Abstract
The multidrug resistance-encoding plasmids belonging to the IncA/C incompatibility group have recently emerged among Escherichia coli and Salmonella enterica strains in the United States. These plasmids have a unique genetic structure compared to other enterobacterial plasmid types, a broad host range, and a propensity to acquire large numbers of antimicrobial resistance genes via their accessory regions. Using E. coli strain DH5α harboring the prototype IncA/C plasmid pAR060302, we sought to define the baseline transcriptome of IncA/C plasmids under laboratory growth and in the face of selective pressure. The effects of ampicillin, florfenicol, or streptomycin exposure were compared to those on cells left untreated at logarithmic phase using Illumina platform-based RNA sequencing (RNA-Seq). Under growth in Luria-Bertani broth lacking antibiotics, much of the backbone of pAR060302 was transcriptionally inactive, including its putative transfer regions. A few plasmid backbone genes of interest were highly transcribed, including genes of a putative toxin-antitoxin system and an H-NS-like transcriptional regulator. In contrast, numerous genes within the accessory regions of pAR060302 were highly transcribed, including the resistance genes floR, blaCMY-2, aadA, and aacA. Treatment with ampicillin or streptomycin resulted in no genes being differentially expressed compared to controls lacking antibiotics, suggesting that many of the resistance-associated genes are not differentially expressed due to exposure to these antibiotics. In contrast, florfenicol treatment resulted in the upregulation of floR and numerous chromosomal genes. Overall, the transcriptome mapping of pAR060302 suggests that it mitigates the fitness costs of carrying resistance-associated genes through global regulation with its transcriptional regulators.
INTRODUCTION
Antimicrobial resistance is a growing public health concern, as the emergence and rapid dissemination of multidrug resistance (MDR) phenotypes among human and animal pathogens present limited options for disease treatment. The spread of MDR-encoding genetic determinants has been attributed to indiscriminate use of antimicrobial compounds in animal agriculture and human health, although it is still unclear if there are factors aside from antimicrobial selection pressure that drive the persistence of these resistance genes (1, 32, 34). Often, these genes reside on mobile genetic elements that have been circulating among bacteria for far longer than antibiotics have been used by humans (19, 23). Horizontal gene transfer of these elements provides a mechanism for rapid spread of MDR among bacteria; thus, the factors driving the persistence and dissemination of these mobile elements play a large role in the circulation of resistance genes in the absence of selection pressure.
Horizontal gene transfer plays a significant role in the introduction of new genes into microbial populations (28). Conjugative plasmids, integrons, and transposons all contribute as vehicles for this process. Plasmids are extrachromosomal genetic elements that orchestrate their own replication. In some cases, they are able to move via conjugative transfer. The sizes of plasmids can vary widely (1.5 kb to >600 kb), and plasmids are generally considered modular in nature. Core components of plasmids are required for their replication, stability, and, in some cases, transfer. The acquisition of accessory elements by plasmids can result in a wide variety of phenotypes of their host recipient, such as antimicrobial and heavy metal resistance, metabolism of organic compounds, and virulence (11, 13, 21, 25). Plasmids belonging to the A/C incompatibility plasmid group (IncA/C) are a mosaic of a highly conserved core backbone with variable accessory elements. These are large (ca. 140- to 160-kb) broad-host-range plasmids and have been isolated from diverse groups of Proteobacteria found in the environment, food animals, food products, and human pathogens (14, 25, 29, 40). IncA/C plasmids are of particular concern because they carry genes that encode resistance to many different antimicrobial compounds, including blaCMY-2, which encodes an AmpC-like β-lactamase that confers resistance to extended-spectrum cephalosporins (20). The physical linkage of this gene to other resistance genes carried on IncA/C plasmids presumably allows it to persist in the absence of cephalosporin use. The ability of IncA/C plasmids to spread blaCMY-2 and cotransfer resistance genes to a diverse range of bacteria poses a threat to both human and animal health.
Numerous IncA/C plasmids have been fully sequenced (6, 10, 12), though little is known about their basic biology. One such plasmid, pAR060302, is an ∼166-kb plasmid carrying genes for MDR that was isolated from an Escherichia coli strain (strain AR060302) from a dairy cow in 2003 (6). The backbone of this plasmid appears to contain genes required for replication, maintenance, and conjugative transfer. Additionally, three distinct accessory elements are encoded on this plasmid: an ISCR2-containing element that carries the floR, tetA, strBA, and sul2 genes; an ISEcp1-containing element that carries the blaCMY-2 gene; and a Tn21-like, class I integron-containing element that carries several different gene cassettes, including aadA, which confers aminoglycoside resistance. Although many of the phenotypes have been described for this plasmid, the extent to which these genes are transcribed and regulated remains unknown. Similar to many other plasmids, the transcriptional pattern of the plasmid backbone genes and accessory elements is likely regulated by a combination of plasmid-encoded transcriptional regulators and host factors (42). Environmental conditions could also play a role in this regulation. The magnitude of the transcription of these genes can impact phenotypes encoded on these plasmids (30). Due to the presence of multiple antimicrobial resistance genes on IncA/C plasmids, their propensity to acquire new genes, their broad host range, and their rapid dissemination, there is an immediate need to understand the underlying mechanisms of this plasmid's regulation.
