Abstract
Chitin is the second most abundant polysaccharide, present, e.g., in insect and arthropod exoskeletons and fungal cell walls. In some species or under specific conditions, chitin appears to be enzymatically de-N-acetylated to chitosan—e.g., when pathogenic fungi invade their host tissues. Here, the deacetylation of chitin is assumed to represent a pathogenicity mechanism protecting the fungus from the host's chitin-driven immune response. While highly specific chitin binding lectins are well known and easily available, this is not the case for chitosan-specific probes. This is partly due to the poor antigenicity of chitosan so that producing high-affinity, specific antibodies is difficult. Also, lectins with specificity to chitosan have been described but are not commercially available, and our attempts to reproduce the findings were not successful. We have, therefore, generated a fusion protein between a chitosanase inactivated by site-directed mutagenesis, the green fluorescent protein (GFP), and StrepII, as well as His6 tags for purification and detection. The recombinant chitosan affinity protein (CAP) expressed in Escherichia coli was shown to specifically bind to chitosan, but not to chitin, and the affinity increased with decreasing degree of acetylation. In vitro, CAP detection was possible either based on GFP fluorescence or using Strep-Tactin conjugates or anti-His5 antibodies. CAP fluorescence microscopy revealed binding to the chitosan exposing endophytic infection structures of the wheat stem rust fungus, but not the chitin exposing ectophytic infection structures, verifying its suitability for in situ chitosan staining.
INTRODUCTION
Biotrophic pathogens need to prevent triggering of active defense reactions of their host tissues as they depend on living host cells for growth and development. Typically, fungal cell walls contain chitin as a fibrillar element, but many fungi are known to produce chitin deacetylases during specific developmental stages (15). Some plant-pathogenic fungi are known to produce chitin deacetylase to convert the chitin on the surface of their infection structures into chitosan to evade the chitin-driven immune response of their hosts (12). On the one hand, chitinases and β-1,3-glucanases act as antifungal enzymes in most plants (14). By converting chitin into chitosan when invading the plant tissue, the fungus avoids the degradation of its cell walls by host enzymes. On the other hand, the non-self-surveillance machinery of plants is geared toward recognizing chitin (41). Conversion of chitin into chitosan prevents the generation of elicitor active chitin oligomers which would reveal the presence of the pathogen to the plant, triggering active defense responses. Hence, the deacetylation of surface-exposed chitin into chitosan acts as a molecular disguise strategy, and, consequently, chitin deacetylases may constitute crucial pathogenicity factors. Pathogenicity factors are perfect targets for broad-spectrum antifungal agents as they can be expected to be important for different types of pathogens but may not be present in nonpathogenic endophytic or mutualistic fungi.
A similar disguise strategy likely acts as a pathogenicity mechanism in fungal pathogens of human tissues too. Chitin deacetylase was shown to be present at the plasma membrane of the early stages of the life cycle in the human-pathogenic fungus Encephalitozoon cuniculi (9). Cryptococcus neoformans, the causal agent of cryptococcosis in humans, was shown to possess four putative chitin deacetylase genes, and two of its chitin deacetylases have been reported to induce immune responses in the host, indicating that they are expressed during host tissue colonization (3, 7, 24). Deletion mutants of the chitin deacetylases in this opportunistic fungal pathogen showed that chitosan is important during vegetative growth; it appears to help in maintaining cell integrity and to aid in bud separation (3). Paracoccidioidomycosis, a disease affecting 10 million people in Latin America, is caused by the fungus Paracoccidioides brasiliensis, and transcriptional analysis showed that a chitin deacetylase gene is upregulated in the pathogenic yeast form of the fungus (13). Similarly, the cyst wall of Entamoeba invadens, a reptile-pathogenic relative of the human pathogen Entamoeba histolytica, was shown to contain chitosan, consistent with the presence of a functional chitin deacetylase (11).
