Abstract
The β-1,4-endoglucanase (EC 3.2.1.4) from the hyperthermophilic archaeon Pyrococcus horikoshii (EGPh) has strong hydrolyzing activity toward crystalline cellulose. When EGPh is used in combination with β-glucosidase (EC 3.2.1.21), cellulose is completely hydrolyzed to glucose at high temperature, suggesting great potential for EGPh in bioethanol industrial applications. The crystal structure of EGPh shows a triosephosphate isomerase (TIM) (β/α)8-barrel fold with an N-terminal antiparallel β-sheet at the opposite side of the active site and a very short C-terminal sequence outside of the barrel structure. We describe here the function of the peripheral sequences outside of the TIM barrel core structure. Sequential deletions were performed from both N and C termini. The activity, thermostability, and pH stability of the expressed mutants were assessed and compared to the wild-type EGPh enzyme. Our results demonstrate that the TIM barrel core is essential for enzyme activity and that the N-terminal β-sheet is critical for enzyme thermostability. Bioinformatics analyses identified potential key residues which may contribute to enzyme hyperthermostability.
INTRODUCTION
First-generation bioethanol has been produced with corn starch or simple sugars derived from sugar cane and beets, but use of these sources for bioethanol production competes directly with food supply. Second-generation bioethanol makes use of lignocellulosic agricultural waste, wood, or grass. However, hydrolysis of lignocellulosic biomass to fermentable sugars is a difficult challenge due to the crystallinity of cellulose, and the compact packing of cellulose, hemicelluloses, and lignin in plant material (5).
Currently, pretreatment and hydrolysis processes have to be tailored to suit different feedstocks due to the high variability of lignocellulosic biomasses from different sites or even different seasons. Enzymes from extreme thermophiles might play key roles in the development of biomass conversion technology that is insensitive to feedstock fluctuation and yet robust during the challenging pretreatment and hydrolysis processes (2). Hyperthermophilic cellulase would benefit the industrial process of biomass degradation when hydrolysis is performed at elevated temperatures. Some advantages include increased substrate solubility, increased reaction rate and process yield, and prevention of mesophilic bacterial contamination (4). The β-1,4-endoglucanase (EC 3.2.1.4) from the hyperthermophilic archaeon Pyrococcus horikoshii (EGPh) is one of the few extensively studied hyperthermophilic cellulases. It has strong hydrolyzing activity toward crystalline cellulose (1, 9, 10). In the presence of EGPh and the hyperthermophilic β-glucosidase (EC 3.2.1.21) from Pyrococcus furiosus, cellulose such as Avicel is completely hydrolyzed to glucose at high temperatures (11). Thus, EGPh may be an ideal candidate for use in high-temperature biomass conversion.
The EGPh structure, the first resolved from the family 5 archaeal endoglucanases, consists of a triosephosphate isomerase (TIM) (β/α)8-barrel fold with an N-terminal antiparallel β-sheet sitting on the opposite side of the active site and a very short C-terminal sequence outside of the barrel structure (13, 14). All known (βα)8-barrels are enzymes with only few exceptions. It is the most common enzyme fold and is found in ca. 10% of all proteins whose structures have been determined (19). This superfold occurs in many different groups of enzymes that have neither sequence nor functional similarities (15). Among all six classes of enzymes defined by the Enzyme Commission, (βα)8-barrels constitute five of them. Overall, hydrolases, and especially glycosidases, are the dominant class, comprising about half of the known TIM barrels (19). Understanding the relationship between function and the TIM barrel structure will aid in the design of molecules with new or enhanced properties.
As a hyperthermophilic cellulosic hydrolase, EGPh is a good model for studying the structure-function relationship and for determining factors conferring enzyme thermostability. The EGPh crystal structure was obtained with a C-terminal deletion form (EGPhΔC5) because crystallization of the wild-type enzyme was not successful (14). Kang et al. (8) reported that a 5-amino-acid (aa) deletion mutant from both the N- and C-terminal ends of EGPh not only maintained enzyme thermostability but exhibited increased activity. Alignment of EGPh with other closely related endoglucanases showed that the sequence forming the TIM barrel structure is highly conserved (∼50% identity), but not the N-terminal 34 aa forming the β-sheet. We hypothesized that the nonconserved sequence may not be essential for enzyme activity and tested the possibility of attaining a more compact enzyme with possibly better functionality. Mutants with sequential deletions from both N and C termini were expressed, characterized, and compared to wild-type EGPh.
MATERIALS AND METHODS
Bacterial strains, enzymes, and chemicals.
The Escherichia coli strains used in the present study were XL1-Blue (Stratagene) for gene cloning and BL21(DE3) (Novagen) for protein expression. The enzymes for PCR and DNA manipulation were purchased from New England Biolabs, and the chemicals used were purchased from Sigma-Aldrich.
Construction of plasmids for enzyme expression.
All enzymes were expressed with a C-terminal His6 tag using the expression plasmid pET-22b(+) (Novagen) by inserting the genes between NdeI and XhoI restriction sites. Genes encoding both wild-type EGPh and deletion mutants were amplified using PCR with primers containing NdeI and XhoI restriction sites. The plasmid containing the gene for the full-length EGPh supplied by K. Ishikawa (1) was used as a template. All primers used are listed in Table 1. The primer pairs used for PCR amplification of each corresponding gene were as follows: pEGNdeI5′ and pEGXhoI3′ (wild type), pEGNdeI5′ and pEGXhoI-1 (ΔC10), pEGNdeI5′ and pEGXhoI-2 (ΔC20), pEGNdeI5′ and pEGXhoI-3 (ΔC30), pEGNdeI-1 and pEGXhoI3′ (ΔN10), pEGNdeI-2 and pEGXhoI3′ (ΔN20), pEGNdeI-3 and pEGXhoI3′ (ΔN30), pEGNdeI-34 and pEGXhoI3′ (ΔN34), and pEGNdeI-4 and pEGXhoI3′ (ΔN40).
Table 1.