The present study aimed to characterize the transcriptome of pAR060302, a prototypical blaCMY-2-positive IncA/C plasmid, under laboratory conditions using Illumina platform-based RNA sequencing (RNA-Seq) methods. We hypothesized that the level of transcription would vary between plasmid backbone genes and those carried on accessory elements and that the transcriptional landscape of pAR060302 would differ under antimicrobial selection pressure. Therefore, we mapped the transcriptome of pAR060302 to provide initial insights into the regulation of gene expression of this plasmid.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strain DH5α harboring pAR060302 was grown in 10-ml Difco Luria-Bertani (LB) broth aliquots at 37°C with shaking until an optical density at 600 nm of 0.5 was reached. A total of eight cultures representing two biological replicates per condition tested were independently grown. Six of the cultures, two cultures per antibiotic, were amended with ampicillin (final concentration, 50 μg/ml), florfenicol (final concentration, 30 μg/ml), or streptomycin (final concentration, 50 μg/ml) and allowed to incubate at 37°C with shaking for an additional 30 min. Two cultures were not amended with any antibiotic. Cells were pelleted, and RNA was purified using a commercially available RNA extraction kit (Qiagen). RNA preparations were then subjected to a DNase treatment to eliminate DNA contamination from the sample (Qiagen). A treatment to deplete rRNA using a commercially available kit (MicrobExpress; Ambion) was also included. The two biological replicates for each growth condition were pooled for sequencing.
Ilumina platform-based sequencing for transcriptome mapping.
cDNA libraries with an insert size of 100 bp were generated and sequenced with 76-base cycles of single-end reads, using a Genome Analyzer II platform (Illumina) according to the manufacturer's protocols, at the Biomedical Genomics Center (University of Minnesota, Minneapolis, MN). Approximately 160,000 plasmid-mapped reads each were obtained for the ampicillin- and streptomycin-treated samples, and 260,000 plasmid-mapped reads each were obtained for the control and florfenicol-treated samples. Genome-mapped read counts were as follows: control, ∼6.4 million reads; florfenicol treatment, ∼5.7 million reads; ampicillin treatment, ∼1.7 million reads; and streptomycin treatment, ∼5.2 million reads. We used only those reads uniquely mapped on plasmid or chromosomal DNA for global normalization and further analysis.
RNA-Seq data analysis.
cDNA reads were trimmed so that the quality at each base position was above 30 (∼15 to 20 bp) and then mapped either to the E. coli K-12 MG1655 published genome sequence (GenBank accession no. NC_000913) or to the pAR060302 published sequence (GenBank accession no. NC_092692) using the BOWTIE program (24). E. coli strain DH5α has an incomplete annotation, and for this reason, the E. coli K-12 annotation, representing an estimation of differentially expressed genes due to exposure to antimicrobials, was used. For each condition, graphs representing the number of mapped reads per nucleotide were generated and visualized using the Integrated Genome Viewer (IGV) (31). Images were created using the XplasMap program (Ian York) and IGV. The number of reads mapped per kilobase of gene per million reads (RPKMs) was calculated using either the E. coli chromosome or the pAR060302 annotation and was used for global normalization (27). The per kilobase cDNA length normalized the effect of different lengths of cDNAs, such that the sequence reads have an equal chance to map on the long cDNA regions and the short cDNA regions. After RPKM normalization, each sample is comparable to the others. An R package, DEGseq (39), was used to identify differentially expressed genes between the control and each antibiotic treatment condition. A cutoff of q value of <0.05 and a fold change of >3 were used to measure statistical significance (39).
RESULTS
Transcription of plasmid core regions.
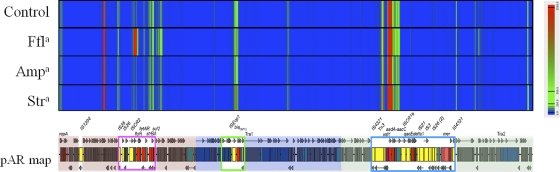
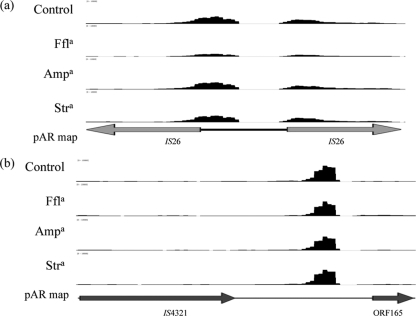
Much of the putative backbone of pAR060302 was transcriptionally inactive under the conditions examined (Fig. 1). Only about 18% of the coding sequences located in core regions were transcribed. The repA replication initiation gene was expressed at a low level (RPKMs, ∼3,700 to 4,700). This gene was used for subsequent comparisons because it is involved with plasmid replication and was assumed to be expressed consistently during exponential growth due to continuous cell division. Just one other locus, the one containing ORF_051-053, was expressed on the backbone of the plasmid in a pattern to similar that for repA (Table 1). BLASTp analysis of ORF_051-053 identified two of these putative proteins as having functions pertaining to stability and maintenance. Two additional regions were transcribed at high levels but had not previously been annotated (Fig. 2). These two regions have short predicted open reading frames (ORFs; <65 amino acids); however, it is unclear which small peptides, if any, are expressed by these transcripts. Nucleotide BLAST (megablast) analysis of the regions showed that they had similarity only to other IncA/C plasmids, which indicates that it is unlikely that these reads were mapped incorrectly. These regions did not have similarity to the host E. coli genomic sequences and therefore did not originate from chromosomal transcripts.