Hence, conversion of the more typical structural polysaccharide chitin present in the exoskeletons and in the cell walls or extracellular matrices of many pathogenic organisms into the less crystalline and, thus, structurally less resilient but also less tale-telling chitosan emerges as a widespread pathogenicity strategy not only of fungal pathogens. However, the evidence so far relies mostly on molecular genetic detection of the expression of a chitin deacetylase gene, while the presence of the product of the enzyme chitin deacetylase, namely, chitosan, has only been achieved in very few instances (8, 12, 22). On the one hand, this is due to the fact that neither lectins (10, 25) nor antibodies with specificity to chitosan (3, 20) are commercially available. On the other hand, widely used tools like calcofluor white or eosin Y are lacking in specificity, so that prior biochemical knowledge of the cell wall composition of the studied organism is required to obtain reliable results. In fact, chitosan is a rather poor antigen, and attempts to generate antisera with reasonable specificity and affinity have failed more often than not. Hence, alternative chitosan-specific probes are needed to test for chitosan on the surfaces of, e.g., pathogenic fungal hyphae (18). One possibility for the specific detection of polysaccharides are polysaccharide hydrolases or other polysaccharide-modifying enzymes with an affinity to the targeted molecule, or specific polysaccharide binding proteins or modules (16, 19, 28, 32, 35), an approach already successfully employed for the detection of chitosan (17). Such proteins can either be detected by using specific antibodies directed against them (12, 33, 38) or by tagging the proteins chemically, e.g., using fluorescence tags such as fluorescein isothiocyanate (FITC) or using gold particles (4–6, 17, 26), or genetically, by generating fusion proteins, e.g., with the green fluorescent protein (GFP) (19) or with a peptide tag against which commercial antibodies are available. We here describe an extension of the latter method, generating by genetic engineering a chitosan affinity protein (CAP) based on a bacterial chitosanase (CSN) that we inactivated using site-directed mutagenesis and to which we fused three different tags, namely, enhanced GFP (eGFP) and the affinity tags StrepII and His6, for purification and detection. This is a versatile, generic technique that can easily be transferred to other substrates and which allows multiple staining of different substrates if GFP variants that absorb and emit light at different wavelengths are used.
MATERIALS AND METHODS
Chemicals.
Wheat germ agglutinin (WGA) lectin coupled to Texas Red or to fluorescein isothiocyanate (FITC) was purchased from Molecular Probes (Eugene, OR). Restriction enzymes, the Rapid DNA ligation kit, and Phusion DNA polymerase were purchased from Fermentas (St. Leon-Rot, Germany).
Bacterial strains, plasmids, and culture conditions.
Escherichia coli DH5α was used as a host for recombinant plasmids. E. coli Rosetta 2(DE3)(pLysSRARE2) was used for recombinant protein expression (purchased from Merck, Darmstadt, Germany). The pET-22b(+) vector was purchased from Merck, Germany. Plasmids were prepared for sequencing and sequenced at MWG-Biotech AG (Ebersberg, Germany). E. coli DH5α and E. coli Rosetta 2(DE3)(pLysSRARE2) with pET-22b(+) constructs were grown in LB at 37°C with 100 μg/ml ampicillin and 100 μg/ml ampicillin plus 34 μg/ml chloramphenicol, respectively, for the selection of transformants. Autoinduction solutions M (50× stock: 1.25 M Na2HPO4, 1.25 M KH2PO4, 2.5 M NH4Cl, 0.25 M Na2SO4) and 5052 (50× stock: 25% [vol/vol] glycerol, 2.5% [wt/vol] d-glucose, 10% [wt/vol] α-lactose monohydrate) (34) were added to induce the cells for expression of the target protein, and cultures were grown at 28°C. For long-term storage, liquid cultures were supplemented with 30% (vol/vol) glycerol and stored at −70°C.
Cloning of the Bacillus CSN.
Details of the primers used in the present study are given in the supplemental material. From soil, a Bacillus sp. strain was isolated that served as the template for CSN gene amplification using CSN_pET_for and CSN_pET_rev. This gene (GenBank accession no. JQ425408) is similar to the known chitosanase (CSN) genes (e.g., Bacillus cereus 95; EEL16702.1) and was cloned into pET-22b(+) via EcoRI and HindIII without the native signal peptide-encoding sequence (nucleotides [nt] 1 to 141), generating pET-22b-CSN. Using this plasmid as a template, the CSN gene was amplified using primer pairs CSN_StrepII_for and CSN_StrepII_rev. In doing so, an amplificate containing an upstream-located StrepII coding sequence was generated and subsequently cloned in pET-22b(+) via NdeI and HindIII sites, resulting in pET-22b-StrepII-CSN.
In the following step, the gene encoding eGFP was amplified from the pEGFP vector (U76561.1; Clontech, Saint-Germain-en-Laye, France) using the eGFP_for and eGFP_rev primers. The amplified eGFP gene was cloned in pET-22b-StrepII-CSN via HindIII and XhoI restriction sites, generating the expression plasmid pET-22b-StrepII-CSN-eGFP-His6.
Site-directed mutagenesis of CSN.
