and is regulated under its native promoter. Other genes in this figure originate from A. mimigardefordensis DPN7 and are regulated under their own promoters. 3MP, 3-mercaptopropionic acid; DTDP, 3,3′-dithiodipropionic acid; 3SP, 3-sulfinopropionic acid; lpdA, disulfide reductase; mdo, thiol dioxygenase; sucCD, succinyl-CoA synthetase; caiA, acyl-CoA dehydrogenase; buk, butyrate kinase; ptb, phosphotransbutyrylase; phaCAm, poly(hydroxyalkanoic acid) synthase; caiB, acyl-CoA transferase; ahpD, alkylhydroperoxidase; rdgC, recombination-associated protein; recA, recombination protein; recX, recombination regulator protein; cfa, cyclopropane-fatty-acyl-phospholipid synthase; phaB, acetoacetyl-CoA reductase; phaR, polyhydroxyalkanoate synthesis repressor protein; PphaCAm, native promoter of phaCAm.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotides used in this study.
Bacterial strains with their relevant characteristics and sources as well as a complete description of the plasmids are listed in Table 1. Oligonucleotide sequences and their applications are presented in Table S1 in the supplemental material.
Table 1.
Strains and plasmids
| Strains and plasmids | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| S17-1 | thi-1 proA hsdR17(rK− mK+) recA1; tra genes of plasmid RP4 integrated into the genome | 46 |
| XL10 Gold | endA1 glnV44 recA1 thi-1 gyrA96 relA1 lac Hte Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 tetR F′ [proAB lacIqZΔM15 Tn10(Tetr Amy Cmr)] | Stratagene |
| A. mimigardefordensis | ||
| DPN7 | Wild-type strain | 54 |
| M1 | DPN7 mutant (Δmdo) | This study |
| M2 | DPN7 mutant (ΔrecA) | This study |
| M3 | DPN7 mutant (Δmdo ΔrecA) | This study |
| M4 | DPN7 mutant (Δmdo recA::buk-ptb) | This study |
| M5 | DPN7 mutant (recA::buk-ptb) | This study |
| M6 | DPN7 mutant (Δmdo recA::buk-ptb ΔphaCAm) | This study |
| M7 | DPN7 mutant (Δmdo ΔrecA ΔphaCAm) | This study |
| SHX1 | DPN7 harboring pBBR1MCS5::BPEC | This study |
| SHX2 | DPN7 harboring pBBR1MCS5 | This study |
| SHX3 | DPN7 harboring pBBR1MCS5::phaEC | This study |
| SHX4 | DPN7 harboring pBBR1MCS5::buk-ptb | This study |
| SHX5 | DPN7 mutant (Δmdo recA::buk-ptb) harboring pBBR1MCS5::phaEC | This study |
| SHX6 | DPN7 mutant (Δmdo recA::buk-ptb) harboring pBBR1MCS5 | This study |
| SHX7 | DPN7 mutant (Δmdo) harboring pBBR1MCS5::BPEC | This study |
| SHX8 | DPN7 mutant (Δmdo) harboring pBBR1MCS5 | This study |
| SHX9 | DPN7 mutant (recA::buk-ptb) harboring pBBR1MCS5::phaEC | This study |
| SHX10 | DPN7 mutant (recA::buk-ptb) harboring pBBR1MCS5 | This study |
| SHX11 | DPN7 mutant (ΔrecA) harboring pBBR1MCS5::BPEC | This study |
| SHX12 | DPN7 mutant (ΔrecA) harboring pBBR1MCS5 | This study |
| SHX13 | DPN7 mutant (Δmdo ΔrecA) harboring pBBR1MCS5 | This study |
| SHX14 | DPN7 mutant (Δmdo ΔrecA ΔphaCAm) harboring pBBR1MCS5 | This study |
| SHX15 | DPN7 mutant (Δmdo recA::buk-ptb ΔphaCAm) harboring pBBR1MCS5 | This study |
| SHX16 | DPN7 mutant (Δmdo recA::buk-ptb ΔphaCAm) harboring pBBR1MCS5::phaEC_colinear | This study |
| SHX17 | DPN7 mutant (Δmdo recA::buk-ptb ΔphaCAm) harboring pBBR1MCS5::phaEC_antilinear | This study |
| SHX18 | DPN7 mutant (Δmdo recA::buk-ptb ΔphaCAm) harboring pBBR1MCS5::phaCAm_colinear | This study |
| SHX19 | DPN7 mutant (Δmdo recA::buk-ptb ΔphaCAm) harboring pBBR1MCS5::phaCAm_antilinear | This study |
| SHX20 | DPN7 mutant (Δmdo recA::buk-ptb ΔphaCAm) harboring pBBR1MCS5::phaC1Re_colinear | This study |
| SHX21 | DPN7 mutant (Δmdo recA::buk-ptb ΔphaCAm) harboring pBBR1MCS5::phaC1Re_antilinear | This study |
| SHX22 | DPN7 mutant (Δmdo recA::buk-ptb) harboring pBBR1MCS5::phaCAm_colinear | This study |
| Plasmids | ||
| pBBR1MCS5 | Broad-host-range vector, 4.9 kb, Gmr with lac promoter in front of MCS for white/blue selection | 23 |
| pBPP1 | Harbors the whole BPEC pathway genes | 29 |
| pBBR1MCS-5::phaEC_colinear | Harbors the operon of phaEC from Thiocapsa pfennigii colinear to lac promoter (Plac) at the locus of EcoRI site | This study |
| pBBR1MCS-5::buk-ptb | Harbors buk-ptb from Clostridium acetobutylicum colinear to Plac at the locus of BamHI site | This study |
| pBBR1MCS-5::BPEC | Harbors the phaEC operon at the locus of EcoRI site and the operon of buk-ptb at the locus of BamHI site together colinear to Plac | This study |
| pBBR1MCS-5::phaEC_antilinear | Harbors the operon of phaEC from Thiocapsa pfennigii antilinear to Plac at the locus of EcoRI site | This study |
| pBBR1MCS5::phaCAm_colinear | Harbors the operon of phaCAm from A. mimigardfordensis DPN7 colinear to Plac at the locus of HindIII site | This study |
| pBBR1MCS5::phaCAm_antilinear | Harbors the operon of phaCAm from A. mimigardfordensis DPN7 antilinear to Plac at the locus of HindIII site | This study |
| pBBR1MCS5::phaC1Re_colinear | Harbors the operon of phaC1 (phaC1Re) gene from Ralstonia eutropha H16 colinear to Plac at the locus of BamHI site | This study |
| pBBR1MCS5::phaC1Re_antilinear | Harbors the operon of phaC1 (phaC1Re) gene from Ralstonia eutropha H16 antilinear to Plac at the locus of BamHI site | This study |
| pJQ200mp18 | Gmr, suicide vector for gene deletion | 38 |
| pJQ200mp18Tc | Tcr, suicide vector for gene deletion | 37 |
| pJQ200mp18Tc::Δmdo | Tcr, suicide vector for mdo gene deletion | This study |
| pJQ200mp18::ΔrecA | Gmr, suicide vector for recA gene deletion | This study |
| pJQ200mp18::ΔrecA::Ωbuk-ptb | Gmr, suicide vector for buk-ptb operon sequence substitute for recA gene | This study |
| pJQ200mp18::ΔphaCAm | Gmr, suicide vector for phaC gene deletion | This study |
Isolation and transfer of DNA.
Genomic DNA from A. mimigardefordensis cells was isolated according to Marmur (34). Plasmid DNA from E. coli and A. mimigardefordensis strains was isolated by using the GeneJET plasmid miniprep kit from Fermentas (St. Leon-Rot, Germany) according to the manufacturer's manual. DNA fragments were isolated from agarose gels by using a peqGOLD gel extraction kit (PeQLab, Biotechnologie GmbH, Erlangen, Germany). For transformation, E. coli competent cells were prepared by using the calcium chloride procedure (41). Plasmid DNA was transferred from E. coli to A. mimigardefordensis strains by conjugation (12). The transfer of plasmids to different A. mimigardefordensis strains was accomplished by electroporation (Easyject electroporators; Equibio). The preparation of electrocompetent cells of A. mimigardefordensis and the parameters applied for electroporation were similar to the method used for E. coli DH5α according to the Bio-Rad manual.
