Abstract
NAD-dependent l- and d-lactate dehydrogenases coexist in Lactobacillus genomes and may convert pyruvic acid into l-lactic acid and d-lactic acid, respectively. Our findings suggest that the relative catalytic efficiencies of ldhL- and ldhD-encoded products are crucial for the optical purity of lactic acid produced by Lactobacillus strains.
TEXT
Lactic acid is a building block widely used in the food, pharmaceutical, and chemical industries (6). The use of lactic acid in the synthesis of polylactic acid has grown over the years, and high optical purity is an inevitable prerequisite for lactic acid polymerization (1, 23). Strains of Lactobacillus, the largest genus of lactic acid bacteria (LAB), are the most frequently used lactic acid producers (2, 4, 10, 19, 22, 24), but the optical purities of lactic acid produced by various Lactobacillus strains are markedly different (Table 1) (3, 15).
Table 1.
Comparison of enantiomeric excess (ee) values of lactic acid produced by the three representative Lactobacillus strains after 24 h of batch fermentation
| Strain | Concn of total lactic acid (g l−1) | ee value of l-lactic acid (%) | ee value of d-lactic acid (%) |
|---|---|---|---|
| L. bulgaricus ATCC 11842 | 18.1 ± 0.5 | 97.2 ± 0.1 | |
| L. plantarum ATCC 14917 | 18.4 ± 0.1 | 3.3 ± 1.7 | |
| L. casei ATCC 334 | 18.3 ± 0.1 | 92.0 ± 0.8 |
The enzymes responsible for l- and d-lactic acid production are NAD-dependent l-lactate dehydrogenases (l-nLDHs) and NAD-dependent d-lactate dehydrogenases (d-nLDHs), respectively, which fall into two different families and are encoded by ldhL and ldhD, respectively (7, 20). Lactate racemase, which converts l-lactic acid into d-lactic acid, has only been reported in a few dl-type Lactobacillus strains. The enzyme was found in only four (L. curvatus, L. paracasei, L. plantarum WCFS1, and L. sakei 23k) of hundreds of Lactobacillus strains that have been sequenced or reported. However, both ldhL and ldhD have been shown to be widely distributed in all sequenced or reported Lactobacillus strains except L. sakei 23k (7, 8, 13) (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). Due to the fact that the optical purities of lactic acid are clearly different and in some cases even completely opposite in various Lactobacillus strains (3, 15), studies focusing on the relationship between the ldhL and ldhD genes and the optical purity of lactic acid are needed.
In this study, Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842, L. plantarum ssp. plantarum ATCC 14917, and Lactobacillus casei ATCC 334 were selected as representative strains for exhaustive analysis. These three species are lactic acid high-producing strains, and their lactic acid products represent three different types (d, dl, and l, respectively) (see the supplemental material for details of batch fermentation) (Table 1) (5, 9, 14, 16, 17, 18, 23). Moreover, both ldhL and ldhD but no lactate racemase genes or homologs were found in all three genomes during sequencing and annotation (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). First, the enzymatic activities of l- and d-nLDHs in these three Lactobacillus strains were determined using whole-cell extracts. Because the substrates for both l- and d-nLDHs are pyruvate and NADH, the reduction activity of these enzymes cannot be measured by monitoring the decrease in NADH. Instead, the reduced products, l- and d-lactic acid, were used to evaluate enzymatic activity (see the supplemental material for details). As shown in Table 2, the relative activities of l- and d-nLDHs were evidently different in the Lactobacillus strains. In L. bulgaricus ATCC 11842 (d-LAB strain), the specific activity of d-nLDH was much higher than that of l-nLDH, about 111-fold; in L. plantarum ATCC 14917 (dl-LAB strain), the specific activity of d-nLDH was similar to that of l-nLDH; and in L. casei ATCC 334 (l-LAB strain), the specific activity of l-nLDH was higher than that of d-nLDH, about 39-fold.
Table 2.