Fig 1.
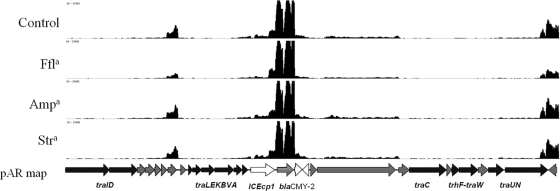
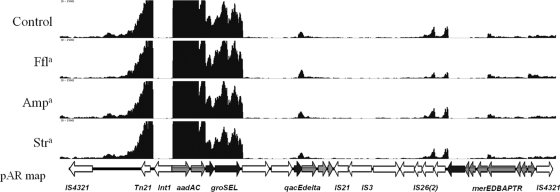
Heat map displaying transcriptome map of pAR060302. Blue indicates no or low expression (0 to 75 reads aligned per nucleotide position), green indicates moderate expression (250 to 500 reads aligned per nucleotide position), and red indicates high expression (>2,500 reads aligned per nucleotide position). In the pAR060302 (pAR) map, the shaded regions indicate the plasmid backbone, as follows: red, IncA/C replicon and hypothetical genes; blue, Tra1 region; and green, hypothetical genes and Tra2 region. Open boxes depict the accessory regions, as follows: pink, flo-tet-sul-containing region (enlarged in Fig. 4); green, blaCMY-2-containing region (enlarged in Fig. 5); and blue, Tn21-like region, which contains a class 1 integron (enlarged in Fig. 6). a, antibiotics are abbreviated as follows: ampicillin, Amp; florfenicol, Ffl; and streptomycin, Str.
Table 1.
RPKM values calculated for transcribed genes of interest
| Gene function | Gene or ORF | Function or predicted function | RPKMa |
|||
|---|---|---|---|---|---|---|
| Control | Ffl | Amp | Str | |||
| Putative maintenance and stability genes | repA | Replication initiation | 4,376 | 3,780 | 4,772 | 4,753 |
| pAR060302_0051 | Putative ParA partition protein | 6,942 | 11,306 | 7,534 | 7,066 | |
| pAR060302_0052 | Putative ParB partition protein | 14,265 | 20,154 | 13,480 | 14,939 | |
| Putative DNA binding genes | pAR060302_0023 | Putative IHFb transcriptional regulator | 5,130 | 2,097 | 5,583 | 5,243 |
| pAR060302_0025 | Putative antitoxin component protein | 206,070 | 361,900 | 201,615 | 203,271 | |
| pAR060302_0026 | Putative toxin component protein | 86,543 | 148,982 | 85,362 | 81,898 | |
| pAR060302_0027 | Putative DNA binding protein | 3,176 | 3,074 | 3,431 | 3,769 | |
| pAR060302_0188 | Putative H-NS-like protein | 23,351 | 14,876 | 26,414 | 27,179 | |
| Resistance genes | floR | Phenicol resistance | 8,543 | 71,346c | 8,463 | 8,845 |
| tetA | Tetracycline resistance | 991 | 1,544 | 958 | 993 | |
| blaCMY-2 | Extended-spectrum β-lactamase | 34,314 | 35,415 | 32,488 | 32,308 | |
| aadA | Aminoglycoside resistance | 323,889 | 217,434 | 334,860 | 330,472 | |
| aacA | Aminoglycoside resistance | 64,841 | 61,992 | 56,338 | 61,287 | |
| Genes within the putative transfer loci | pAR060302_0075 | Hypothetical protein | 8,277 | 3,343 | 8,070 | 8,170 |
| pAR060302_0097 | Hypothetical protein | 13,364 | 6,383 | 14,597 | 13,921 | |
| pAR060302_0183 | Hypothetical protein | 3,452 | 5,266 | 3,985 | 4,104 | |
RPKM is calculated as the number of reads aligned per kilobase of gene per millions total reads. Antibiotics are abbreviated as follows: ampicillin, Amp; florfenicol, Ffl; and streptomycin, Str.
IHF, integration host factor.
A >3-fold change in expression that is statistically significant (q < 0.05).
Fig 2.
Transcription of regions not previously annotated. (a) Region between two IS26 elements upstream of the ISCR2 elements; (b) intergenic transcript downstream of the class I integron. Antibiotic treatment conditions are labeled along the x axis. The x axis has a scale of 0 to 2,500 reads aligned per base. Arrows indicate direction of transcription. a, antibiotics are abbreviated as follows: ampicillin, Amp; florfenicol, Ffl; and streptomycin, Str.
The majority of the genes located in the Tra1 and Tra2 regions of pAR060302 were not transcribed under the conditions that we used for assays. Only three genes within these loci, ORFs 75, 97, and 183, were transcribed under the conditions examined here (Table 1). The expression levels of these genes were comparable to the expression level of repA, and the products of all of them are all annotated as hypothetical proteins. The product of the gene locus pAR060302_183 is annotated as a hypothetical protein. This ORF is conserved among IncA/C plasmids but shares no similarity to sequences of ORFs for proteins with known functions. Its location is similar to the locations of the regulators of the transfer regions of SXT-like integrative conjugative elements (ICEs) that share significant homology with IncA/C plasmids (41). However, no work to characterize this gene has previously been done.