Primers used in this study
| Primer | Sequence (5′–3′)a |
|---|---|
| pEGNdeI5' | ATATACATATGGAAAATACAACATATCAAACACCGACT |
| pEGXhoI3' | GTGGTCTCGAGAGAACTTTTGGAACAACTATCC |
| pEGXhoI-1 | GTGGTCTCGAGCTTCAGGTTATTATACTTATCTTCC |
| pEGXhoI-2 | GTGGTCTCGAGTGTTGTCCAATCATCCTGTAG |
| pEGXhoI-3 | GTGGTCTCGAGGGTATCTCCACTATCTGGATTC |
| pEGNdeI-1 | GCGCTCATATGATTTACTACGAAGTGAGAGGAG |
| pEGNdeI-2 | GCGCTCATATGTACATGATTAATGTCACCAG |
| pEGNdeI-3 | GCGCTCATATGACTCCCATTCATCTCTTTGG |
| pEGNdeI-34 | GCGCTCATATGCTCTTTGGTGTAAACTGG |
| pEGNdeI-4 | GCGCTCATATGTTTGGCTTTGAAACACCTAATC |
Restriction sites are indicated in boldface.
Enzyme purification.
E. coli transformants containing the enzyme expression plasmids were grown overnight at 37°C and 200 rpm in 3 ml of M9 medium with ampicillin (100 μg/ml). A 1-ml portion of the overnight culture was used to inoculate 50 ml of the same medium, followed by growth and 30°C and 200 rpm for ∼5 h to reach an optical density at 600 nm (OD600) of ∼1. Then, 15 ml of this culture was transferred to 1 liter of the same medium and grown at 16°C overnight. On the next day, when the OD600 reached ∼0.8, 110 ml of 10× TB induction medium was added before 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added, and induction was continued for 5 h at 200 rpm. The cells were harvested after centrifugation at 3,200 × g for 15 min at 4°C. Cell pellets were resuspended in 25 mM sodium phosphate buffer (pH 7.4) at 10 ml/g (wet weight) and lysed by three passages through the pressured chamber of an Emulsiflex-C5 (Avestin). The cell extracts were prepared by heat purge at 70°C for 20 min, followed by centrifugation at 11,000 × g for 30 min at 4°C. The supernatants were stored at 4°C for comparative activity analysis or for use in further purification. The heat purge process precipitated most of the cellular bulk proteins and resulted in samples containing enzymes that account for >50% of the total proteins that are used for pH and temperature profiles and stability analyses.
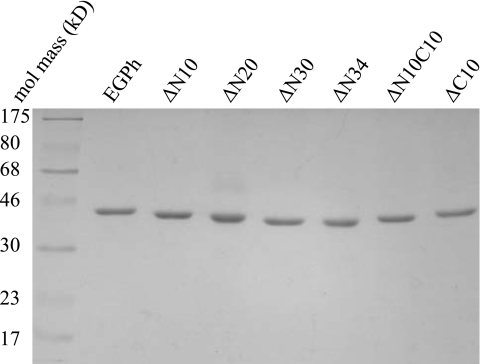
Further purification was performed with an AKTA FPLC apparatus (Amersham Bioscience) using a His-Trap HP (5 ml) column (Amersham Bioscience). Binding buffer was composed of 25 mM sodium phosphate (pH 7.4), 100 mM sodium chloride, and 5 mM imidazole. Elution was performed using a linear gradient of 5 to 400 mM imidazole in the same buffer. The enzymes were eluted at an average concentration of 175 mM imidazole. Purified proteins were quantified using the Bradford protein assay kit (Bio-Rad) and then used for specific-activity and kinetic-parameter analyses. The purities of the enzymes were analyzed by 15% SDS–polyacrylamide gel electrophoresis and Coomassie blue staining. Equal amounts of purified enzymes were loaded into each lane. Electrophoresis was performed at 130 V and 50 mA for 1.5 h using a Bio-Rad mini-Protean II gel set. The ColorPlus Prestained protein marker (broad range) from NEB was used to show the molecular mass.
Enzymatic activity analysis.
A hydrolytic activity assay was performed with 1% low-viscosity carboxymethyl cellulose (CMC; Sigma) or 1% Avicel (Analtech) in 100 mM acetate buffer (pH 5.5). The total reducing sugars were determined by using the modified Somogi-Nelson method (6). Avicel was prepared by swelling in 85% phosphoric acid according to the method of Wood (22). Purified enzymes were used for specific activity and kinetic analyses, and heat-purged partially purified extracts were used for other analyses.
The pH profile was determined at 80°C for EGPh and N-terminal deletion mutants and at 70°C for mutants containing C-terminal deletion with 1% CMC in different buffers ranging from pH 2 to 9. Buffers were prepared at a 100 mM concentration at 1-pH intervals with citric acid and sodium phosphate for pH 2 to 5, potassium phosphate and dipotassium phosphate for pH 6 to 8, and glycine and sodium hydroxide for pH 9.
For the pH stability, enzymes were first incubated at 50°C for 2 h in buffers of different pH levels ranging from pH 2 to 9. Buffer stocks (1 M) with different pH levels with one-unit intervals were prepared using the same chemical pairs as described for the pH profile analysis. The working concentration for enzyme treatment is 430 mM for each buffer. The residual enzymatic activity of the EGPh and mutants was measured by adding 35 μl of pretreated enzymes into 600 μl of 1% CMC dissolved in 250 mM sodium acetate buffer (pH 5.5). The reducing sugars were measured as described above.
The temperature profile was determined by comparing the enzymatic activities of the EGPh and mutants using the heat-purged cell extracts at 60, 70, 80, 90, and 95°C. The hydrolytic activities were analyzed with 1% CMC in 100 mM sodium acetate buffer (pH 5.5) as described above.
For thermostability analyses of EGPh and mutants, enzymes were diluted in 25 mM sodium phosphate buffer (pH 7.4) so that 8 μl would result in an OD420 of <0.9 during the enzyme activity assay. For heat treatment, 125 μl of the diluted enzymes was put in 1.5-ml screw-cap microtubes (Diamed) covered with 200 μl of mineral oil (Pharmacia Biotech) to prevent evaporation and incubated at temperatures of 80, 85, 90, and 95°C in an oil bath (Fisher Thermo Scientific). At different time points, 8-μl portions of the samples were obtained to determine the residual activity of each enzyme. Reactions were carried out at 80°C using 1% CMC in 100 mM sodium acetate buffer (pH 5.5) for 1 h.