The specific activities of l- and d-nLDHs of Lactobacillus strains in exponential phase
| Strain | Sp act (μmol min−1 mg−1) |
Sp act ratio of l-nLDH to d-nLDH | |
|---|---|---|---|
| l-nLDH | d-nLDH | ||
| L. bulgaricus ATCC 11842 | (1.3 ± 0.1) × 10−1 | 14.4 ± 0.7 | 1:111 |
| L. plantarum ATCC 14917 | 2.4 ± 0.2 | 4.3 ± 0.2 | 1:1.8 |
| L. casei ATCC 334 | 12.4 ± 0.7 | (3.2 ± 0.3) × 10−1 | 39:1 |
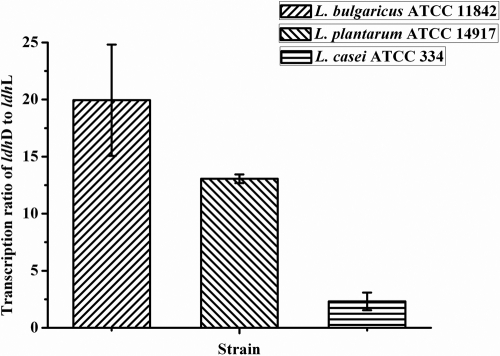
Next, the relative transcriptional levels of ldhL and ldhD in the three representative strains were examined. Little transcriptional analysis of ldhL and ldhD has been reported previously (11). In this study, cells of the three representative strains in the middle of the exponential phase were collected for RNA isolation using the E.Z.N.A. bacterial RNA kit (Omega Bio-Tek). The isolated total RNA was treated with DNase I (Fermentas) and then used as a template for cDNA synthesis with TransScript first-strand cDNA synthesis supermix (TransGen Biotech). The resultant cDNA was used for quantitative PCR (qPCR) analysis. Primers for six different genes were designed with Beacon Designer software (see Table S1 in the supplemental material). The qPCRs were performed using the LightCycler 480 Real-Time PCR system with LightCycler 480 SYBR green I master (Roche) according to the manufacturer's instructions. As shown in Fig. 1, the transcriptional levels of ldhD were higher than those of ldhL, from about 2-fold to 20-fold, in the three representative strains.
Fig 1.
Determination of the relative transcriptional levels of ldhD and ldhL by using qPCR. Error bars represent the standard deviations of the means of three independent experiments.
The catalytic efficiencies of ldhL- and ldhD-encoded products were also measured using purified proteins. The ldhL and ldhD genes of the three strains were heterologously expressed. Six recombinant plasmids containing the various ldhL and ldhD genes were constructed and transformed separately into Escherichia coli Rosetta(DE3) (see Table S1 in the supplemental material). These strains were incubated aerobically in lysogeny broth medium (100 μg ml−1 ampicillin) at 37°C to an optical density of 0.6 at 600 nm. Then, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to induce protein expression, and cultures were grown at 25°C for a further 5 h. Cells were harvested by centrifugation and washed with 0.85% (wt/vol) sodium chloride solution. Cell pellets were subsequently suspended in a binding buffer (20 mM sodium phosphate, 500 mM sodium chloride, and 20 mM imidazole [pH 7.4]) and then disrupted by sonication. Thereafter, intact cells and cell debris were removed by centrifugation, and the resultant supernatant was filtered and loaded onto a HisTrap HP 5-ml column (GE Healthcare). Purification was performed with gradient elution by using an elution buffer (20 mM sodium phosphate, 500 mM sodium chloride, and 500 mM imidazole [pH 7.4]). All nLDHs were purified to electrophoretic homogeneity for the activity assay. The catalytic efficiencies of recombinant nLDHs were calculated by kcat/Km (Table 3). For L. bulgaricus ATCC 11842 (d-LAB strain), d-nLDH showed a high catalytic efficiency toward pyruvate, whereas l-nLDH showed no detectable activity; for L. plantarum ATCC 14917 (dl-LAB strain), d- and l-nLDHs showed similar catalytic efficiencies toward pyruvate; for L. casei ATCC 334 (l-LAB strain), l-nLDH showed approximately 166-fold higher catalytic efficiency than d-nLDH. On the other hand, d-nLDH of L. bulgaricus ATCC 11842 (d-LAB strain) exhibited the highest catalytic efficiency of the three d-nLDHs, and l-nLDH of L. casei ATCC 334 (l-LAB strain) exhibited the highest catalytic efficiency of the three l-nLDHs.
Table 3.
Kinetic parameters of purified heterologously expressed His-tagged l- and d-nLDHs of the three representative Lactobacillus strains
| Strain |
l-nLDHa |
d-nLDH |
Catalytic efficiency ratio of l-nLDH to d-nLDH | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Km (mM) | Vmax (U mg−1) | kcat (s−1) | kcat/Km (M−1 s−1) | Km (mM) | Vmax (U mg−1) | kcat (s−1) | kcat/Km (M−1 s−1) | ||
| L. bulgaricus ATCC 11842 | ND | ND | ND | ND | 1.1 ± 0.2 | 2771.9 ± 307.2 | 1947.2 ± 215.8 | (1.8 ± 0.2) × 106 | 0 |
| L. plantarum ATCC 14917 | 1.7 ± 0.2 | 27.5 ± 2.1 | 18.0 ± 1.4 | (1.0 ± 0.0) × 104 | 3.1 ± 0.4 | 371.4 ± 32.2 | 264.5 ± 22.9 | (8.7 ± 0.5) × 104 | 1:8.7 |
| L. casei ATCC 334 | 1.0 ± 0.1 | 969.1 ± 49.4 | 655.7 ± 33.4 | (6.3 ± 0.6) × 105 | 3.5 ± 0.2 | 18.7 ± 0.6 | 13.2 ± 0.4 | (3.8 ± 0.1) × 103 | 166:1 |
ND, not detected.