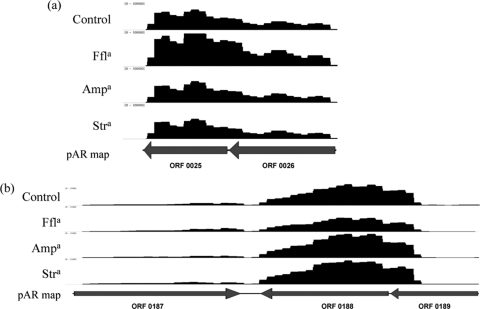
A few genes on the backbone of pAR060302 were expressed at high levels. Genes in one set, pAR060302_25 and pAR060302_26, were transcribed at levels 47- and 20-fold higher than repA, respectively (Table 1; Fig. 3a), and display BLAST similarity to genes of a toxin-antitoxin system, although this system has not been experimentally characterized as having toxin-antitoxin function. pAR060302_25 is a putative XRE-like transcriptional regulator and the putative antitoxin component, while pAR060302_26 is a putative toxin component of the system. These predicted proteins share amino acid similarity (70%) to a chromosomal locus found in Salmonella enterica serovar Paratyphi B strain SPB7. A third gene, pAR060302_188, was transcribed at a level 5-fold higher than repA (Table 1; Fig. 3b) and is annotated as a putative H-NS-like DNA-binding protein. Analysis of the predicted amino acid sequence of pAR060302_0188 showed 52% sequence similarity to its closest match and did not show any significant similarity to other H-NS proteins encoded on plasmids from other Gram-negative bacteria; however, it is conserved among all sequenced IncA/C plasmids (10).
Fig 3.
Transcriptome maps of plasmid backbone genes of interest. (a) pAR060302_0025 and pAR060302_0026 are transcribed at extremely high levels. The x axis has a scale of 0 to 30,000 reads aligned per nucleotide position. Arrows indicate direction of transcription. (b) pAR060302_0188 is transcribed as a single gene transcript. The x axis has a scale of 0 to 2,500 reads aligned per nucleotide position. Arrows indicate direction of transcription. a, antibiotics are abbreviated as follows: ampicillin, Amp; florfenicol, Ffl; and streptomycin, Str.
Transcription pattern of pAR060302 accessory regions.
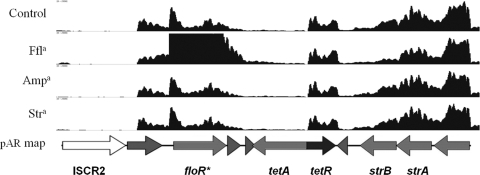
There are at least three hot spots for insertion of mobile genetic elements on IncA/C plasmids (6, 10, 12). In pAR060302, the first hot spot carries an ISCR2-containing mobile genetic element that contains the floR, tetRA, strBA, and sul2 resistance gene cluster, as well as several putative genes with predicted regulator capabilities. This inserted module is highly conserved among blaCMY-2-positive IncA/C plasmids (6). The floR gene was transcribed on a transcriptional unit that includes three ORFs, and this transcript was apparently promoted by a dual-promoter system (Fig. 4). A transcriptional start site (TSS) was located upstream of ORF_0040, which encodes a conserved hypothetical protein, and an inducible promoter was located just upstream of the floR ORF. The transcription of floR from this promoter site was increased ∼8-fold (q < 0.05) after exposure to florfenicol for 30 min. Immediately downstream of floR, pAR060302_0042 was also upregulated. This gene likely resides on the same transcript, given the pattern of aligned reads that was observed (Fig. 4). The product of pAR060302_0042 is annotated as a hypothetical protein. BLASTp analysis of the predicted amino acid sequence of this ORF showed a predicted helix-turn-helix domain and showed that it shares homology with the LysR family of transcriptional regulators. No changes in transcription of this region were observed in the presence of ampicillin or streptomycin. Located downstream of the floR locus on the opposite strand is the tetRA gene cluster, one of the most abundant genetic factors for antimicrobial resistance in Gram-negative bacteria. The transcriptional regulation of tetRA has been previously characterized (17), and similar phenomena were observed here. That is, only the repressor gene tetR was transcribed while the tetA gene was under repression. This gene cluster has previously been shown to be upregulated in response to tetracycline exposure (17) but was unaffected by the antimicrobial compounds that we tested here. The strBA and sul2 genes, encoding streptomycin and sulfonamide resistance, respectively, are located on the ISCR2 element upstream of tetRA and were transcribed together on the same transcript, originating with a TSS located just upstream of sul2. Florfenicol, ampicillin, or streptomycin treatment had no effect on the abundance of this transcript.
Fig 4.
Transcriptome map of the sul2 containing region of pAR060302. Antibiotic treatment conditions are labeled along the y axis. The x axis has a scale of 0 to 2,500 reads aligned per base. Arrows indicate direction of transcription. a, antibiotics are abbreviated as follows: ampicillin, Amp; florfenicol, Ffl; and streptomycin, Str. *, a statistically significant greater or less than log2 (fold change) in expression (q < 0.05).