For short-term thermostability analysis, purified enzymes were incubated at 85°C. At short time intervals (15 or 30 min), the samples were taken to perform enzyme activity assays at 80°C with 1% CMC in 100 mM acetate buffer (pH 5.5).
To monitor the accumulated product release, purified enzymes (1 μg for the wild type and the ΔN30 and ΔN34 mutants and 7 μg for the ΔC10 and ΔN10C10 mutants) were added to 1 ml of 1% Avicel in 100 mM sodium citrate (pH 6.0), followed by incubation at 85°C. At selected time points, 100 μl was removed to detect the total reducing sugar, as described previously. After 1 h, the reaction volume was reduced to 600 μl. At this time point, to maintain the linear range of OD420 reading during the reducing sugar analysis, an equal volume of fresh 1% Avicel was added to the reaction tube, and hydrolysis was continued at the same temperature. The same volume of sample was taken for sugar analysis at a later time point. These experiments were repeated three times.
Kinetic parameters were determined using p-nitrophenyl cellobiose (G2-PNP) (Sigma, St. Louis, MO) as a substrate. Reactions were performed at 70°C for 15 min with 2.5 μg of each purified enzyme in 500-μl reaction volume in 100 mM sodium citrate buffer (pH 6.0). Released p-nitrophenol (PNP) was measured with a spectrometer at 420 nm and quantified by comparison to the PNP (Sigma) standard curve. Initial velocities and substrate concentrations were used to determine the Km and kcat values by the nonlinear least-squares method with Prism version 3.03 for Windows (GraphPad Software, San Diego, CA).
For specific activity analysis, purified enzymes (2.5 μg) were incubated at 85°C in 600 μl of 1% CMC (Sigma; low viscosity) that had been dissolved in100 mM sodium citrate buffer (pH 6.0). After 15 min, a 25-μl sample was used for the colorimetric process of the modified Somogyi-Nelson method, followed by measuring the OD420. The quantity of released glucose was determined by comparison to the glucose standard curve. The specific activity was defined as micromoles of glucose (equivalent)/minute/micromoles of enzyme.
For homology modeling and energy minimization, a starting structure of the N-terminal 10-aa deletion mutant was obtained using the Swiss-Model workspace (http://swissmodel.expasy.org/workspace/). The primary sequence was submitted in the Automatic Modeling Mode, and the resulting output structure was modeled based on the known structure of EGPh (PDB ID 2ZUM).
Energy minimization was performed using the AMBER v.11 suite of programs (3). The ff99SB atomic force field was used (7). Counter ions were added to neutralize the molecular system, followed by solvation by a truncated octagonal TIP3P water box that is at least 8.0 Å from the surface of the protein. During energy minimization, nonbonded interactions were computed for up to a distance of 12.0 Å. The molecular system was first energy minimized by using the steepest decent method for 50 cycles and then the conjugate gradient method to a maximum of 50,000 cycles. Default convergence criteria were met before the maximum cycles were reached. Energy-minimized structures were rendered using the program PyMOL (Delano Scientific, San Carlos, CA).
RESULTS
Minimum sequence required for EGPh activity.
Based on the genomic DNA sequence, the originally submitted full-length EGPh (Swiss-Prot accession number O58925) contains 458 aa with a signal peptide sequence of 28 residues and a C-terminal region involved in membrane anchoring (4, 6). Deletion of the C-terminal region facilitates expression of soluble protein and has no effect on enzyme activity (6). Therefore, for the purposes of the present study, wild-type EGPh (389 aa) was expressed with both N (28 residues)- and C (42 residues)-terminal truncations (Fig. 1A). Given that shorter versions of EGPh maintain thermostability and may possess increased hydrolytic activity compared to the wild-type enzyme (8), we sought to determine whether enzymatic activity could be increased by further deletions. EGPh exhibits 43% sequence identity with EGAc, another member of the family 5 endoglucanase from the thermophilic and acidophilic bacterium Acidothermus cellulolyticus (1). Sequence alignment between these two proteins revealed that the internal 332 amino acids (from aa 37 to 368) of EGPh are 48% identical to EGAc, whereas the N-terminal 36 amino acids have only 17% identity and the C-terminal 32 aa have 34% identity. The EGPh sequence was also aligned with other closely related enzyme sequences. A protein BLAST search was carried out with the EGPh sequence, and five sequences with the highest amino acid identity relative to EGPh were selected for further analyses. These include EGSh (gene ID 9234087 from Staphylothermus hellenicus DSM 12710), EGPa (gene ID 1495295 from Pyrococcus abyssi GE5), EGXf (gb/EFF43630.1 from Xanthomonas fuscans subsp. aurantifolii strain ICPB), EGXax (gene ID 1154683 engXCA from Xanthomonas axonopodis pv. citri strain 306), and EGXal (gene ID 8702198 XALc_2969 from Xanthomonas albilineans). Since the first two organisms are hyperthermophilic and Xanthomonas spp. are aerobic bacteria, these five proteins cover both hyperthermophilic and mesophilic enzymes. The corresponding identities with EGPh are as follows: EGSh (56%), EGPa (54%), EGXf (49%), EGXax (49%), and EGXal (52%). Sequence alignment clearly demonstrated that indeed the peripheral sequences are not as conserved as the internal sequences (data for the whole alignment are not shown), especially the 30 aa from the N terminus and about 15 residues from the C terminus (Fig. 1B and C).
Fig 1.