Because the l- and d-nLDHs of the different strains exhibited considerable sequence identity (40% to 70%) (see Fig. S1 and S2 in the supplemental material) but various catalytic efficiencies (Tables 2 and 3), multiple alignment of the cloned l- and d-nLDH sequences was undertaken by using Clustal X2 (see Fig. S1 and S2) (12). The analysis revealed that certain pivotal residues are mutated in l-nLDH of L. bulgaricus ATCC 11842 (d-LAB) and d-nLDH of L. casei ATCC 334 (l-LAB) (see the supplemental material for details). These mutations may affect the activities of l- and d-nLDHs in different Lactobacillus strains and result in the different optical purities of lactic acid.
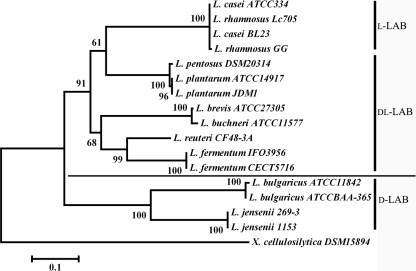
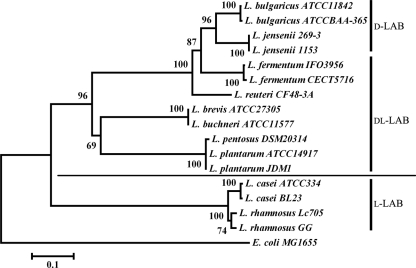
In addition to l- and d-nLDHs from the three representative strains, a series of other l- and d-nLDHs from other Lactobacillus strains was selected for phylogenetic analysis by using MEGA4 with the neighbor-joining (NJ) analytic approach (see Table S2 in the supplemental material) (21). The NJ tree showed that l-nLDHs of d-LAB strains (Lactobacillus bulgaricus and Lactobacillus jensenii) form a monophyletic group, whereas l-nLDHs of dl-LAB and l-LAB strains cluster to another group (Fig. 2). This topology was consistent with the activity assay results shown in Tables 2 and 3, where the l-nLDH catalytic efficiency of d-LAB strains was distinctly different from that of dl-LAB and l-LAB strains. Similarly, d-nLDHs of l-LAB strains (L. casei and Lactobacillus rhamnosus) also form a monophyletic group, whereas d-nLDHs of dl-LAB and d-LAB strains cluster together (Fig. 3). It may be inferred that, because of holistic changes, l-/d-nLDHs of different Lactobacillus strains formed separate clades, resulting in different catalytic efficiencies; these different catalytic efficiencies of l- and d-nLDHs resulted in different optical purities of lactic acid and different types of Lactobacillus strains.
Fig 2.
NJ tree based on l-nLDH amino acid sequences from various Lactobacillus strains, rooted with Xylanimonas cellulosilytica. Numbers at nodes indicate the percentage of NJ bootstrap analyses with 1,000 replicates. The scale bar indicates the level of amino acid sequence divergence.
Fig 3.
NJ tree based on d-nLDH amino acid sequences from various Lactobacillus strains, rooted with E. coli. Numbers at nodes indicate the percentage of NJ bootstrap analyses with 1,000 replicates. The scale bar indicates the level of amino acid sequence divergence.
In summary, the relative catalytic efficiencies of l- and d-nLDHs displayed evident differences, although the transcription of ldhL and ldhD showed no obvious distinctions among various Lactobacillus strains. As the optical purity of the lactic acid monomer is pivotal for polymerization, the production of optically pure lactic acid is of significant importance. The observation that the relative catalytic efficiencies of ldhL- and ldhD-encoded products are crucial for lactic acid optical purity may provide useful guidance for lactic acid production processes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grants 31000014 and 31170052), the Research Fund for the Doctoral Program of Higher Education of China (grant 20090131110036), and the China Postdoctoral Science Special Foundation (grant 201104262).