Modification and amplification of DNA.
DNA was digested with restriction endonucleases under conditions described by the manufacturer or according to Sambrook and Russell (41). PCRs were carried out in an Omnigene HBTR3CM DNA thermal cycler (Hybaid, Heidelberg Germany) using Pfx DNA polymerase (Invitrogen, Karlsruhe, Germany), Taq DNA polymerase (Fermentas, St. Leon-Rot, Germany), or Phusion high-fidelity DNA polymerase (New England Biolabs). T4 DNA ligase was purchased from Invitrogen (Karlsruhe, Germany). Primers were synthesized by MWG-Biotech (Ebersberg, Germany).
DNA sequencing and sequence data analysis.
DNA sequences were determined according to Sanger et al. (43). Sequencing was done using an ABI Prism 3730 capillary sequencer at the Universitäts-Klinikum Münster (UKM) with a BigDye Terminator v3.1 cycle sequencing kit according to the manufacturer's manual (Applied Biosystems, Darmstadt, Germany). The program BlastX (National Center for Biotechnology Information; http://www.ncbi.nml.nih.gov) was used for determination of nucleotide identity (1). The program BioEdit was used for multiple sequence alignments (16).
Gene cloning and plasmid construction.
A DNA fragment containing the phaEC operon of Thiocapsa pfennigii was obtained from plasmid pBPP1 by digestion with EcoRI. A DNA fragment containing the buk-ptb operon was obtained from pBPP1 by digestion with BamHI. These fragments were purified and ligated with the cloning vector pJet1.2 (Fermentas, St. Leon-Rot, Germany). They were cut from respective plasmids and ligated with pBBR1MCS5, which was digested by the same enzyme. Plasmids pBBR1MCS5::phaEC_colinear, pBBR1MCS5::phaEC_antilinear, pBBR1MCS5::buk_ptb, and pBBR1MCS5::BPEC were obtained (23). From the genome of A. mimigardefordensis strain DPN7T the phaCAm promoter sequence (553 bp upstream of the ORF sequence) was amplified by using the Phusion polymerase. The PCR product was subsequently purified and cloned into the vector pJet1.2. After sequence analysis, the fragment was cut by HindIII, and the purified fragment was cloned into linearized pBBR1MCS5 DNA yielding the hybrid plasmids pBBR1MCS5::phaCAm_colinear and pBBR1MCS5::phaCAm_antilinear. In addition, a DNA fragment harboring phaC1Re with its promoter sequence was amplified from the genome of R. eutropha strain H16 by using Phusion polymerase and subsequently ligated to the same vector, yielding pBBR1MCS5::phaC1Re_colinear and pBBR1MCS5::phaC1Re_antilinear, respectively.
Construction of different gene deletion suicide plasmids.
The 798-bp and 832-bp DNA fragments upstream and downstream of mdo in the genome of A. mimigardefordensis strain DPN7T were amplified employing the oligonucleotides mdo_UFR_Fr and mdo_UFR_Rev or mdo_DFR_Fr and mdo_DFR_Rev, respectively (see Table S1 in the supplemental material). The resulting fragments were EcoRI digested and cloned to yield a 1,630-bp fragment. The latter was amplified, and the resulting PCR product was cloned into the XbaI site of pJQ200mp18Tc to yield pJQ200mp18Tc::Δmdo (37).
In a similar way, the suicide vectors pJQ200mp18::ΔrecA and pJQ200mp18::ΔphaC were generated (38). The fragment harboring the buk-ptb operon was obtained from pBPP1. This fragment and the suicide vector pJQ200mp18::ΔrecA were further digested by EcoRI, and both fragments were ligated, yielding pJQ200mp18::ΔrecAΩbuk-ptb with the buk-ptb operon colinear to the recA gene.
Construction of deletion mutants using the sacB system.
Precise-deletion gene replacement plasmids were used to generate the corresponding deletion mutants M1, M2, M3, M4, M5, M6 and M7 of A. mimigardefordensis (Table 1). Standard protocols using the plasmids pJQ200mp18Tc and pJQ200mp18 were adapted to generate these deletion mutants (37, 38). The donor strain E. coli S17-1 was transformed with the plasmids to subsequently mobilize these plasmids into A. mimigardefordensis (46). Identification of mutants was carried out on LB agar plates supplemented with 150 g/liter sucrose and on mineral salts medium agar plates containing 25 μg/ml tetracycline or 20 μg/ml gentamicin. The correct gene replacement strains were confirmed by PCR analysis and DNA sequencing by employing primers which bind beyond the primers used for construction of the deletion gene replacement plasmids (see Table S1 in the supplemental material).
Reverse transcription-PCR (RT-PCR) analysis of total RNA isolated from A. mimigardefordensis.
DNA-free total RNA from cells of the early exponential growth phase of A. mimigardefordensis strain SHX5 and strain SHX8 was prepared with an RNeasy RNA purification kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. For identification of transcription of relevant genes participating in BPEC pathway in the cells, RT-PCR was applied by using the indicated oligonucleotides (Table 1). RT-PCR was carried out using a Qiagen OneStep RT-PCR kit (Qiagen, Hilden, Germany) and 0.5 ng RNA as the template. To confirm the absence of contamination with DNA, which could serve as the template for PCR, template RNA was added in a control experiment after inactivation of reverse transcriptase for 15 min at 95°C in the presence of Taq polymerase. The absence of PCR products indicated that the RT-PCR products did not derive from DNA.
Growth conditions and PMP formation.
E. coli strains were cultivated in Luria-Bertani (LB) medium at 37°C with the addition of applicable antibiotics, if necessary. E. coli XL10 Gold and the broad-host-range vector pBBR1MCS5 were used for DNA cloning. E. coli S17-1 was used as the donor for mobilization. Cells of A. mimigardefordensis were cultured aerobically in mineral salt (MS) medium or LB medium at 30°C with appropriate antibiotics for normal growth. Carbon sources were supplied from filter-sterilized stock solutions as indicated below. Strains of A. mimigardefordensis were used as receptors for mobilization. Antibiotics were added to the medium in the following concentrations: ampicillin (Ap), 100 μg/ml; kanamycin (Km), 50 μg/ml; gentamicin (Gm), 20 μg/ml; and tetracycline (Tc), 12.5 μg/ml.
Two different batch cultivation processes for PMP production were used in this study. In the one-stage cultivation process, 1% (vol/vol) inoculum from LB medium was transferred to MS medium containing 5 g/liter carbon source and 0.5 g/liter DTDP if not indicated otherwise. In the two-stage cultivation process, 1% (vol/vol) inoculum from a preculture in LB medium was first transferred into MS medium with a defined carbon source at the first stage. All cells were then collected, washed, and transferred into fresh MS medium containing the same carbon source plus DTDP as a precursor for cultivation in the second stage. If not indicated otherwise, the culture scale was 50 ml medium in a 300-ml Erlenmeyer flask without baffles. Additionally, a modified MS medium was also used in some of the experiments containing 2 g/liter NH4Cl and 0.4 g/liter yeast extract. The incubation time for the first cultivation stage was varied in different trials, as indicated below.
Analysis of PTE or PHA by GC and GC/MS.
Cells of A. mimigardefordensis were harvested by centrifugation (15 min, 6,000 × g, 4°C), washed three times in saline, and then lyophilized for 24 h. The PTE or PHA contents of the cells were determined upon methylation of 5 to 10 mg lyophilized cells in the presence of 85% (vol/vol) methanol and 15% (vol/vol) sulfuric acid for 4 h at 100°C. The resulting methyl esters of 3HB, 3HV, or 3MP were analyzed by gas chromatography (GC) as described previously (3, 51). The chemical structures of the substances in the various peaks were further confirmed by GC-mass spectrometry (GC/MS) analysis.