The second hot spot in pAR060302 contains the blaCMY-2 gene, which is part of an ISEcp1-containing mobile genetic element also containing blc1 and sugE1. It is inserted among plasmid backbone genes predicted to be involved with conjugative transfer (Fig. 5). There are 22 ORFs in the previously defined Tra1 gene cluster (10). In all blaCMY-2-positive IncA/C plasmids that have been sequenced thus far, the ISEcp1 mobile element containing blaCMY-2 is inserted in this region (6). In pAR060302, this insertion is located downstream of traA, but this site of insertion is not conserved among all IncA/C plasmids. The ISEcp1-containing element of pAR060302 appeared to carry its own strong promoter just downstream of ISEcp1 conferring a high level of transcription of blaCMY-2, which was not changed in the presence of ampicillin, florfenicol, or streptomycin. Downstream of blaCMY-2, both blc1 and sugE1 were minimally transcribed compared to the level of blaCMY-2 transcription, indicating that there may be a transcription stop site between blaCMY-2 and these two genes.
Fig 5.
Transcriptome map of the blaCMY-2-containing Tra1 region of pAR060302. Antibiotic treatment conditions are labeled along the x axis. The x axis has a scale of 0 to 2,500 reads aligned per base. Arrows indicate direction of transcription. a, antibiotics are abbreviated as follows: ampicillin, Amp; florfenicol, Ffl; and streptomycin, Str.
Nearly all sequenced blaCMY-2-positive IncA/C plasmids carry a Tn21-like class I integron region in their third accessory hot spot (6). The class I integron of pAR060302 contains aminoglycoside resistance genes, aadA and aacC; heat shock chaperones groSEL; the qacEΔl and sul1 genes; and the merEDBAPTR mercury resistance operon. Our data suggest that a transcript started from an apparently strong promoter upstream of aadA and continued until just downstream of groEL, where transcription seemingly halted. aadA was transcribed at a level 74-fold higher than repA, while aacC and groSEL were transcribed at decreasing levels (Fig. 6). This position-dependent transcription of integrons has been previously observed (7). The transcription of this region was not affected by treatment with ampicillin, florfenicol, or streptomycin. The merDBAPTR gene cassette, previously shown to be activated in the presence of mercuric compounds (36), was unaffected by treatment with the three antibiotics used here.
Fig 6.
Transcriptome map of the class I integron-containing region of pAR060302. Antibiotic treatment conditions are labeled along the y axis. The x axis has a scale of 0 to 2,500 reads aligned per base. Arrows indicate direction of transcription. a, antibiotics are abbreviated as follows: ampicillin, Amp; florfenicol, Ffl; and streptomycin, Str.
E. coli chromosomal transcriptional response to exposure to antimicrobials.
Reads obtained from each treatment were also mapped to the chromosome of the E. coli DH5α host. Neither the ampicillin nor the streptomycin treatment yielded differential gene transcription on the chromosome that was statistically significant. However, the florfenicol treatment resulted in 286 chromosomal genes which were transcribed at statistically significantly different levels compared to the control cells. Most of these genes (∼250) were downregulated, while the remaining genes were upregulated. Among the genes that had increased expression were rbfA, deaD, cspA, and cspG, which are annotated as being induced upon cold shock. Two transmembrane transporters, proP and mdtK, were also upregulated. Several genes annotated as part of the σs-directed stress response were downregulated. These include the dps gene, which encodes a histone-like protein that forms clusters with DNA; the acid response genes gadC and hdeAB; and the cfu gene, which is involved with alterations of the cell membrane (35).
DISCUSSION
IncA/C plasmids have been increasingly associated with the dissemination of antimicrobial resistance genes in populations of enteric bacteria from humans and animals. To gain insights on the biology of these plasmids, we conducted RNA sequencing experiments, resulting in the first report of the transcriptional landscape of these large plasmids. IncA/C plasmids are distributed globally yet remain highly similar, aside from their accessory elements (6, 10, 12). Data generated in this study, therefore, may be applied broadly across the many sequenced IncA/C derivatives. Illumina-based RNA-Seq provided a robust approach to mapping the transcriptional levels down to the single base pair. This provided an opportunity to amend the current annotation of pAR060302 and gain insights into the genetic determinants of plasmid maintenance and stability.
When studying plasmid biology, the host carrying any given plasmid is inherently important. Plasmids rely on their host cell for aspects of DNA replication and gene expression. In the present study, E. coli strain DH5α was used as the host cell. This common lab strain is well adapted to growth in lab media. It is probable that the extent to which DH5α is able to respond to external stimuli is severely muted compared to that of wild-type bacteria. In the present study, we observed that only florfenicol treatment altered chromosomal transcription, which was unexpected. No clear pattern emerged from the analysis of the differentially expressed chromosomal genes. This may be evidence that E. coli DH5α is ill equipped to respond to environmental stresses. Another likely possibility could be that our strain and plasmid have a naïve host-plasmid relationship and this interaction may be confounding the effects of antimicrobial exposure. Indeed, other hosts would likely have a larger impact on plasmid transcriptional profiles due to the overlap of their spectra of regulatory networks. The ability of chromosomal regulatory networks to interact with plasmid-encoded regulatory networks ultimately governs plasmid gene expression. Therefore, while the present study sought to define IncA/C transcription in the absence of complex interactions between the host chromosome and plasmid, it will be necessary in future work to study the effects of IncA/C transcription in additional bacterial hosts and under additional growth conditions to fully appreciate the plasmid-host relationship.