In order to obtain a mutein of CSN which binds to chitosan with high affinity and specificity but is devoid of hydrolytic activity, a bioinformatics approach was chosen to identify an amino acid which is essential for CSN catalytic activity. At position 122 of the polypeptide chain, an amino acid was identified that represents a catalytically active glutamate (E) of the CSN that was then replaced by the structurally related but chemically distinct glutamine (Q) (1). Therefore, the plasmid pET-22b-StrepII-CSN-eGFP-His6 was subjected to site-directed mutagenesis using the 5′-phosphorylated primers CSN_E122Q_for and CSN_E122Q_rev to obtain pET-22b-StrepII-CSN-eGFP-His6 E122Q (encoding CAP).
Expression and purification of CAP.
Both CAP and its active form were synthesized in E. coli Rosetta 2(DE3)(pLysSRARE2). Cell suspensions in 20 mM triethanolamine (pH 8) containing 400 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 1 mM MgCl2, and 10 U/ml benzonase were incubated for 30 min at room temperature and then sonicated briefly. Crude extracts (50 ml from 15 g net biomass) were obtained by centrifugation and applied to a Strep-Tactin column (Strep-Tactin superflow Plus, 1-ml bed volume; Qiagen, Hilden, Germany), which was then washed with 20 mM triethanolamine (pH 8) containing 400 mM NaCl, and finally eluted in the same buffer additionally containing 2.5 mM d-desthiobiotin. The eluted fractions were concentrated in ultrafiltration devices (Sartorius Stedim Biotech, Göttingen, Germany) with a molecular mass cutoff at 10 kDa, supplemented with 10% (vol/vol) glycerol, and stored at 4°C.
Preparation of chitosans.
Chitosan (originating from shrimp shell chitin, with an average degree of acetylation [DA] of 3% and average degree of polymerization [DP] of 2,124) was generously provided by Dominique Gillet, Mahtani Chitosan, Veraval, India. This raw chitosan was dissolved in an aqueous acetic acid solution and purified by successive filtration and extensive washing steps involving repeated precipitation and centrifugation, before chitosans with different DAs were prepared by partial re-N-acetylation using acetic anhydride in 1,2-propanediol, as described previously (39). The DA of the resulting chitosans was determined using 1H-nuclear magnetic resonance (NMR) spectroscopy (21).
Analyzing CAP enzyme activity.
In a dot activity assay, the hydrolytic activity of CAP toward chitosan was assessed. The crude extracts of E. coli Rosetta 2(DE3)(pLysSRARE2) bearing pET-22b(+), pET-22b-StrepII-CSN-eGFP-HIS6, or pET-22b-StrepII-CSN-eGFP-HIS6 E122Q were applied to a polyacrylamide gel containing chitosan with a DA of 35% (0.1 mg/ml) as a substrate. The gel was incubated at 37°C overnight. Later, the gel was stained with 0.01% (wt/vol) calcofluor white (Sigma, Steinheim, Germany) in 0.5 M Tris (pH 8.9) for 5 min; the gel was washed in deionized water for 1 h, and dark spots indicating enzyme activity were visualized on a UV transilluminator (29).
Binding specificity of CAP.
Fusions to eGFP and two affinity tags allow the usage of CAP for several detection strategies, namely, by chemiluminescence under UV light and by transilluminator (MoBiTech Dark Reader) and fluorescence microscopy. The excitation wavelength for eGFP is 488 nm, and the emission wavelength is 510 nm. In order to analyze the affinity of CAP, different chitosans with DA ranging from 10% to 56% as well as glycol-chitin were spotted in dilution series (from 1,000 ng to 2 ng) onto a nitrocellulose membrane (GE Healthcare, Munich, Germany). Membranes were then incubated at 70°C for 30 min to allow the different chitosans and glycol-chitin to stick firmly to the membrane. The membrane was blocked using 5% (wt/vol) bovine serum albumin (BSA) in 1× Tris-buffered saline (TBS) buffer for 1 h at room temperature; later, the membrane was washed with 1× TBS buffer for 15 min. After washing, the membrane was incubated with either CAP or WGA-FITC as a positive control for the detection of glycol chitin (0.1 mg/ml in TBS containing 5% [wt/vol] BSA) for 1 h at room temperature. Washing steps were continued with 1× TBS plus 0.05% (vol/vol) of both Tween 20 and Triton X-100 twice for 15 min each, and then the membrane was incubated with Strep-Tactin–horseradish peroxidase (HRP) conjugate (IBA, Göttingen, Germany). In an independent experiment, the same procedure was followed as explained above, but anti-His5 antibody (Qiagen, Hilden, Germany) was used for detection. In both cases, the signal was detected by chemiluminescence. Later, membranes were used to observe fluorescence on a transilluminator (Dark Reader; MoBiTech, Goettingen, Germany) or under a UV lamp to check for fluorescence. Although the excitation wavelength of a UV transilluminator is not optimal to excite eGFP, the fluorescence was also observed when using such a device instead of the Dark Reader.