Supernatant analysis by HPLC.
DTDP, 3MP, and other carbon sources in the medium supernatant were analyzed by high-performance liquid chromatography (HPLC) employing a LaChrom Elite HPLC apparatus (VWR-Hitachi International GmbH, Darmstadt, Germany). Every sample was filtered through a 0.2-μm filter membrane (Whatman GmbH, Dassel, Germany) before being applied to the HPLC device. The Metacarb 67H advanced C column (Varian, Palo Alto, CA; Bio-228 Rad Aminex equivalent) (300 mm by 6.5 mm) consists of a sulfonated polystyrene resin in the protonated form. The 2350 VWR-Hitachi column oven temperature was kept at 30°C. Detection of the samples was done by an L-2490 VWR-Hitachi refractive index detector. Aliquots of 50 μl were injected and eluted with 5 mM sulfuric acid in MilliQ pure water at a flow rate of 0.8 ml/min. For integration and analysis of the data the EZ Chrome Elite Software (VWR, International GmbH, Darmstadt, Germany) (45) was used.
RESULTS
PMP production in A. mimigardefordensis SHX1.
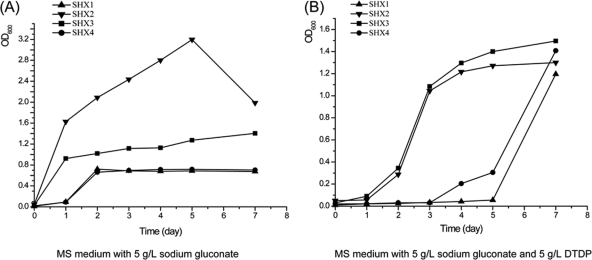
Because plasmid pBBR1MCS5 could be successfully transferred into A. mimigardefordensis strain DPN7T, plasmid pBBR1MCS5::BPEC (Table 1) was constructed. It comprised the genes for the nonnatural and synthetic BPEC pathway that was successfully used in the past for synthesis of PHA and PTE in E. coli (28, 30). Cells of E. coli XL10 Gold/pBBR1MCS5::BPEC accumulated PMP up to 22.3% of the CDW under similar cultivation conditions, as previously described for E. coli JM109/pBPP1 (31). This result proved the correct construction of this plasmid and showed that the enzymes encoded on this plasmid constitute a functional pathway. Plasmid pBBR1MCS5::BPEC was then mobilized into A. mimigardefordensis strain DPN7T, yielding strain SHX1. At the same time, vector pBBR1MCS5 was also transferred into the same strain, yielding strain SHX2 as the control (Table 1). Only few and small colonies of the recombinant strain SHX1 appeared on Gmr agar plates. Attempts to produce PMP with SHX1 in a one-stage cultivation process failed, since its growth was severely repressed (Fig. 2).
Fig 2.
Growth of different strains in MS medium containing sodium gluconate or DTDP. All four strains were incubated in different media. (A) MS medium with 5 g/liter sodium gluconate. (B) MS medium with 5 g/liter DTDP. The OD600 was set to 0.02 after inoculation and before growth started. OD600 profiles of different samples were measured in daily intervals.
Therefore, a modified two-stage cultivation process was employed using MS medium with normal (1 g/liter NH4Cl) or limited (0.5 g/liter NH4Cl) nitrogen in the second stage. The results showed that under normal nitrogen conditions, strain SHX1 could produce only about 2% (wt/wt) PMP (see Table S2 in the supplemental material) at most. Growth of strain SHX1 in MS medium was also slow in comparison to the control strain SHX2. Both SHX1 and SHX2 produced poly(3HB-co-3HV) under the conditions with a limited nitrogen supply. The growth curves were normal, and plasmid pBBR1MCS5 was stable at all times in cells of strain SHX2, whereas pBBR1MCS5::BPEC showed a structural instability in strain SHX1 (see Fig. S2 in the supplemental material).
Growth inhibition of strain SHX3 and SHX4.
To identify the factor which causes the growth repression of A. mimigardefordensis cells, the phaEC operon and the buk-ptb operon were separately cloned into vector pBBR1MCS5, yielding the plasmids pBBR1MCS5::phaEC and pBBR1MCS5::buk-ptb, respectively. Each plasmid was then mobilized into A. mimigardefordensis DPN7T, yielding strains SHX3 and SHX4, respectively (Table 1). Analyses of cell growth revealed that A. mimigardefordensis SHX4 showed repressed growth similar to that of A. mimigardefordensis SHX1, whereas A. mimigardefordensis SHX3 showed growth similar to that of A. mimigardefordensis SHX2 (Fig. 2). It was therefore concluded that the growth inhibition was apparently caused by the proteins encoded by the buk-ptb operon or by these genes themselves.
Systematic optimization to avoid growth inhibition and to enhance the PMP content of the cells.
To obtain further information on the problems caused by Buk/Ptb or buk-ptb for A. mimigardefordensis, different carbon sources were used, and the buk-ptb operon was inserted into the genome. When 5 g/liter sodium succinate instead of 5 g/liter sodium gluconate in MS medium was used as the carbon source, growth of the cells was still negatively affected. Evidently, the problem was caused by the buk-ptb operon itself, and it was not possible to prevent this growth repression by modifying the cultivation conditions. As reported before, recA+ could cause plasmid multimerization and plasmid rearrangement, which are two important factors leading to plasmid segregational instability, owing to RecA's central role in homologous recombination (24, 44, 56). Both plasmid multimerization and plasmid DNA rearrangement were also observed in recA+ strains of A. mimigardefordensis (see Fig. S2 in the supplemental material). Accordingly, the buk-ptb operon was inserted into the recA locus in the genome of A. mimigardefordensis. The deletion of mdo, which encodes the 3MP dioxygenase catalyzing the conversion of 3MP into 3SP, was also considered for the improvement of PMP production. At first, the mdo was deleted in strain A. mimigardefordensis DPN7T to generate strain M1. Subsequently, the buk-ptb operon was inserted into the locus of recA in strain M1 or in the wild-type strain to generate strains M4 and M5, respectively. M2 and M3 were simultaneously constructed by deletion of recA in the wild-type strain and in M1 as control genotypes. Finally, plasmid pBBR1MCS5::phaEC was transferred into strain M4 and M5 to generate SHX5 and SHX9, respectively. The vector pBBR1MCS5 was transferred into strains M1, M2, M3, M4, and M5 to yield SHX8, SHX12, SHX6, SHX13, and SHX10, respectively. Plasmid pBBR1MCS5::BPEC was transferred into strains M1 and M2 to yield strains SHX7 and SHX11, respectively (Table 1).
To verify if the two operons were transcribed properly, four pairs of primers were designed for reverse transcription-PCR assays. The assays for the transcripts of buk, ptb, phaC, and phaE were all positive when RNA samples from strain SHX5 were used (see Fig. S3 in the supplemental material). As a control, the assays with RNA samples from strain SHX8 were all negative as predicted (data not shown).