Wild-type EGPh, sequence alignment, and enzyme activity analyses. (A) Illustration of the full-length EGPh with an N-terminal signal peptide (SP, 28 aa) and C-terminal membrane anchoring region (MA, 42 aa) that have been truncated to obtain the wild-type EGPh. (B and C) N-terminal (B) and C-terminal (C) sequence alignments of wild-type EGPh with the highest-identity proteins. EGSh (gene ID 9234087 from Staphylothermus hellenicus DSM 12710), EGPa (gene ID 1495295 from Pyrococcus abyssi GE5), EGXf (gb/EFF43630.1 from Xanthomonas fuscans subsp. aurantifolii strain ICPB), EGXax (gene ID 1154683 engXCA from Xanthomonas axonopodis pv. citri strain 306), and EGXal (gene ID 8702198 XALc_2969 from Xanthomonas albilineans). The deleted, nonconserved N-terminal 34 aa and C-terminal 10 aa are underlined. Identical residues are indicated by asterisks, and conserved residues are indicated by single or double dots. The alignment was performed using the CLUSTAL 2.1 multiple sequence alignment program. (D) Drawing showing the deletion locations and qualitative analysis of wild-type EGPh and deletion mutants that maintain (+) or lose (−) activity. The enzyme activities were determined with CMC at 80 and 60°C.
Sequential deletion analyses from both N and C termini, removing 10 amino acids at a time, were carried out to determine whether a shorter enzyme with improved characteristics could be created. The enzyme activity was assessed at 80°C, a lower-than-optimum temperature, to ensure that enzymes having compromised thermostability would still be active. Among the three C-terminal deletions, only the C10 mutant, which misses the last 10 amino acids, maintained activity. In contrast, all N-terminal deletion mutants (ΔN10, ΔN20, and ΔN30) retained activity except ΔN40. Detailed analysis of the aligned sequences revealed that the highly conserved sequence begins at L35, after the 34 N-terminal residues of EGPh. Therefore, a mutant with a deletion between ΔN30 and ΔN40 (ΔN34) was prepared and expressed. Interestingly, this mutant retained enzymatic activity like ΔN30, suggesting that the critical residues for enzyme activity start from L35. Each of the active N-terminal deletions was combined with the C10 deletion to establish the minimal sequence required for activity. When enzyme function was tested in the presence of CMC, only the N10C10 mutant exhibited activity, suggesting that the loss of activity may be due to the loss of thermostability. To test this, enzymatic activity assays were performed at 60°C. At this lower temperature, the ΔN20C10, ΔN30C10, and ΔN34C10 mutants all exhibited carboxymethyl cellulase (CMCase) activity, but the ΔN40, ΔC20, and ΔC30 mutants did not (Fig. 1D). Therefore, the shortest EGPh sequence maintaining hydrolytic activity is ΔN34C10, representing an 11% reduction in amino acid residues.
Expression and purification of EGPh and mutants.
Shorter version mutants exhibiting activity at 80°C were expressed in E. coli with a C-terminal His6 tag to facilitate purification using immobilized-metal affinity chromatography. Both the wild type and the deletion mutants were highly expressed in E. coli after IPTG induction. However, under regular growth and induction conditions (i.e., LB medium, 37°C, 0.5 mM IPTG, and 2-h induction), the majority of the recombinant proteins were not soluble. Protein expression was optimized through the use of minimal culture medium, decreasing growth temperature and IPTG concentration, and increasing induction time. Under these conditions, more than 50% of each protein was expressed in soluble form. Whole-cell extracts were heated for 20 min at 70°C to eliminate the majority of bacterial proteins to facilitate further purification of the recombinant enzymes. Even though the calculated pIs for each enzyme are slightly different (Table 2), the same phosphate buffer (pH 7.4) was found to be suitable for the preparation of all enzyme extracts. Enzymes were eluted at ∼175 mM imidazole using a linear gradient, following binding to a Ni affinity column. Protein samples were all highly pure (Fig. 2), as assayed by Coomassie blue staining, and protein migration in the gel corresponded well to their expected molecular mass (Table 2). In addition, no obvious degradation products were detected.
Table 2.
Properties and kinetic parameters of EGPh and mutants with G2-PNP
| Enzyme | Mol mass (Da) | pI | Mean ± SD |
||
|---|---|---|---|---|---|
| Km (mM) | kcat (1/s) | kcat/Km | |||
| EGPh | 45,561 | 5.64 | 0.37 ± 0.027 | 0.55 ± 0.046 | 1.51 ± 0.023 |
| EGPhΔN10 | 44,468 | 5.73 | 0.20 ± 0.009 | 0.48 ± 0.016 | 2.44 ± 0.042 |
| EGPhΔN20 | 43,257 | 5.82 | 0.46 ± 0.054 | 0.44 ± 0.019 | 0.96 ± 0.079 |
| EGPhΔN30 | 42,133 | 5.99 | 0.32 ± 0.040 | 0.34 ± 0.020 | 1.07 ± 0.074 |
| EGPhΔN34 | 41,685 | 5.95 | 0.23 ± 0.015 | 0.25 ± 0.006 | 1.08 ± 0.058 |
| EGPhΔN10C10 | 43,372 | 5.64 | 0.36 ± 0.029 | 0.28 ± 0.012 | 0.78 ± 0.045 |
| EGPhΔC10 | 44,466 | 5.55 | 0.44 ± 0.011 | 0.50 ± 0.025 | 1.13 ± 0.057 |
Fig 2.
Purification of EGPh and deletion mutants. A Coomassie blue-stained SDS-polyacrylamide gel is shown. The first lane is the broad-range ColorPlus Prestained protein marker (NEB) with the corresponding molecular mass labeled on the left side.
Characterization of recombinant proteins.
Purified enzymes were next characterized based on kinetic analyses. The molecular mass, pI, and kinetic parameters for each enzyme are shown in Table 2. To facilitate the manual kinetic parameter analysis process, a lower temperature (70°C) was selected, allowing for assays to be carried out over a 15-min period when the product release is still within linear range. The kinetic parameters were determined using pNPG2, a water-soluble substrate analogue commonly used for cellulase activity center analysis (16). The molecular mass and pIs for the His-tagged EGPh and mutants were calculated using the program Clone Manager (Scientific and Educational Software). The Km, kcat, and kcat/Km values of the mutants are similar to the wild type, indicating that the enzymatic active center and catalytic function of EGPh are not negatively affected by deletion of the peripheral sequences. Interestingly, even though the ΔN10 mutant showed a slightly increased kcat/Km and ΔC10 showed parameters similar to those for wild-type EGPh, ΔN10C10 showed a significantly decreased value (Table 2). The double deletion mutation seems to affect the catalytic efficiency.