In situ staining of chitosan using CAP.
In a further experiment, CAP was employed for specific staining of chitosan in the cell walls of a plant-pathogenic fungus, in analogy to the chitosan-specific antibody used by El Gueddari et al. (12). Urediniospores of the wheat stem rust fungus Puccinia graminis (Pers.) f. sp. tritici (Eriks. & E. Henn) (race 32) were used for the experiment. Differentiation of infection structures was induced by a mild shock (2 h, 30°C) starting 2 h after sowing urediniospores into polystyrene petri dishes (diameter, 5 cm) and adding distilled water (5 ml), as previously described (12). Then, incubation was continued at 23°C overnight for the development of infection structures.
After allowing the spores to produce different infection structures, fungal germlings were incubated with 2% (wt/vol) BSA in 1× phosphate-buffered saline buffer for 2 h at room temperature. Incubation was followed by three washing steps with 1× PBS plus 0.05% (vol/vol) Tween 20 for 15 min each. Spores were further incubated with the chitin-specific lectin WGA coupled to Texas Red (Life Technologies GmbH, Darmstadt, Germany) and CAP (both at 0.1 mg/ml in TBS containing 5% [wt/vol] BSA) for 1 h at room temperature. After incubation, washing was done with 1× PBS–Tween 20 three times for 15 min each. GFP and Texas Red were monitored with a confocal laser scanning microscope (Leica TCS SP5 X; Leica, Wetzlar, Germany) with excitation/emission wavelengths of 488/595 nm and 500 to 545/608 to 700 nm, respectively.
RESULTS
Cloning, site-directed mutagenesis, heterologous expression, and purification of CAP.
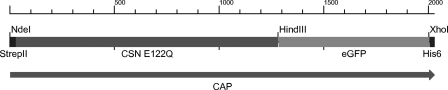
An enzymatically inactive construct of a bacterial chitosanase was engineered (Fig. 1) using site-directed mutagenesis as described above. The wild-type chitosanase gene was amplified from genomic DNA of a Bacillus sp. strain isolated from soil, using primers derived from conserved regions of Bacillus chitosanases. The chitosanase gene was amplified with an upstream StrepII coding sequence. An eGFP-encoding sequence was cloned downstream from the chitosanase gene before the His6-encoding sequence in the expression vector. This construct was termed the wild-type chitosanase fusion protein. After creating the whole construct, site-directed mutagenesis was done to exchange the catalytically active glutamic acid residue for glutamine at position 122 (1). This second construct was termed chitosan affinity protein (CAP).
Fig 1.
Schematic representation of the CAP gene. The scale above indicates the nucleotide position.
Both the wild-type chitosanase fusion protein and CAP were synthesized in E. coli Rosetta 2(DE3)(pLysSRARE2). Cultures were incubated at 37°C for 48 h. Recombinant proteins were purified by affinity chromatography. SDS-PAGE analysis (Fig. 2A) showed both proteins at the expected size of 75 kDa. This was confirmed by Western blot analysis (Fig. 2B). A minor band was observed at a lower size in both the SDS-PAGE and Western blot, potentially indicating partial degradation of the fusion protein.
Fig 2.
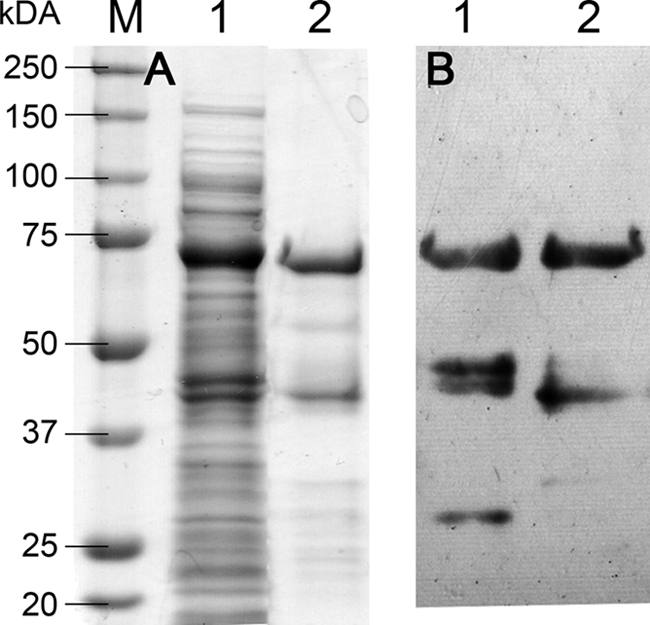
SDS-PAGE of the crude extract from E. coli Rosetta 2(DE3)(pET-22b-StrepII-CSN-eGFP-HIS6 E122Q, pLysSRARE2); (60 μg, lane 1) and CAP purified using affinity chromatography (6 μg; lane 2), either stained using Coomassie brilliant blue G-250 (A) or Western blotted and detected by chemiluminescence using HRP-coupled StrepII affinity protein (B). The band at ca. 75 kDa represents CAP. M, molecular mass markers.