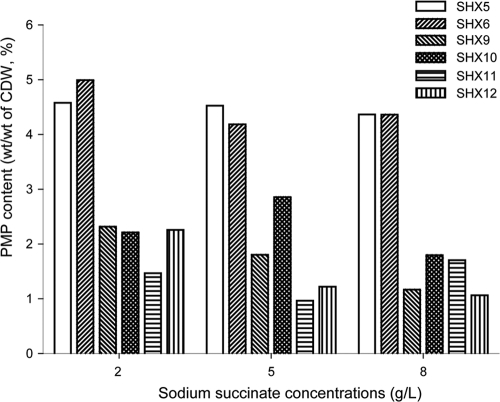
The growth behavior of strains SHX5, SHX6, SHX7, and SHX8 was compared. Strains SHX5 and SHX6, which both harbored the buk-ptb operon in the genome, exhibited growth similar to that of the control strain SHX8. In contrast, growth of strain SHX7 was still severely repressed. Therefore, growth repression caused by the buk-ptb operon or the corresponding proteins was relieved by insertion into the chromosome. Strains SHX5, SHX6, SHX9, SHX10, SHX11, and SHX12 were investigated with regard to PMP production in MS medium containing various concentrations of sodium succinate (2, 5, and 8 g/liter) as the carbon source (Fig. 3). Interestingly, cells of strains SHX5 and SHX6, which harbor the buk-ptb operon inserted into recA, accumulated more polymer than all other strains. This demonstrated that the chromosomal integration of the buk-ptb operon enhanced accumulation of PMP in the cells. Unexpectedly, little PMP was also accumulated in strain SHX12 (Fig. 3). This showed that A. mimigardefordensis obviously harbors a native PMP biosynthesis pathway which is independent of the heterologously expressed genes. Based on these conclusions, two strategies for further PMP optimization were chosen: one was the optimization of the cultivation conditions, and the second was further metabolic engineering.
Fig 3.
PMP production of indicated strains in MS medium using sodium succinate as the carbon source. All indicated strains were first incubated in MS medium containing 5 g/liter sodium succinate as the carbon source. In the late exponential phase, all of them were transferred into fresh medium containing 5 g/liter DTDP. After 4 days of incubation, PMP contents from different samples were analyzed.
Cultivation optimization for PMP production.
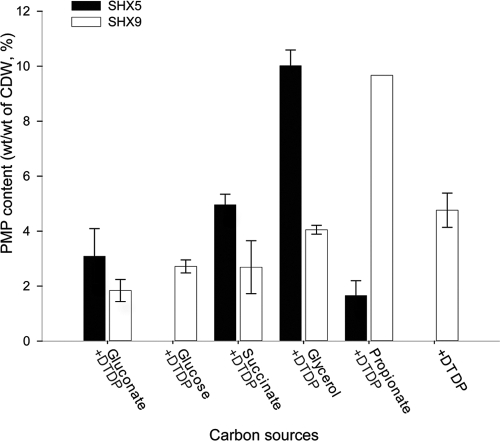
To identify the most suitable carbon source for PMP production by A. mimigardefordensis, the recombinant strains SHX5 and SHX9 were cultivated in MS medium containing glucose, glycerol, sodium gluconate, sodium propionate, or sodium succinate alone or supplemented with DTDP or in medium containing DTDP alone (control) (Fig. 4). Cells of strain SHX5 with glycerol as the carbon source contained about 10% (wt/wt [CDW]) PMP, which was the highest content compared to other applied carbon sources. Up to 10% (wt/wt [CDW]) PMP was also obtained in experiments in which 0.2% sodium propionate was used as the carbon source with strain SHX9. However, accumulation of PMP with sodium propionate varied considerably, and in some experiments, much lower PMP contents (3%, wt/wt [CDW]) were also recorded. The other problem was that the cells were unable to tolerate concentrations of sodium propionate higher than 2 g/liter for growth. In conclusion, the results of these studies indicated that glycerol was the best carbon source, and it was therefore employed in all further experiments.
Fig 4.
PMP production in A. mimigardefordensis SHX5 and SHX9 by using different carbon sources. The cells were first incubated in MS medium containing different carbon sources at a concentration of 5 g/liter except sodium propionate, which was supplied at a concentration of only 2 g/liter, in the first cultivation stage. When sodium gluconate, sodium propionate, or sodium succinate was used as the carbon source, the weight of sodium was eliminated. In the late exponential phase, all cells were transferred into the same MS medium containing 5 g/liter of the carbon sources (or 2 g/liter sodium propionate) plus 5 g/liter DTDP as precursor substrate. After 4 days of incubation, the PMP content of each sample was analyzed. The error bars represent the standard deviations from three independent experiments.
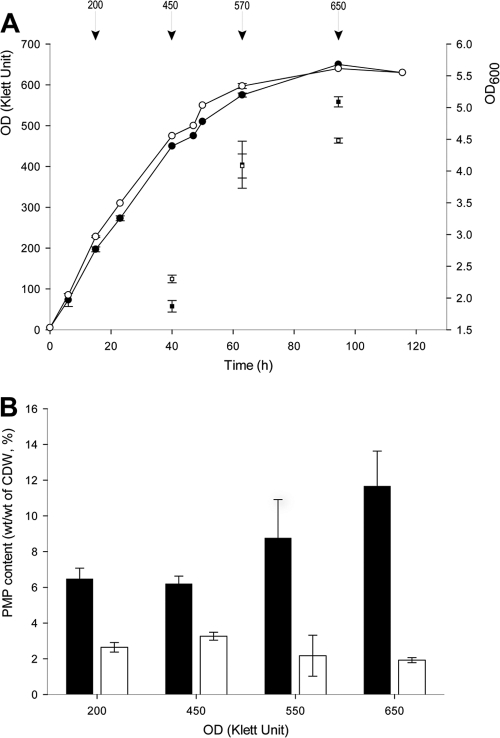
Afterwards, the relation between cell mass formation in the first growth stage and PMP production in the second growth stage were studied, using glycerol as the carbon source (Fig. 5). Cells of strains SHX5 and SHX13 were first incubated in modified MS medium containing 2 g/liter NH4Cl, 10 g/liter glycerol, and 0.4 g/liter yeast extract in 2-liter flasks for the first growth stage, and the optical density was measured at defined intervals during the time course of the experiment (Fig. 5A). The cells were taken from different growth phases, when the optical density reached 200, 450, 570, and 650 Klett units (Fig. 5A), and were then transferred into the same volume of fresh medium containing in addition 6 g/liter DTDP as a precursor for PMP biosynthesis. After 4 days of incubation, the cells were analyzed for the PMP contents by GC (Fig. 5B). It was found that the PMP contents were higher if the cultures were inoculated with high-density cells (Fig. 5B), thereby revealing a positive correlation between cell mass and PMP accumulation. The optimal time point for cell transfer from the first stage to the second stage was also defined and applied in further experiments.
Fig 5.
Relationship between cell amount and PMP production. (A) Growth curves of different strains. Symbols: ●, growth curve of strain SHX5 in the first cultivation stage; ○, growth curve of strain SHX13 in first cultivation stage; ■, OD600 of strain SHX5; □, OD600 of strain SHX13. Arrowheads indicate the time points when the cells were transferred from the first to the second stage. (B) PMP content relative to cell growth in the first stage. Black bars, strain SHX5; white bars, strain SHX13. The error bars represent the standard deviations from three independent experiments.