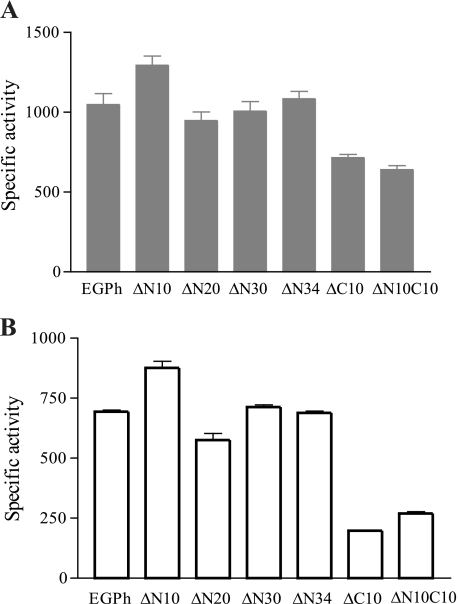
Using the mutant enzymes, the specific activities were measured with either the soluble cellulosic substrate CMC or the insoluble microcrystalline cellulose substrate Avicel at 85°C. This temperature was selected as a compromise temperature closer to the optimum temperature but not too high for the functionality of C-terminal deletion mutants. The N10 deletion mutant showed increased activities toward both substrates, whereas the N20, N30, and N34 deletion mutants showed similar activities compared to the wild-type enzyme (Fig. 3). Mutants with a C10 deletion and a N10C10 deletion showed decreased activities toward both substrates. When using CMC as the substrate, the C10 deletion and N10C10 deletion mutants retained 68 and 61% of activities compared to the wild type, while only 38 and 39% of activities remained using Avicel as a substrate.
Fig 3.
Specific activity analyses of EGPh and mutants using CMC (A) and Avicel (B). Activities were determined at 85°C using purified enzymes in 100 mM sodium citrate buffer (pH 6.0). Enzyme activity units were defined as the release of micromoles of glucose (equivalent) per minute by each micromole of enzyme. Each assay was repeated three times. Means and standard deviations were used to plot the graphs.
Effect of pH and temperature on enzyme activity.
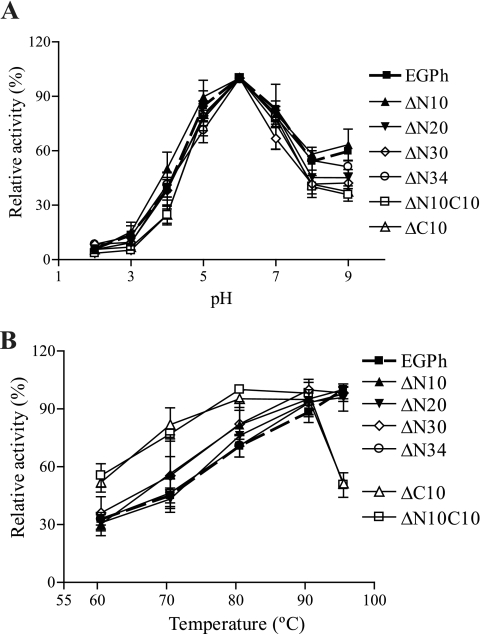
The effect of pH and temperature on the activities of EGPh wild-type and deletion mutants toward CMC was characterized. Both wild-type and shorter versions of EGPh were optimally active at pH 6 (Fig. 4A). To determine the optimal temperature, the enzyme activity at different temperatures was determined after a 1 h reaction with 1% CMC in 100 mM acetate buffer (pH 5.5). Under our testing conditions, the wild-type and N-terminal deletion mutants were optimally active at 95°C or higher, whereas the N10C10 and C10 deletion mutants showed a decreased optimal temperature of 90°C (Fig. 4B). These results suggest that the microenvironment for the residues involved in the catalytic reaction is not affected by deletion of peripheral sequences, even though the optimum temperature was decreased. The decreased optimal activity of the C-terminal deletion mutants may be due to compromised thermostability as described below.
Fig 4.
Effect of pH and temperature on the activities of EGPh and mutants. Enzyme activities were tested with 1% CMC for 1 h. (A) pH optima were measured at 80°C for EGPh and N-terminal deletion mutants and at 70°C for mutants containing C-terminal deletions. (B) Temperature optima were determined with 100 mM acetate buffer (pH 5.5). Each point represents the mean ± the standard deviation of triplicate experiments. The data were normalized to the maximum enzymatic activity.
Effect of pH on enzyme stability.
Even though the optimal pHs for the mutants were the same as for the wild type, the calculated pI values for deletion mutants did change (Table 2). To verify whether pI changes and sequence deletions affect the pH stability of the mutants, enzymes were incubated for 2 h at 50°C at different pHs and then tested for activity at a normal reaction pH. The wild-type EGPh and deletion mutants were all stable over a wide pH range; >70% of the enzymatic activity was retained between a pH range of 4 to 9. The enzymes showed decreased activity at pH 2, but the enzyme activity was not affected at pH 9 (data not shown). Overall, all of the mutants maintained a similar pH stability that is comparable to that of the wild-type EGPh.
Importance of peripheral sequences for thermostability.