Footnotes
Published ahead of print 17 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Auras R, Harte B, Selke S. 2004. An overview of polylactides as packaging materials. Macromol. Biosci. 4:835–864 [DOI] [PubMed] [Google Scholar]
- 2. Cui FJ, Li YB, Wan CX. 2011. Lactic acid production from corn stover using mixed cultures of Lactobacillus rhamnosus and Lactobacillus brevis. Bioresour. Technol. 102:1831–1836 [DOI] [PubMed] [Google Scholar]
- 3. De Vos P, et al. 2009. Bergey's manual of systematic bacteriology, 2nd ed, vol 3 The Firmicutes. Springer-Verlag, New York, NY [Google Scholar]
- 4. Dumbrepatil A, Adsul M, Chaudhari S, Khire J, Gokhale D. 2008. Utilization of molasses sugar for lactic acid production by Lactobacillus delbrueckii subsp. delbrueckii mutant Uc-3 in batch fermentation. Appl. Environ. Microbiol. 74:333–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fu WG, Mathews AP. 1999. Lactic acid production from lactose by Lactobacillus plantarum: kinetic model and effects of pH, substrate, and oxygen. Biochem. Eng. J. 3:163–170 [Google Scholar]
- 6. Gao C, Ma CQ, Xu P. 2011. Biotechnological routes based on lactic acid production from biomass. Biotechnol. Adv. 29:930–939 [DOI] [PubMed] [Google Scholar]
- 7. Garvie EI. 1980. Bacterial lactate dehydrogenases. Microbiol. Rev. 44:106–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goffin P, et al. 2005. Lactate racemization as a rescue pathway for supplying d-lactate to the cell wall biosynthesis machinery in Lactobacillus plantarum. J. Bacteriol. 187:6750–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ha M-Y, Kim S-W, Lee Y-W, Kim M-J, Kim S-J. 2003. Kinetic analysis of growth and lactic acid production in pH-controlled batch cultures of Lactobacillus casei KH-1 using yeast extract/corn steep liquor/glucose medium. J. Biosci. Bioeng. 96:134–140 [PubMed] [Google Scholar]
- 10. John RP, Nampoothiri KM, Pandey A. 2007. Fermentative production of lactic acid from biomass: an overview on process developments and future perspectives. Appl. Microbiol. Biotechnol. 74:524–534 [DOI] [PubMed] [Google Scholar]
- 11. Kylä-Nikkilä K, Hujanen M, Leisola M, Palva A. 2000. Metabolic engineering of Lactobacillus helveticus CNRZ32 for production of pure l-(+)-lactic acid. Appl. Environ. Microbiol. 66:3835–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 13. Liu SQ. 2003. Practical implications of lactate and pyruvate metabolism by lactic acid bacteria in food and beverage fermentations. Int. J. Food Microbiol. 83:115–131 [DOI] [PubMed] [Google Scholar]
- 14. Lu ZD, Lu M, He F, Yu LJ. 2009. An economical approach for d-lactic acid production utilizing unpolished rice from aging paddy as major nutrient source. Bioresour. Technol. 100:2026–2031 [DOI] [PubMed] [Google Scholar]
- 15. Manome A, Okada S, Uchimura T, Komagata K. 1998. The ratio of l-form to d-form of lactic acid as criteria for the identification of lactic acid bacteria. J. Gen. Appl. Microbiol. 44:371–374 [DOI] [PubMed] [Google Scholar]
- 16. Okano K, et al. 2009. Homo-d-lactic acid fermentation from arabinose by redirection of the phosphoketolase pathway to the pentose phosphate pathway in l-lactate dehydrogenase gene-deficient Lactobacillus plantarum. Appl. Environ. Microbiol. 75:5175–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Okano K, et al. 2009. Improved production of homo-d-lactic acid via xylose fermentation by introduction of xylose assimilation genes and redirection of the phosphoketolase pathway to the pentose phosphate pathway in l-lactate dehydrogenase gene-deficient Lactobacillus plantarum. Appl. Environ. Microbiol. 75:7858–7861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Okano K, et al. 2009. Efficient production of optically pure d-lactic acid from raw corn starch by using a genetically modified l-lactate dehydrogenase gene-deficient and α-amylase-secreting Lactobacillus plantarum strain. Appl. Environ. Microbiol. 75:462–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Plessas S, et al. 2008. Lactic acid production by mixed cultures of Kluyveromyces marxianus, Lactobacillus delbrueckii ssp. bulgaricus and Lactobacillus helveticus. Bioresour. Technol. 99:5951–5955 [DOI] [PubMed] [Google Scholar]
- 20. Taguchi H, Ohta T. 1991. d-Lactate dehydrogenase is a member of the d-isomer-specific 2-hydroxyacid dehydrogenase family. Cloning, sequencing, and expression in Escherichia coli of the d-lactate dehydrogenase gene of Lactobacillus plantarum. J. Biol. Chem. 266:12588–12594 [PubMed] [Google Scholar]
- 21. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 22. Wang LM, et al. 2010. Efficient production of l-lactic acid from cassava powder by Lactobacillus rhamnosus. Bioresour. Technol. 101:7895–7901 [DOI] [PubMed] [Google Scholar]
- 23. Wee Y-J, Kim J-N, Ryu H-W. 2006. Biotechnological production of lactic acid and its recent applications. Food Technol. Biotechnol. 44:163–172 [Google Scholar]
- 24. Yu B, et al. 2011. Genome sequence of Lactobacillus rhamnosus strain CASL, an efficient l-lactic-acid producer from cheap substrate cassava. J. Bacteriol. 193:7013–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.