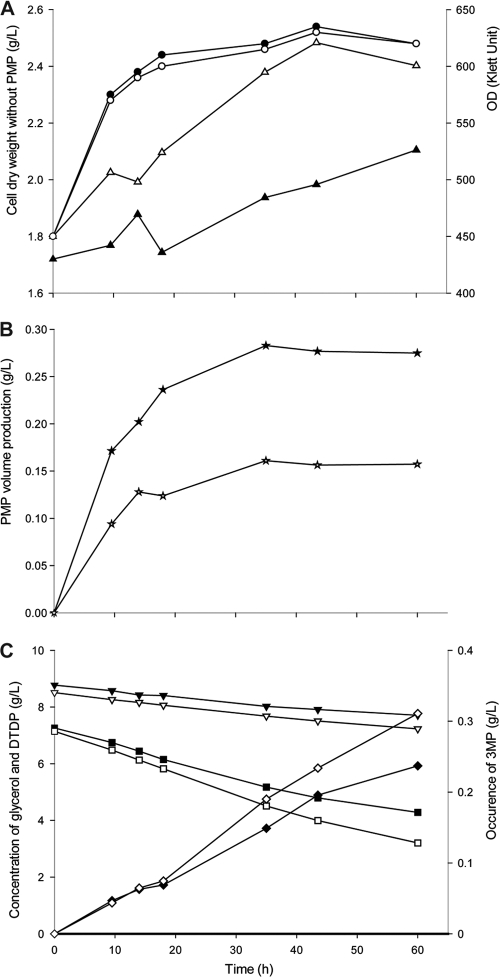
PMP formation was also studied according to time scale (Fig. 6). Both strains were cultivated in modified MS medium containing 2 g/liter NH4Cl and 7 g/liter glycerol until the cells entered the late exponential growth phase at an optical density of about 500 Klett units. The cells were then collected and transferred into the same fresh medium supplied with 9 g/liter DTDP as a precursor. Cell mass, optical density at 600 nm (OD600), PMP content, and concentrations of glycerol, DTDP, and 3MP in the supernatant were analyzed at defined intervals. As expected, the rate of PMP production was higher in cells of strain SHX5 than in cells of strain SHX13. 3MP continuously accumulated extracellularly in the supernatants of both cultures. DTDP and glycerol were continuously consumed. PMP accumulated for only approximately 30 h during the experiment. From this, it was concluded that 3MP was formed in excess, and since the concentration continued to increase at a low rate, it could not be a limiting factor for PMP formation. The cessation of PMP accumulation after 30 h cultivation was most probably caused by other, not-yet-identified factors.
Fig 6.
PMP content, OD, cell dry weight, and supernatant analysis during the time course of the cultivation experiment. A. mimigardefordensis strains SHX5 and SHX13 were first incubated in modified MS medium containing 2 g/liter NH4Cl and 7 g/liter glycerol at the first cultivation stage. After their OD had reached 500 Klett units, all cells were transferred into the same MS medium containing 7 g/liter glycerol plus 9 g/liter DTDP as a precursor. (A) Increase of OD (Klett unit) and cell dry weight in the second cultivation stage. Symbols: ● and ○, turbidity increase of SHX5 and SHX13; ▲ and △, cell dry weight increase of SHX5 and SHX13. (B) PMP content over time in the second-stage cultivation stage. Symbols: ★ and ☆, strains SHX5 and SHX13. (C) Concentrations of DTDP, 3MP, and glycerol in different culture supernatants in the second-stage cultivation stage. Symbols: ▼ and ▽, consumption of DTDP in cultures of SHX5 and SHX13; ■ and □, degradation of glycerol in cultures of SHX5 and SHX13; ♦ and ♢, occurrence of 3MP in cultures of strains SHX5 and SHX13.
Genetic optimization for enhanced PMP accumulation.
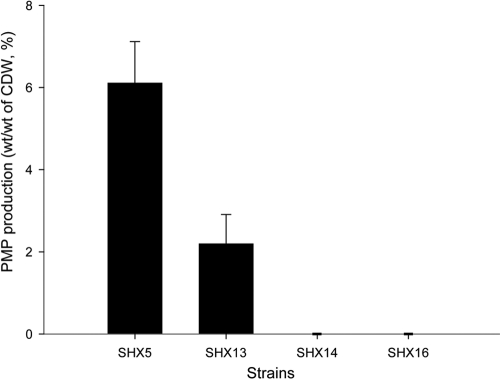
To determine which enzyme could be the limiting factor in A. mimigardefordensis, the native PHA synthase gene of the host, phaCAm, was deleted in strains M3 and M4, yielding strains SHX14 and SHX16, respectively (Table 1). These new strains were then compared with SHX5 and SHX13 with regard to their ability to form PMP (Fig. 7). Strain SHX5 accumulated approximately three times more PMP than strain SHX13. In contrast, strains SHX14 and SHX16 lost the ability to produce PMP homopolymer. The performance of phaEC was inefficient in A. mimigardefordensis strain SHX16, although the RT-PCR showed positive signals for phaC and phaE (see Fig. S3 in the supplemental material). However, phaCAm from A. mimigardefordensis itself was obviously an alternative candidate to enhance PMP accumulation. In addition to and in accordance with the results described above, A. mimigardefordensis strain DPN7T encodes its own as-yet-unknown enzymes, which are able to convert 3MP into 3MP-CoA at a comparably low rate (Fig. 3).
Fig 7.
PMP production in engineered strains. Cells were first incubated in modified MS medium containing 2 g/liter NH4Cl, 10 g/liter glycerol, and 0.4 g/liter yeast extract when their OD600 had reached about 3 to 4 (late exponential phase). Then the cells were collected and transferred into fresh medium supplied with 6 g/liter DTDP as precursor. After 4 days of incubation, all samples were analyzed by GC to determine the PMP content. The error bars represent the standard deviations from three independent experiments.
Three different phaC genes with their own promoters (phaC1Re from R. eutropha H16, phaCAm from A. mimigardefordensis, and phaEC from Thiocapsa pfennigii) were compared for their ability to confer and enhance PMP production in A. mimigardefordensis. Suitable plasmids were constructed for this purpose (Table 1) and were transferred to A. mimigardefordensis M6, yielding strains SHX15, SHX16, SHX17, SHX18, SHX19, SHX20, and SHX21 (Table 1). In view of a potential effect of the exogenetic promoter (lac promoter) or the operon orientation in the vector pBBR1MCS5, the operon was arranged in two directions, which were colinear or antilinear to the lac promoter (18, 29). In addition, strain SHX15, which harbors the vector pBBR1MCS5, was used as a negative control. All these strains were investigated for their ability for PMP production in comparison to positive-control strain SHX6 (Table 2).
Table 2.
Relationships between PMP production and phaCs from different sourcesa
| Strain | PMP content (%) (wt/wt [CDW]) | CDW without PMP wt (g/liter) | Concn (g/liter) in supernatant of: |
||
|---|---|---|---|---|---|
| Glycerol | 3MP | DTDP | |||
| SHX6 | 7.02 ± 0.06 | 4.34 ± 0.41 | 0.6 ± 0.9 | — | 3.8 ± 0.3 |
| SHX15 | 0 | 3.10 ± 0.17 | 6 ± 0.8 | 0.48 ± 0.00 | 3.7 ± 0.9 |
| SHX18 | 13.38 ± 0.14 | 4.21 ± 0.08 | — | — | 3.6 ± 0.1 |
| SHX19 | 12.76 ± 1.26 | 4.35 ± 0.32 | — | — | 3.9 ± 0.8 |
| SHX20 | 4.49 ± 0.40 | 3.30 ± 0.35 | 4.9 ± 0.5 | 0.4 ± 0.01 | 4.4 ± 0.2 |
| SHX21 | 4.31 ± 0.23 | 3.53 ± 0.15 | 4.9 ± 0.5 | 0.34 ± 0.06 | 4.7 ± 0.3 |
The strains were first incubated in modified MS medium containing 2 g/liter NH4Cl, 10 g/liter glycerol, and 0.4 g/liter yeast extract. A 1 mM final concentration of IPTG was added when the OD600 reached 0.6 (early exponential phase). After the cells arrived at late exponential phase (OD600 of approximately 4.3), they were collected and transferred into the same medium supplied with 7 g/liter DTDP as a precursor. After 4 days of incubation, all samples were analyzed for PMP content by GC analysis. CDW, OD600, and supernatant of the final cultures were also measured. Values are means and standard deviations from three independent experiments. —, below detection limit.
The highest PMP accumulation occurred in strains SHX18 and SHX19, with up to about 14% (wt/wt [CDW]) PMP. Both recombinant strains possessed phaCAm (Table 2). PMP production in strain SHX18 and SHX19 was almost doubled in comparison to that in strain SHX6. These results showed that the native PHA synthase PhaCAm conferred the highest accumulation of PMP if it was present in the vector instead of the chromosome (Table 2). Furthermore, higher cell dry weight, higher cell density, and enhanced glycerol consumption were also achieved in comparison to the control strain SHX6. Data for strains SHX16 and SHX17 are not shown, since the cells did not accumulate any PMP. Obviously, the defect of the phaEC operon is not altered by its orientation in the vector.