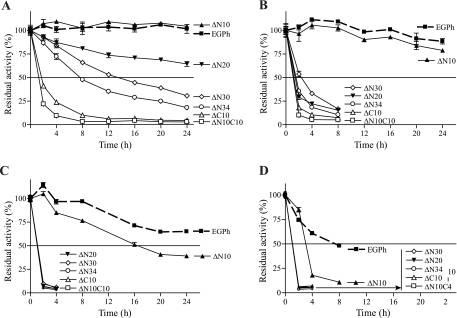
In addition to the decreased optimal temperature of ΔC mutants (Fig. 4B), ΔN and ΔC combination mutants (except ΔN10C10) were active at 60°C but not at 80°C (Fig. 1D). This suggests that peripheral sequences may be important for thermostability. To explore this possibility, enzymes were incubated at different temperatures during a 24-h period, and samples were tested for any residual activity at different time points in the presence of CMC. At 80°C, the wild type and the ΔN10 mutant were stable, whereas ΔN20, ΔN30, and ΔN34 showed gradually decreasing activity. A longer deletion led to a more severe decrease in activity, and the ΔN34 mutant exhibited the shortest t1/2 of 8 h. Both ΔC10 and ΔN10C10 lost >50% activity in less than 2 h at 80°C, suggesting that the decreased activity of C-terminal deletion mutants was due to decreased thermostability. At 85°C, the wild-type and ΔN10 strains demonstrated the same stability and only showed a slight decrease after 24 h of incubation. The other mutants dramatically lost activity at 85°C, with the C-terminal deletion mutants showing the most sensitivity to activity loss. At 90°C, the ΔN10 mutant became slightly less stable than the wild type with a t1/2 of 16 h; all of the other mutants lost activity quickly. At 95°C, the wild-type enzyme began to lose activity gradually with a t1/2 of ∼8 h, while ΔN10 had a t1/2 of <4 h. All of the other enzyme mutants completely lost activity before 2 h (Fig. 5). These results clearly show that the deletion of peripheral sequences decreases enzyme thermostability, particularly at higher temperatures.
Fig 5.
Thermostability of EGPh and mutants. Enzymes were incubated at 80°C (A), 85°C (B), 90°C (C), and 95°C (D). At different time points, samples were removed, and the remaining enzyme activities were tested at 80°C with 1% CMC in 100 mM acetate buffer (pH 5.5). Each point represents the mean ± the standard deviation of triplicate assays. Enzyme activity was normalized to that at 0 min of incubation.
Decreased thermostability did not correspond to the enzyme activity of ΔN mutants.
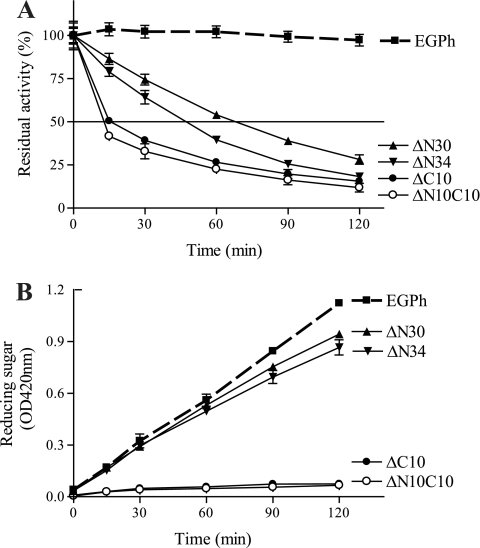
All active mutants except ΔN10 lost activity quickly when incubated at 85°C (Fig. 5B). In addition, at this temperature, the specific activities of all active N-terminal deletion mutants (ΔN10, ΔN20, ΔN30, and ΔN34) were not significantly different from the wild-type EGPh. In contrast, active mutants with a C-terminal deletion (ΔC10 and ΔN10C10) exhibited dramatically decreased specific activities (Fig. 3). These results suggest that the active ΔN mutants, but not ΔC mutants, might be stabilized by the presence of substrate. To confirm this possibility, we selected both ΔC mutants and the two ΔN mutants, ΔN30 and ΔN34, which contain the longest deletion, to investigate the influence of temperature and substrate on enzyme activity in more detail. Thermostability analyses were performed at shorter incubation time intervals for both wild-type and mutant enzymes, without substrate, at 85°C (Fig. 6A). As expected, all four deletion mutants showed greatly decreased stability compared to the stable, wild-type enzyme. At 120 min, EGPh maintained 97% residual activity (Fig. 6A), while ΔN30, ΔN34, ΔN10C10, and ΔC10 retained only 28, 18, 12, and 16% residual activity, respectively. However, in the presence of substrate at 120 min (Fig. 6B), both ΔN mutants (ΔN30 and ΔN34) showed a product accumulation pattern similar to the wild-type enzyme (Fig. 6B). In contrast, neither of the ΔC mutants (ΔC10 and ΔN10C10) showed significant product accumulation, suggesting that these mutants were not stabilized by substrate. Therefore, it seems that the active N-terminal deletion mutants may be stabilized by the cellulosic substrate.
Fig 6.
Short-term thermostability and product release of Avicel hydrolysis by EGPh and mutants. (A) Half-life time determination. Purified enzymes were incubated at 85°C for different times, and the remaining enzyme activity was analyzed at 80°C. The enzyme activity was normalized to that at 0 min of incubation. (B) Reducing sugar release through hydrolysis of 1% Avicel in 100 mM sodium citrate (pH 6.0) at 85°C. Each point represents the mean ± the standard deviation of triplicate assays.
Identification of putative atomic interactions between EGPh N terminus and the core structure using molecular modeling.
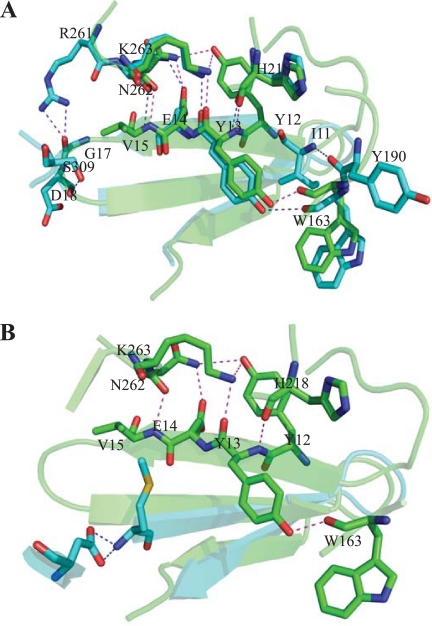
Using the solved structure of EPGh, careful investigation of all of the possible hydrogen bond interactions between residues in the N terminus and the core identified four consecutive N-terminal residues (YYEV) involved in interactions with four residues in the barrel (Fig. 7). These interactions are between Y12-N262, Y12-K263, Y13-K263, Y13-H218, Y13-W163, E14-N262, and V15-N262. These four residues (i.e., Y12, Y13, E14, and V15) are all located in the first β-strand at the N terminus and have multiple interactions with four other residues located just outside the N termini of three of the eight β-barrel-forming β-strands [β-4(W163), β-5(H218), and β-6(N262, K263)]. Molecular modeling of the ΔN10 mutant showed that these hydrogen bonds are most likely not disrupted. However, in the ΔN20 mutant, the first β-strand had been removed, resulting in the loss of these interactions.