Two of the three accessory regions of pAR060302 were transcribed at high levels compared to the backbone of the plasmid, while the floR-tetRA-strAB-sul2 region was not. Evidence of transcription of these genes supports phenotypic observations of their ability to confer resistance against high concentrations of antimicrobial compounds. However, there could also be consequences associated with their expression at high levels in the absence of antibiotic selection pressure. A recent study by Call et al. (6) showed that the blaCMY-2 region of pAR060302 is prone to stochastic excision and, in the absence of selection pressure, populations of E. coli harboring blaCMY-2 were outcompeted by blaCMY-2-lacking plasmids over many generations. Other studies have shown a significant cost of carrying other variants of AmpC-like β-lactamase genes (18, 26). It was hypothesized that this cost is a result of unregulated expression and that this cost of carriage results in reduced dissemination. It is also possible that IncA/C plasmids mitigate the fitness cost conferred by blaCMY-2, to some extent, to allow its persistence and spread.
Call et al. (6) also showed that the flo-tet region was prone to apparent excision over time but required significantly more generations to be lost from pAR060302-containing bacterial populations. We show here that the floR gene is transcribed at relatively low levels compared to the other resistance genes of pAR060302 but that its transcription is seemingly induced by the presence of florfenicol. Florfenicol has previously been shown to increase expression of other genes, and this was found to be a result of the increased half-life of mRNA due to the presence of florfenicol (5). Still, the transcript containing floR was the only plasmid-encoded mRNA that was significantly upregulated in response to antibiotic treatment. Other mechanisms resulting in the induction of expression of resistance genes by exposure to florfenicol have been described before (22). The putative transcriptional regulator immediately downstream of floR may represent a control mechanism for the regulated transcription of floR. No previous studies have addressed this. However, since florfenicol was the only treatment that resulted in altered chromosomal expression, this may instead be a result of chromosome-plasmid cross talk. Given that many genes on the chromosome were differentially transcribed, why only a specific mRNA on the plasmid was affected remains in question. It is unclear at this time what the molecular basis for the increased floR transcripts that we observed might be. Others have shown that the tetRA region is induced by tetracycline (17), which is likely the case in pAR060302, given our results. While strAB was transcribed and its transcription was not induced by the addition of streptomycin to the growth medium in our experiments, it was transcribed at a much lower level than blaCMY-2, aadA, or aacC. Thus, the increased stability of the flo-tet region observed by others could be correlated to a lower fitness cost for the carriage due to its apparent regulation. This is in contrast to the apparently uncontrolled transcription of blaCMY-2 and genes within the Tn21-like region, which were highly transcribed under all of our study conditions. The stability of the Tn21-like region over long periods of time in the absence of antimicrobial selection pressure has yet to be measured, although we expect that portions of this region would also be deleted over time due to the relative cost of high transcription. This could, in part, explain why some sequenced IncA/C plasmids, such as pAM04528 (6, 10), possess only the mer operon and lack other Tn21-associated genes. Further analysis is required to verify the impact of gene expression on the evolution of IncA/C plasmids.
Several genes that might be key to this plasmid's success have been identified here, including several putative plasmid-encoded transcriptional regulators that were transcribed at high levels compared to other backbone components. In other plasmid systems, similar genes have been shown to have a wide array of effects. For example, the IncP-7 plasmid group encodes an H-NS-like protein, Pmr, and recent studies have shown that Pmr alters the transcriptional landscape of the host chromosome (42). R27, an IncH plasmid isolated from Salmonella enterica serovar Typhimurium, encodes Sfh, another H-NS-like protein. Deletion of this gene resulted in a nearly 4-fold increase in the number of chromosomal genes that were differentially expressed due to plasmid acquisition (8). These H-NS-like proteins have been termed “stealth-like” and have been hypothesized to silence the effects of plasmid acquisition on the host chromosome. A recent survey of sequenced plasmids found that similar genes reside on many known broad-host-range plasmids (37). All sequenced IncA/C variants encode a putative H-NS-like protein that could represent a genetic factor enabling transfer and/or stability among diverse host backgrounds. H-NS orthologs might provide alleviation of the cost of plasmid carriage incurred by the host cell. However, the putative H-NS-like regulator of pAR060302 shares low amino acid similarity to its closest known match (10), so it is still necessary to elucidate the role, if any, that this gene plays in the success of IncA/C plasmids in their diverse hosts.
The putative toxin-antitoxin system of pAR060302 was also highly transcribed under the conditions studied. Such systems are well-known for their involvement in postsegregational killing in plasmid-free daughter cells (38). The transcription of this system in pAR060302 suggests that IncA/C plasmids ensure their stability in bacterial populations by potentially killing plasmid-free progeny that may arise. However, no previous work has been done to describe the function of these genes found in IncA/C plasmids.