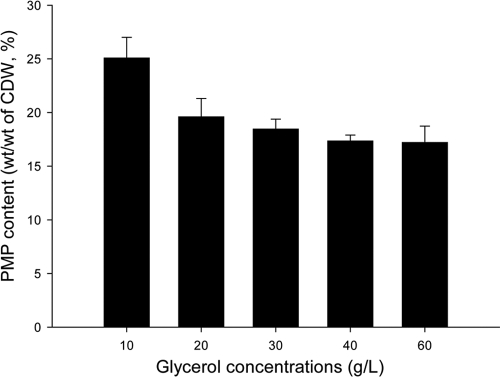
To further increase PMP accumulation, A. mimigardefordensis SHX22 (Δmdo recA::buk-ptb pBBR1MCS5::phaCAm_colinear) was constructed. This strain harbors phaCAm on the vector pBBR1MCS5 in addition to its copy in the genome. Glycerol concentrations of 10, 20, 30, 40, and 60 g/liter were used in the second stage to further optimize PMP accumulation (Fig. 8). The highest PMP content, about 25% (wt/wt [CDW]), occurred at the lowest glycerol concentration (10 g/liter). Interestingly, the PMP content in cells of SHX22 was almost equal to the combined PMP contents of the cells of strains SHX6 and SHX18 (see Fig. S4 in the supplemental material). This demonstrates that even higher PMP contents can probably be obtained by inserting additional copies of phaC into the genome or by enhancing the expression of the present copies. Although PMP production decreased when the glycerol concentration was enhanced to more than 10 g/liter, the volumetric production levels among different samples were actually similar owing to the increase of the cell mass in the samples with more than 10 g/liter glycerol (Table 3; also, see Fig. S5 in the supplemental material). Considering that 10 g/liter glycerol was completely consumed, it is obvious that the reason for low cell mass accumulation in 10 g/liter glycerol samples is due to the shortage of glycerol (Table 3). To keep glycerol abundant for further optimization, an application of more than 10 g/liter glycerol is recommended.
Fig 8.
PMP production in cells of A. mimigardefordensis strain SHX22 cultivated in the presence of different glycerol concentrations. The strains were first incubated in modified MS medium with 2 g/liter NH4Cl, 10 g/liter glycerol, and 0.4 g/liter yeast extract. After the cells had reached the early stationary growth phase (OD600 of approximately 4.2 to 4.4), they were collected and transferred to fresh medium supplied with different concentrations of glycerol (10, 20, 30, 40, and 60 g/liter) and 6 g/liter DTDP as a precursor. After 4 days of incubation, all samples were analyzed for PMP content by GC analysis.
Table 3.
| Glycerol concn (g/liter) | CDW without PMP wt (g/liter) | Concn (g/liter) in supernatant of: |
|||
|---|---|---|---|---|---|
| Glycerol | 3MP | DTDP | NH4Cl | ||
| 10 | 3.18 ± 0.14 | — | — | 5.6 ± 0.3 | 0.96 ± 0.03 |
| 20 | 4.09 ± 0.41 | 2.3 ± 2.0 | 0.16 ± 0.14 | 3.8 ± 1.1 | 0.76 ± 0.2 |
| 30 | 4.34 ± 0.16 | 10.3 ± 1.9 | 0.04 ± 0.07 | 4.4 ± 0.9 | 0.69 ± 0.15 |
| 40 | 4.16 ± 0.25 | 18.3 ± 0.2 | 0.05 ± 0.08 | 4.7 ± 0.3 | 0.68 ± 0.15 |
| 50 | 4.19 ± 0.50 | 35.1 ± 1.9 | — | 5.0 ± 0.7 | 0.61 ± 0.13 |
Cells of A. mimigardefordensis strain SHX22 were first incubated in modified MS medium with 2 g/liter NH4Cl, 10 g/liter glycerol, and 0.4 g/liter yeast extract. After the optical density indicated that the early stationary phase (OD600 of approximately 4.3) had been reached, cells were collected and transferred into the same medium supplied with different concentrations of glycerol (10, 20, 30, 40, and 60 g/liter) and 6 g/liter DTDP as a precursor. After 4 days of incubation, CDW, OD600, and supernatant of the final cultures were also measured. The values are means and standard deviations from three independent experiments. —, below detection limit.
DISCUSSION
The species A. mimigardefordensis as well as the entire genus Advenella is not very well studied as yet. So far, only a few publications are available; however, scientific interest in these bacteria has begun to increase (5, 8, 13, 14, 19, 45, 53–55). Recently, the complete genome of Advenella kashmirensis WT001T was sequenced (14). Until now, there were no reports on applications in biotechnology for members of this genus. This study unraveled a potential use of A. mimigardefordensis for the efficient production of PMP homopolymer by applying one of its special metabolic features: the utilization of the nontoxic PMP precursor substrate DTDP (Fig. 1B).
During this study, a growth repression caused by the buk-ptb operon became obvious, which has never been reported before. Some studies applied these two enzymes heterologously in recombinant strains of E. coli without an observed growth repression effect (10, 29, 49). One explanation may be the broad substrate specificity of these enzymes (17, 52). This may allow the enzymes to convert some important metabolic intermediates, which usually support regular growth, into not-required products, thereby causing metabolic turbulences and finally growth inhibition. By integrating the buk-ptb operon into the genome of A. mimigardefordensis, inhibition of growth was finally relieved. Actually, integration of genes into the genome is a common strategy to avoid plasmid instability and/or to obtain a stable expression of proteins encoded by foreign genes, especially when the expressed product is toxic to cells (40). It causes the cells of A. mimigardefordensis to accumulate more PMP, most probably due to the decrease of transcripts of buk and ptb, which finally decrease the enzyme activities to an endurable level.
The deletion of mdo was another important step to enhance PMP accumulation (Fig. 3). The mdo deletion mutant could not use DTDP as the carbon source for growth, which is in accordance with previous results (4). Since the further catabolism of 3MP is disrupted in the mdo deletion mutant, sufficient 3MP should now be available in the cells for PMP synthesis (Fig. 1). 3MP could be exported out of the cells but cannot be transported back into the cells (53). 3MP, as a reactive thiol compound, can cause serious problems for cell metabolism if it is accumulated in high concentrations in or outside the cells (11, 33, 36). Considering these consequences, the in vivo effects of an mdo deletion mutant were hard to predict. Finally, the analysis of the Δmdo mutant and its comparison with the wild type showed that the amount of surplus 3MP and its conversion are bottlenecks in the strains of A. mimigardefordensis possessing an intact mdo, which is basically due to the fact that the high Mdo enzyme activity allows a strong metabolic flux of 3MP toward 3SP (4).
The use of glycerol further increased the accumulation of PMP in comparison to all other applied carbon sources (Fig. 4). The pH stability in the cultivation medium with glycerol as the carbon source was probably the main reason. When sodium gluconate, sodium succinate, or sodium propionate was used as the carbon source, the pH value of the respective medium increased to up to 9 within 2 days. This effect is caused by the decreasing amount of the anionic carbon source consumed by the cells. Accordingly, residues of sodium in the respective medium caused an increase of the pH. Glucose acted in the opposite way. The pH value of the medium containing glucose as the sole carbon source decreased to 3 to 4 in less than 8 h. Only during cultivation of the cells with glycerol did the pH of the medium remain more or less stable at around 7 even after 4 days of incubation. As reported before, acetate is a main by-product when excess glucose or glycerol is used as the carbon source in E. coli even under aerobic conditions (50). However, utilization of glycerol resulted in less acetate formation, mostly because of the lower rate of transport into the cells, which led to the decrease of carbon flux into glycolysis, as also observed before in E. coli (25). Consequently, a similar process most probably occurred in A. mimigardefordensis. The maintenance of a stable pH in the medium is actually an important factor during the cultivation of bacteria. The cells, which are used to produce PMP, have to endure serious disulfide stress and oxidative stress from DTDP and 3MP, respectively (Fig. 6) (26, 36). Therefore, it is probably disadvantageous to expose the cells also to acid stress at the same time. Since glycerol is at present abundantly available, it is also practically viable (9, 50).