Fig 7.
H-bond interactions between N-terminal β-sheet and residues located in the (βα)8-barrel of EGPh. The wild-type structure (green) was overlaid with deletion models (blue) of ΔN10 (A) or ΔN20 (B). The image was rendered by using PyMOL software (PyMOL Molecular Graphics System, v1.2r3pre; Schrödinger, Inc). The wild-type structure H bonds are indicated in magenta, and the deletion structure H bonds are indicated in blue.
DISCUSSION
Catalytic activity is retained within the α/β-barrel core.
Kim and Ishikawa (12) recently reported a detailed study describing the catalytic mechanisms of the hyperthermophilic EGPh. However, much less is known regarding its structure-function relationship. Our sequential deletion analysis showed that the N-terminal nonconserved 34 aa can be deleted without a significant effect on enzyme activity, but deletion of the immediately following 6 aa abolished enzyme activity. Inspection of the available crystal structure of EGPh (13) reveals that the 34 residues fold into the N-terminal antiparallel β-sheet, which is spatially separated from the α/β-barrel core structure. Our results suggest that the conserved internal (β/α)8-barrel is the catalytic core responsible for the enzymatic activity of EGPh.
All known (βα)8-barrel enzymes, including EGPh, contain the canonical (βα)8-barrel with eight β-strands to form the central barrel surrounded by the α-helices. The catalytically active residues are located at the C-terminal ends of the β-strands and in the βα-loops to form the “catalytic face” (19). In EGPh, eight conserved residues located in the catalytic cleft were identified as critical for enzyme activity (8, 13), with the exception of R102 located within β2; all other residues are located at the C-terminal of the β-sheets. Perhaps due to the distance from the catalytic center, deletion of the 34 aa which form the N-terminal β-sheet structure caused no significant reduction in enzymatic activity. However, thermostability is significantly affected in the absence of these N-terminal residues.
Factors contributing to the EGPh hyperthermostability.
The intrinsic differences between hyperthermophilic enzymes and their mesophilic counterparts are still not well defined. Some of the factors proposed to contribute to protein hyperthermostability include amino acid composition, disulfide bridges, hydrophobic interaction, helix dipole stabilization, terminus and loop stabilization, aromatic interaction, hydrogen bonds, ion pairs, and decreases in number and size of surface loops (20, 21). A significant reduction of the thermostability in both active mutants ΔC10 and ΔN10C10 is likely the result of deletion of the C-terminal 10 aa, which includes some residues forming the last α-helix and a cysteine, C384, that is critical in maintaining the hyperthermostability of EGPh (9). The structure of EGPh (PDB 2ZUM) has been resolved using a ΔC5 mutant (13), but the mechanisms involved in its hyperthermostability remain elusive. Based on a chemical process, C384 may not be involved in the formation of a disulfide bond (9) even though the sequence similarity to EGAc indicates it may form a disulfide bond with C344 (10). However, since ΔC5 mutant has C384 deleted, it has not been confirmed whether C384 forms a disulfide bond in the actual structure. In all known (βα)8-barrel enzymes, residues maintaining the fold stability are located in the core, on the end of the barrel opposite the catalytic center and in the αβ loops. These residues are described to form the “stability face” (19). Even though the “stability face” may function as a structural element for the (βα)8-barrel enzyme stability, this does not imply that these proteins use a common mechanism for their stability. Instead, each class of protein appears to have its own mechanism because different insertions, peripheral sequences, or domains are attached to the core sequences in different enzymes. The function of the N-terminal peripheral structure in the context of protein stability has not been well established. In fact, the N-terminal structure is not conserved. For example, instead of an N-terminal β-sheet, the recently characterized hyperthermophilic endoglucanase Cel5A from Thermotoga maritima contains a short α-helix that is proposed to seal the bottom of the barrel from solvent (17). The β-strands in the EGPh N-terminal β-sheet are considerably longer than those in the mesophilic counterpart, EGAc (13, 18). Although a longer N-terminal β-sheet may contribute to hyperthermostability, this remains to be tested.
To our knowledge, the present deletion study of EGPh may be the first demonstration of an N-terminal β-sheet involved in enzyme hyperthermostability of a (βα)8-barrel enzyme. In the wild-type structure (Fig. 7), four conservative N-terminal residues, YYVE, are involved in hydrogen bonding with four residues from the core structure. The latter four residues are all located on the end of the barrel within the “stability face.” Therefore, all of these interactions occur between the N-terminal peripheral structure and the “stability face” of the (βα)8-barrel structure. Since the four N-terminal residues all reside in the first peripheral β-strand, it is possible that deletion of this strand would significantly affect stability. In contrast, deletion of the first 10 residues eliminated the N-terminal unstructured residues but not the four critical residues. Therefore, most of the interactions between the N terminus and the barrel structure are most likely to be conserved (Fig. 7A). This may explain why the ΔN10 mutant showed very similar thermostability compared to wild-type EGPh. In addition, in the ΔN10 mutant, two new potential interactions (I11-Y190 and D18-S09) may be forming to help stabilize the mutant protein. However, in the ΔN20 and other mutants, with more extensive deletion of the N terminus, the first β-strand containing the YYEV residues is completely deleted (Fig. 7B), eliminating the stabilizing interactions and leading to decreased stability, even at lower temperatures. Therefore, it seems that the interactions of the first β-strand with the rest of the molecule may help to preserve a stable enzyme structure at high temperatures. Future experiments using single amino acid substitution analysis would help to confirm this possibility. The observations that the ΔN5 (8) and the ΔN10 (Table 2 and Fig. 3) mutants have slightly increased enzymatic activity suggest deletion of the N-terminal unstructured residues may be beneficial to enzyme activity.