Within the putative conjugal transfer regions of pAR060302, only a few genes of unknown function were transcribed in our study. pAR060302 has been shown by our group and by others to be transferrable (6). The apparent conjugal transfer machinery of IncA/C plasmids is unlike that of any other known plasmids but is highly syntenic to the transfer loci of SXT/R391 ICEs (41). These ICEs have a highly regulated transfer mechanism, suggesting that the same is true for IncA/C plasmids, yet the key regulators of R391-like ICEs are missing from IncA/C plasmids, pointing to potentially novel mechanisms of regulation (2–4). Several scenarios could explain the regulation of the transfer region in IncA/C plasmids. One possibility is that the region may simply be unregulated; however, this is unlikely due to the inability to observe transcription of the tra genes. Another possibility is that the transfer loci may be under host cell control, as is the case for plasmids that undergo zygotic induction; this, too, seems unlikely for a plasmid with such a broad host range, since relying on host-specific factors would result in an evolutionary dead-end if the plasmid were to transfer to a host unfit for such regulation. A third possibility is that IncA/C plasmids may encode their own genes for regulation of the transfer loci. This seems to be the most likely scenario, since these plasmids harbor an array of putative regulators, some of which might regulate the expression of the transfer loci. Other plasmid systems rely on a signal from suitable recipients to initiate plasmid transfer, and although this has not been described for plasmids in Gram-negative bacteria, it remains to be tested for IncA/C plasmids (9). Given the role that IncA/C plasmids play in the dissemination of resistance genes among strains of the Enterobacteriaceae, the mechanisms by which they are transferred deserve further study.
The data generated in this study afforded us the opportunity to examine the annotation of pAR060302 on the basis of experimental evidence. The two short transcripts in areas not previously annotated could represent small peptides encoded on pAR060302. They could, alternatively, represent small noncoding RNAs (ncRNAs). In E. coli, ncRNAs have previously been shown to be up to 300 bp in length (15). Although the experimental design of our study was not specifically created to isolate ncRNAs, these transcripts are of appropriate length to be observed under our conditions. Further work is needed to identify the nature of these transcripts.
A limitation to this work is that the data represent only a fraction of pAR060302 transcription potential, since we analyzed only a single growth point of E. coli harboring pAR060302 in a single growth medium. This provides insight into the minimum genetic components required for IncA/C plasmid maintenance under laboratory growth conditions. Our first hypothesis about transcriptional differences between plasmid accessory and backbone elements held true; it is remarkable that much of the backbone of pAR060302, including regions predicted to be involved in plasmid maintenance and stability, was transcriptionally inactive. This could imply the possibility of extensive regulation and cross talk between plasmid and host or self-regulation of the plasmid itself. We also hypothesized that the transcriptional profile of the plasmid would respond to antibiotic treatment, which was true only in the case of florfenicol treatment. It is probable that changes could occur if different classes of antibiotics were used, but it is interesting that many of the resistance genes were transcribed at high levels in the absence of antibiotics.
Overall, RNA-Seq offers a robust approach to assess global-scale transcription. In our data we can observe a polarity to the transcripts, which can offer clues to the location of the transcriptional start site. However, an RNA-Seq experimental design different from that used here would be needed to accurately map the start of nascent transcripts (16, 33). Findings from further studies such as these will move us toward a global understanding of the expression of the genes on IncA/C plasmids and the implication of such expression on their overall biology. Host strain-dependent transcriptional profiles of other plasmids, such as IncP, have been previously reported (8, 42). Although we expect differences in transcription of pAR060302 in its wild-type host, E. coli strain DH5α was used in this study to obtain a baseline transcriptional profile of pAR060302. Our observations of pAR060302 transcription indicate that it is likely highly regulated, allowing its dissemination to a broad range of bacterial hosts and its stability within these hosts, despite the likely costs of encoding highly transcribed accessory regions encoding antibiotic resistance. Future efforts will use these data to better understand the impacts of host background and plasmid-borne genes on plasmid transcription and regulation.
ACKNOWLEDGMENTS
We thank Stefan Schwartz, Friedrich-Loeffler-Institut, for providing insights on the effects of florfenicol on mRNA and Randall Singer, University of Minnesota, for providing pAR060302 and its wild-type strain.
Data analysis was carried out using tools available through the Minnesota Supercomputing Institute.