As reported earlier (2), phaC usually plays a dominant role in controlling PHA production. Higher activities or higher expression levels of PhaC could lead to higher PHA and/or PMP accumulation. Although the heterologous expression of PhaEC from T. pfennigii has been successfully performed in R. eutropha, Pseudomonas putida, and E. coli (48), its expression in A. mimigardefordensis was completely insufficient (Table 2; Fig. 6). Most probably, the low expression level of PhaEC or the low activity of this recombinant enzyme in A. mimigardefordensis was the reason, since the two corresponding genes were insufficiently transcribed (see Fig. S3 in the supplemental material). In contrast, the expression cassette of phaC1Re from R. eutropha H16 and the native phaCAm of A. mimigardefordensis were both active. Moreover, the copies of phaC in the cells strongly influenced PMP production (Table 2; also, see Fig. S4); phaCAm was more effective than phaC1Re for PMP production in A. mimigardefordensis. The amino acid sequence of PhaCAm is 58% identical to that of PhaC from Bordetella petrii (highest identity) and 52% identical to that of PhaC1Re, and all these enzymes belong to the class I PHA synthases. The low degree of homology indicates that the properties of PhaCAm could be quite different from those of PhaC1Re.
In comparison to the establishment of PTE heteropolymer synthesis from DTDP in R. eutropha, production of the PMP homopolymer was easier to achieve in A. mimigardefordensis (32, 33). The major and most relevant difference between these two bacteria is the inherent PHB production ability. R. eutropha possesses a strong metabolic flux toward PHB that can hardly be completely suppressed (32) if an active PHA synthase is present. In contrast, the recombinant A. mimigardefordensis strains did not produce any PHA under normal nitrogen conditions when glycerol was used as the carbon source (Fig. 8).
In summary, PMP production in A. mimigardefordensis is controlled by two factors: (i) the supply of applicable monomers and (ii) an effective PHA synthase, as described before (2, 39, 48). Deletion of mdo and integration of buk-ptb into the chromosome are considered two key steps to allow the cells to synthesize appropriate amounts of 3MP-CoA for the PHA synthase. In addition, the increase in the number of copies of phaC resulted in a higher in vivo PhaC activity in the cells. All these steps contributed to the accumulation of PMP in A. mimigardefordensis whereby the PMP content in the cells could be increased in a stepwise manner. The fact that the exported 3MP cannot be reimported into the cells (53) makes the PMP production in this strain different from that in other bacteria. In contrast, 3MP was imported freely into E. coli cells, since PMP can be produced in E. coli by supplying 3MP as a precursor (31). Finally, to overcome the efflux of 3MP and to further optimize the entire process, cells of A. mimigardefordensis require a strong enzyme to convert 3MP into 3MP-CoA. Alternatively, an appropriate transport system could be employed to reimport released 3MP.
For the first time, a novel PMP production pathway based on the nontoxic precursor DTDP was established in a recombinant strain of A. mimigardefordensis. The key points in the pathway for the PMP production are discussed and illustrated (Fig. 1). Cellular contents of almost 25% (wt/wt [CDW]) PMP were obtained with this system.
Supplementary Material
ACKNOWLEDGMENT
We thank the China Scholarship Council for providing a fellowship to Yongzhen Xia.
Footnotes
Published ahead of print 17 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bhubalan K, et al. 2011. Characterization of the highly active polyhydroxyalkanoate synthase of Chromobacterium sp. strain USM2. Appl. Environ. Microbiol. 77:2926–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brandl H, Gross RA, Lenz RW, Fuller RC. 1988. Pseudomonas oleovorans as a source of poly (β-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl. Environ. Microbiol. 54:1977–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bruland N, Wübbeler JH, Steinbüchel A. 2009. 3-Mercaptopropionate dioxygenase, a cysteine dioxygenase homologue, catalyzes the initial step of 3-mercaptopropionate catabolism in the 3,3-thiodipropionic acid-degrading bacterium Variovorax paradoxus. J. Biol. Chem. 284:660–672 [DOI] [PubMed] [Google Scholar]
- 5. Coenye T, et al. 2005. Advenella incenata gen. nov., sp. nov., a novel member of the Alcaligenaceae, isolated from various clinical samples. Int. J. Syst. Evol. Microbiol. 55:251–256 [DOI] [PubMed] [Google Scholar]
- 6. Dam B. 2011. A type Ib plasmid segregation machinery of the Advenella kashmirensis plasmid pBTK445. Plasmid 65:185–191 [DOI] [PubMed] [Google Scholar]
- 7. Dam B, Ghosh W, Das Gupta SK. 2009. Conjugative type 4 secretion system of a novel large plasmid from the chemoautotroph Tetrathiobacter kashmirensis and construction of shuttle vectors for Alcaligenaceae. Appl. Environ. Microbiol. 75:4362–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dam B, Mandal S, Ghosh W, Das Gupta SK, Roy P. 2007. The S4-intermediate pathway for the oxidation of thiosulfate by the chemolithoautotroph Tetrathiobacter kashmirensis and inhibition of tetrathionate oxidation by sulfite. Res. Microbiol. 158:330–338 [DOI] [PubMed] [Google Scholar]
- 9. Da Silva GP, Mack M, Contiero J. 2009. Glycerol: a promising and abundant carbon source for industrial microbiology. Biotechnol. Adv. 27:30–39 [DOI] [PubMed] [Google Scholar]
- 10. Diao J, Hasson MS. 2009. Crystal structure of butyrate kinase 2 from Thermotoga maritima, a member of the ASKHA superfamily of phosphotransferases. J. Bacteriol. 191:2521–2529 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11. Feeney MA, et al. 2011. Repurposing lipoic acid changes electron flow in two important metabolic pathways of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 108:7991–7996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Friedrich B, Hogrefe C, Schlegel HG. 1981. Naturally occurring genetic transfer of hydrogen-oxidizing ability between strains of Alcaligenes eutrophus. J. Bacteriol. 147:198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ghosh W, Bagchi A, Mandal S, Dam B, Roy P. 2005. Tetrathiobacter kashmirensis gen. nov., sp. nov., a novel mesophilic, neutrophilic, tetrathionate-oxidizing, facultatively chemolithotrophic betaproteobacterium isolated from soil from a temperate orchard in Jammu and Kashmir, India. Int. J. Syst. Evol. Microbiol. 55:1779–1787 [DOI] [PubMed] [Google Scholar]
- 14. Ghosh W, et al. 2011. Whole-genome shotgun sequencing of the sulfur-oxidizing chemoautotroph Tetrathiobacter kashmirensis. J. Bacteriol. 193:5553–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gibello A, et al. 2005. 2009 Reclassification of the members of the genus Tetrathiobacter Ghosh et al. 2005 to the genus Advenella Coenye et al. Int. J. Syst. Evol. Microbiol. 59:1914–1918 [DOI] [PubMed] [Google Scholar]
- 16. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41:95–98 [Google Scholar]
- 17. Hartmanis MG. 1987. Butyrate kinase from Clostridium acetobutylicum. J. Biol. Chem. 262:617–621 [PubMed] [Google Scholar]
- 18. Hein S, Söhling B, Gottschalk G, Steinbüchel A. 1997. Biosynthesis of poly(4-hydroxybutyric acid) by recombinant strains of Escherichia coli. FEMS Microbiol. Lett. 153:411–418 [DOI] [PubMed] [Google Scholar]