In the presence of cellulosic substrate, N-terminal deletion mutants maintained full activity compared to the wild-type enzyme, whereas C-terminal deletion mutants only retained residual activities during the 2-h incubation (Fig. 6B). However, when assays were performed for a short time during specific activity analysis, ΔC10 mutants showed decreased but significant activity compared to the wild-type enzyme (Fig. 3). These results suggest that the mutants were only active at a very early stage and lost activity quickly due to structural instability. The loss of stability may be due to the loss of some residues involved in the formation of the last α-helix in the C10 deletion mutants, which may destabilize the core barrel structure. In contrast, the active N-terminal deletion mutants maintain the complete sequence that is involved in the formation of the barrel structure. We speculate that substrate binding may help to maintain an intact core barrel structure in these mutants. The crystal structures of the deletion mutants in the presence or absence of substrate, when available, would help to determine whether the binding of substrates can indeed stabilize the structure, thereby helping retain optimal enzyme activity.
ACKNOWLEDGMENTS
This project is supported in part by ABIP-CBioN, AAFC-NRCan-NRC NBP1, and Naturally Advanced Technologies.
We thank Peter Lau for critical reading of and helpful comments on the manuscript, Juan Gao for technical advice, and Mark Wood and Fang Huang for technical support.
Footnotes
Published ahead of print 17 February 2012
REFERENCES
- 1. Ando S, Ishida H, Kosugi Y, Ishikawa K. 2002. Hyperthermostable endoglucanase from Pyrococcus horikoshii. Appl. Environ. Microbiol. 68:430–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MW, Kelly RM. 2008. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr. Opin. Biotechnol. 19:210–217 [DOI] [PubMed] [Google Scholar]
- 3. Case DA, et al. 2010. AMBER 11. University of California, San Francisco, CA [Google Scholar]
- 4. Haki GD, Rakshit SK. 2003. Developments in industrially important thermostable enzymes: a review. Bioresource Technol. 89:17–34 [DOI] [PubMed] [Google Scholar]
- 5. Himmel ME, et al. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804–807 [DOI] [PubMed] [Google Scholar]
- 6. Hiromi K, Takahashi Y, Ono S. 1963. Kinetics of hydrolytic reaction catalyzed by crystalline bacterial α-amylase: the influence of temperature. Bull. Chem. Soc. Jpn. 36:563–569 [Google Scholar]
- 7. Hornak V, et al. 2006. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65:712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kang HJ, Ishikawa K. 2007. Analysis of active center in hyperthermophilic cellulase from Pyrococcus horikoshii. J. Microbiol. Biotechnol. 17:1249–1253 [PubMed] [Google Scholar]
- 9. Kang HJ, Uegaki K, Fukada H, Ishikawa K. 2007. Improvement of the enzymatic activity of the hyperthermophilic cellulase from Pyrococcus horikoshii. Extremophiles 11:251–256 [DOI] [PubMed] [Google Scholar]
- 10. Kashima Y, Mori K, Fukada H, Ishikawa K. 2005. Analysis of the function of a hyperthermophilic endoglucanase from Pyrococcus horikoshii that hydrolyzes crystalline cellulose. Extremophiles 9:37–43 [DOI] [PubMed] [Google Scholar]
- 11. Kim HW, Ishikawa K. 2010. Complete saccharification of cellulose at high temperature using endocellulase and beta-glucosidase from Pyrococcus sp. J. Microbiol. Biotechnol. 20:889–892 [DOI] [PubMed] [Google Scholar]
- 12. Kim HW, Ishikawa K. 2011. Functional analysis of hyperthermophilic endocellulase from Pyrococcus horikoshii by crystallographic snapshots. Biochem. J. 437:223–230 [DOI] [PubMed] [Google Scholar]
- 13. Kim HW, Ishikawa K. 2010. Structure of hyperthermophilic endocellulase from Pyrococcus horikoshii. Proteins 78:496–500 [DOI] [PubMed] [Google Scholar]
- 14. Kim HW, Mino K, Ishikawa K. 2008. Crystallization and preliminary X-ray analysis of endoglucanase from Pyrococcus horikoshii. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64:1169–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Orengo CA, Jones DT, Thornton JM. 1994. Protein superfamilies and domain superfolds. Nature 372:631–634 [DOI] [PubMed] [Google Scholar]
- 16. Percival Zhang YH, Himmel ME, Mielenz JR. 2006. Outlook for cellulase improvement: screening and selection strategies. Biotechnol. Adv. 24:452–481 [DOI] [PubMed] [Google Scholar]
- 17. Pereira JH, et al. 2010. Biochemical characterization and crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima. J. Struct. Biol. 172:372–379 [DOI] [PubMed] [Google Scholar]
- 18. Sakon J, Adney WS, Himmel ME, Thomas SR, Karplus PA. 1996. Crystal structure of thermostable family 5 endocellulase E1 from Acidothermus cellulolyticus in complex with cellotetraose. Biochemistry 35:10648–10660 [DOI] [PubMed] [Google Scholar]
- 19. Sterner R, Hocker B. 2005. Catalytic versatility, stability, and evolution of the (βα)8-barrel enzyme fold. Chem. Rev. 105:4038–4055 [DOI] [PubMed] [Google Scholar]
- 20. Unsworth LD, van der Oost J, Koutsopoulos S. 2007. Hyperthermophilic enzymes: stability, activity, and implementation strategies for high temperature applications. FEBS J. 274:4044–4056 [DOI] [PubMed] [Google Scholar]
- 21. Vieille C, Zeikus GJ. 2001. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65:1–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wood TM. 1988. Preparation of crystalline, amorphous, and dyed cellulase substrates. Methods Enzymol. 160:19–25 [Google Scholar]