Footnotes
Published ahead of print 17 February 2012
REFERENCES
- 1. Alonso A, Sánchez P, Martínez JL. 2001. Environmental selection of antibiotic resistance genes. Environ. Microbiol. 3:1–9 [DOI] [PubMed] [Google Scholar]
- 2. Beaber JW, Hochhut B, Waldor MK. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beaber JW, Hochhut B, Waldor MK. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72–74 [DOI] [PubMed] [Google Scholar]
- 4. Beaber JW, Waldor MK. 2004. Identification of operators and promoters that control SXT conjugative transfer. J. Bacteriol. 186:5945–5949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blickwede M, Goethe R, Wolz C, Valentin-Weigand P, Schwarz S. 2005. Molecular basis of florfenicol-induced increase in adherence of Staphylococcus aureus strain Newman. J. Antimicrob. Chemother. 56:315–323 [DOI] [PubMed] [Google Scholar]
- 6. Call DR, et al. 2010. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 54:590–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Depardieu F, Podglajen I, Leclercq R, Collatz E, Courvalin P. 2007. Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 20:79–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doyle M, et al. 2007. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science 315:251–252 [DOI] [PubMed] [Google Scholar]
- 9. Dunny GM, Johnson CM. 2011. Regulatory circuits controlling enterococcal conjugation: lessons for functional genomics. Curr. Opin. Microbiol. 14:174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fernández-Alarcón C, Singer RS, Johnson TJ. 2011. Comparative genomics of multidrug resistance-encoding IncA/C plasmids from commensal and pathogenic Escherichia coli from multiple animal sources. PLoS One 6:e23415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fricke WF, et al. 2009. Antimicrobial resistance-conferring plasmids with similarity to virulence plasmids from avian pathogenic Escherichia coli strains in Salmonella enterica serovar Kentucky isolates from poultry. Appl. Environ. Microbiol. 75:5963–5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fricke WF, et al. 2009. Comparative genomics of the IncA/C multidrug resistance plasmid family. J. Bacteriol. 191:4750–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722–732 [DOI] [PubMed] [Google Scholar]
- 14. Galimand M, et al. 1997. Multidrug resistance in Yersinia pestis mediated by a transferable plasmid. N. Engl. J. Med. 337:677–680 [DOI] [PubMed] [Google Scholar]
- 15. Gottesman S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms*. Annu. Rev. Microbiol. 58:303–328 [DOI] [PubMed] [Google Scholar]
- 16. Güell M, Yus E, Lluch-Senar M, Serrano L. 2011. Bacterial transcriptomics: what is beyond the RNA horiz-ome? Nat. Rev. Microbiol. 9:658–669 [DOI] [PubMed] [Google Scholar]
- 17. Hillen W, Berens C. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345–369 [DOI] [PubMed] [Google Scholar]
- 18. Hossain A, Reisbig MD, Hanson ND. 2004. Plasmid-encoded functions compensate for the biological cost of AmpC overexpression in a clinical isolate of Salmonella typhimurium. J. Antimicrob. Chemother. 53:964–970 [DOI] [PubMed] [Google Scholar]
- 19. Hughes VM, Datta N. 1983. Conjugative plasmids in bacteria of the “pre-antibiotic” era. Nature 302:725–726 [DOI] [PubMed] [Google Scholar]
- 20. Jacoby GA. 2009. AmpC β-lactamases. Clin. Microbiol. Rev. 22:161–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson TJ, et al. 2010. Sequence analysis and characterization of a transferable hybrid plasmid encoding multidrug resistance and enabling zoonotic potential for extraintestinal Escherichia coli. Infect. Immun. 78:1931–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kehrenberg C, Schwarz S. 2004. fexA, a novel Staphylococcus lentus gene encoding resistance to florfenicol and chloramphenicol. Antimicrob. Agents Chemother. 48:615–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koonin EV, Wolf YI. 2008. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 36:6688–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lindsey RL, Fedorka-Cray PJ, Frye JG, Meinersmann RJ. 2009. Inc A/C plasmids are prevalent in multidrug-resistant Salmonella enterica isolates. Appl. Environ. Microbiol. 75:1908–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marciano DC, Karkouti OY, Palzkill T. 2007. A fitness cost associated with the antibiotic resistance enzyme SME-1 β-lactamase. Genetics 176:2381–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5:621–628 [DOI] [PubMed] [Google Scholar]
- 28. Norman A, Hansen LH, Sørensen SJ. 2009. Conjugative plasmids: vessels of the communal gene pool. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:2275–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pan J-C, et al. 2008. Vibrio cholerae O139 multiple-drug resistance mediated by Yersinia pestis pIP1202-like conjugative plasmids. Antimicrob. Agents Chemother. 52:3829–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramos JL, Marqués S, Timmis KN. 1997. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu. Rev. Microbiol. 51:341–373 [DOI] [PubMed] [Google Scholar]
- 31. Robinson JT, et al. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Salyers AA, Amábile-Cuevas CF. 1997. Why are antibiotic resistance genes so resistant to elimination? Antimicrob. Agents Chemother. 41:2321–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sharma CM, et al. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–255 [DOI] [PubMed] [Google Scholar]
- 34. Singer RS, Ward MP, Maldonado G. 2006. Can landscape ecology untangle the complexity of antibiotic resistance? Nat. Rev. Microbiol. 4:943–952 [DOI] [PubMed] [Google Scholar]
- 35. Storz G, Hengge-Aronis R. 2000. Bacterial stress responses. ASM Press, Washington, DC [Google Scholar]
- 36. Summers AO. 1986. Organization, expression, and evolution of genes for mercury resistance. Annu. Rev. Microbiol. 40:607–634 [DOI] [PubMed] [Google Scholar]
- 37. Takeda T, Yun C-S, Shintani M, Yamane H, Nojiri H. 2011. Distribution of genes encoding nucleoid-associated protein homologs in plasmids. Int. J. Evol. Biol. 2011:685015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Van Melderen L. 2010. Toxin-antitoxin systems: why so many, what for? Curr. Opin. Microbiol. 13:781–785 [DOI] [PubMed] [Google Scholar]
- 39. Wang L, Feng Z, Wang X, Wang X, Zhang X. 2010. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138 [DOI] [PubMed] [Google Scholar]
- 40. Welch TJ, et al. 2009. IncA/C plasmid-mediated florfenicol resistance in the catfish pathogen Edwardsiella ictaluri. Antimicrob. Agents Chemother. 53:845–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wozniak RAF, et al. 2009. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 5:e1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yun C-S, et al. 2010. Pmr, a histone-like protein H1 (H-NS) family protein encoded by the IncP-7 plasmid pCAR1, is a key global regulator that alters host function. J. Bacteriol. 192:4720–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]