- 19. Hunter WJ, Manter DK. 2008. Bio-reduction of selenite to elemental red selenium by Tetrathiobacter kashmirensis. Curr. Microbiol. 57:83–88 [DOI] [PubMed] [Google Scholar]
- 20. Kato M, Toshima K, Matsumura S. 2005. Preparation of aliphatic poly(thioester) by the lipase-catalyzed direct polycondensation of 11-mercaptoundecanoic acid. Biomacromolecules 6:2275–2280 [DOI] [PubMed] [Google Scholar]
- 21. Kawada J, Lütke-Eversloh T, Steinbüchel A, Marchessault RH. 2003. Physical properties of microbial polythioesters: characterization of poly(3-mercaptoalkanoates) synthesized by engineered Escherichia coli. Biomacromolecules 4:1698–1702 [DOI] [PubMed] [Google Scholar]
- 22. Kim DY, Lütke-Eversloh T, Elbanna K, Thakor N, Steinbüchel A. 2005. Poly(3-mercaptopropionate): a nonbiodegradable biopolymer? Biomacromolecules 6:897–901 [DOI] [PubMed] [Google Scholar]
- 23. Kovach ME, et al. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 24. Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Mol. Biol. Rev. 58:401–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee SY. 1996. High cell-density culture of Escherichia coli. Trends Biotechnol. 14:98–105 [DOI] [PubMed] [Google Scholar]
- 26. Leichert LI, Scharf C, Hecker M. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lenz RW, Marchessault RH. 2005. Bacterial polyesters: biosynthesis, biodegradable plastics and biotechnology. Biomacromolecules 6:1–8 [DOI] [PubMed] [Google Scholar]
- 28. Lindenkamp N, Peplinski K, Volodina E, Ehrenreich A, Steinbüchel A. 2010. Impact of multiple β-ketothiolase deletion mutations in Ralstonia eutropha H16 on the composition of 3-mercaptopropionic acid-containing copolymers. Appl. Environ. Microbiol. 76:5373–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu SJ, Steinbüchel A. 2000. A novel genetically engineered pathway for synthesis of poly(hydroxyalkanoic acids) in Escherichia coli. Appl. Environ. Microbiol. 66:739–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lütke-Eversloh T, Bergander K, Luftmann H, Steinbüchel A. 2001. Identification of a new class of biopolymer: bacterial synthesis of a sulfur-containing polymer with thioester linkages. Microbiology 147:11–19 [DOI] [PubMed] [Google Scholar]
- 31. Lütke-Eversloh T, et al. 2002. Biosynthesis of novel thermoplastic polythioesters by engineered Escherichia coli. Nat. Mater. 1:236–240 [DOI] [PubMed] [Google Scholar]
- 32. Lütke-Eversloh T, Steinbüchel A. 2004. Microbial polythioesters. Macromol. Biosci. 4:166–174 [DOI] [PubMed] [Google Scholar]
- 33. Lütke-Eversloh T, Steinbüchel A. 2003. Novel precursor substrates for polythioesters (PTE) and limits of PTE biosynthesis in Ralstonia eutropha. FEMS Microbiol. Lett. 221:191–196 [DOI] [PubMed] [Google Scholar]
- 34. Marmur J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208–218 [Google Scholar]
- 35. Marvel CS, Kotch A. 1951. Polythiolesters. J. Am. Chem. Soc. 73:1100–1102 [Google Scholar]
- 36. Munday R. 1989. Toxicity of thiols and disulphides: involvement of free-radical species. Free Radic. Biol. Med. 7:659–673 [DOI] [PubMed] [Google Scholar]
- 37. Pötter M, Müller H, Steinbüchel A. 2005. Influence of homologous phasins (PhaP) on PHA accumulation and regulation of their expression by the transcriptional repressor PhaR in Ralstonia eutropha H16. Microbiology (SGM) 151:825–833 [DOI] [PubMed] [Google Scholar]
- 38. Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15–21 [DOI] [PubMed] [Google Scholar]
- 39. Rehm BH. 2003. Polyester synthases: natural catalysts for plastics. Biochem. J. 376:15–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rehm H-J, Reed G, Pühier A, Stadler P. 1993. Strategies for fermentation with recombinant organisms, p 283–294 In Biotechnology: a multi-volume comprehensive treatise, vol. 3 Bioprocessing. VCH, Weinheim, Germany [Google Scholar]
- 41. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 42. Sandler SR. 1996. Polythioesters, p. 46ff In Wasserman HH. (ed), Organic chemistry, vol 29-III: polymer syntheses. Academic Press Inc, San Diego, CA [Google Scholar]
- 43. Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schuch R, Maurelli AT. 1997. Virulence plasmid instability in Shigella flexneri 2a is induced by virulence gene expression. Infect. Immun. 65:3686–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schürmann M, Wübbeler JH, Grote J, Steinbüchel A. 2011. Novel reaction of succinyl coenzyme A (succinyl-CoA) synthetase: activation of 3-sulfinopropionate to 3-sulfinopropionyl-CoA in Advenella mimigardefordensis strain DPN7T during degradation of 3,3′-dithiodipropionic acid. J. Bacteriol. 193:3078–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat. Biotech. 1:784–791 [Google Scholar]
- 47. Steinbüchel A. 2005. Non-biodegradable biopolymers from renewable resources: perspectives and impacts. Curr. Opin. Biotechnol. 16:607–613 [DOI] [PubMed] [Google Scholar]
- 48. Steinbüchel A, Lütke-Eversloh T. 2003. Metabolic engineering and pathway construction for biotechnological production of relevant polyhydroxyalkanoates in microorganisms. Biochem. Eng. J. 16:81–96 [Google Scholar]
- 49. Tessmer N, et al. 2007. Heat-shock protein HspA mimics the function of phasins sensu stricto in recombinant strains of Escherichia coli accumulating polythioesters or polyhydroxyalkanoates. Microbiology (SGM) 153:366–374 [DOI] [PubMed] [Google Scholar]
- 50. Thakor N, Lütke-Eversloh T, Steinbüchel A. 2005. Application of the BPEC pathway for large-scale biotechnological production of poly(3-mercaptopropionate) by recombinant Escherichia coli, including a novel in situ isolation method. Appl. Environ. Microbiol. 71:835–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Veit A, Polen T, Wendisch VF. 2007. Global gene expression analysis of glucose overflow metabolism in Escherichia coli and reduction of aerobic acetate formation. Appl. Microbiol. Biotechnol. 74:406–421 [DOI] [PubMed] [Google Scholar]
- 52. Wiesenborn DP, Rudolph FB, Papoutsakis ET. 1989. Phosphotransbutyrylase from Clostridium acetobutylicum ATCC 824 and its role in acidogenesis. Appl. Environ. Microbiol. 55:317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wübbeler JH, Bruland N, Kretschmer K, Steinbüchel A. 2008. Novel pathway for catabolism of the organic sulfur compound 3,3′-dithiodipropionic acid via 3-mercaptopropionic acid and 3-sulfinopropionic acid to propionyl-coenzyme A by the aerobic bacterium Tetrathiobacter mimigardefordensis strain DPN7T. Appl. Environ. Microbiol. 74:4028–4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wübbeler JH, Lütke-Eversloh T, Van Trappen S, Vandamme P, Steinbüchel A. 2006. Tetrathiobacter mimigardefordensis sp. nov., isolated from compost, a betaproteobacterium capable of utilizing the organic disulfide 3,3′-dithiodipropionic acid. Int. J. Syst. Evol. Microbiol. 56:1305–1310 [DOI] [PubMed] [Google Scholar]
- 55. Wübbeler JH, Raberg M, Brandt U, Steinbüchel A. 2010. Dihydrolipoamide dehydrogenases of Advenella mimigardefordensis and Ralstonia eutropha catalyze cleavage of 3,3′-dithiodipropionic acid into 3-mercaptopropionic acid. Appl. Environ. Microbiol. 76:7023–7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhao JB, Wei DZ, Tong WY. 2007. Identification of Escherichia coli host cell for high plasmid stability and improved production of antihuman ovarian carcinoma x antihuman CD3 single-chain bispecific antibody. Appl. Microbiol. Biotechnol. 76:795–800 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.