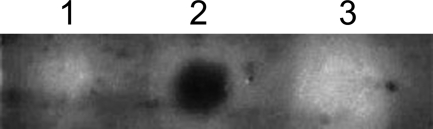
Both the wild-type chitosanase fusion protein and CAP were checked for enzymatic activity in a dot activity assay using a polyacrylamide gel containing chitosan with a DA of 35% as a substrate. Activity, revealed as a dark spot in the calcofluor white-stained gel due to the degradation of the chitosan embedded in the gel, was seen with the wild-type chitosanase fusion protein, but not with CAP (Fig. 3).
Fig 3.
Dot activity assay to check the enzymatic activity of wild-type chitosanase and CAP. Lane 1, crude extract from E. coli Rosetta 2(DE3)(pET-22b(+), pLysSRARE2); lane 2, purified wild-type chitosanase; lane 3, purified CAP. Chitosanase activity is revealed as a dark spot in the calcofluor white-stained gel.
Binding specificity of CAP.
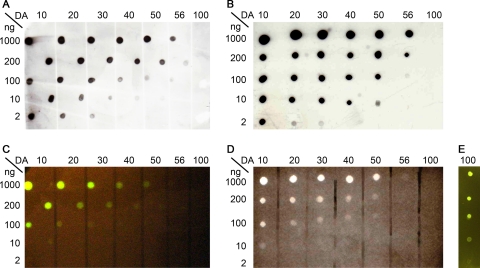
To assess the binding specificity of CAP, different chitosans with DAs ranging from 10% to 56% as well as glycol-chitin (DA, 100%) were spotted in a dilution series (1,000 ng to 2 ng) onto a nitrocellulose membrane. CAP binding to the chitosans can be detected using eGFP fluorescence or by any of the tags attached to it (Fig. 4). eGFP fluorescence was visualized using a transilluminator or under a UV lamp, with StrepII and His6 tags, for example, using the appropriate antibodies coupled to horseradish peroxidase and a chemiluminescence assay. All three detection methods clearly showed that the affinity of CAP decreases with increasing DA and that CAP has no affinity to fully acetylated glycol-chitin; the presence of glycol-chitin on the membrane was verified using WGA-FITC (Fig. 4E).
Fig 4.
Binding specificity of CAP. Different chitosans (DAs from 10% to 56%) and glycol-chitin (DA of 100%) were spotted in dilution series (starting with 1,000 ng) onto nitrocellulose membrane, and the membrane was blocked with BSA, washed, and incubated with CAP (0.1 mg ml−1 in TBS containing 5% [wt/vol] BSA) for 1 h at room temperature before excess CAP was washed off. Bound CAP was detected using StrepII affinity protein (A) or anti-His5 antibody (B) or by the fluorescence of eGFP on a transilluminator (Dark Reader, excitation wavelength of 420 to 500 nm; MoBiTech, Germany) (C) or under a UV lamp (AlphaImager, set to 365 nm; Alpha Innotech Corp., United States) (D). Glycol-chitin (DA of 100%) was detected by fluorescence (Dark Reader) after binding to WGA-FITC conjugate to show that it was not lost during fixation (E).
In situ staining of chitosan using CAP.
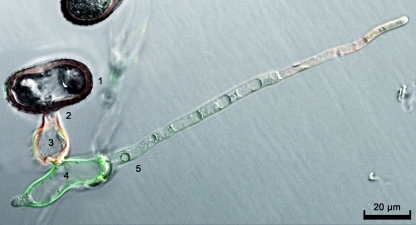
Uredospores of the wheat stem rust fungus were germinated in a petri dish, and differentiation of infection structures was induced using a mild heat shock. It has been shown previously that the ectophytic infection structures, namely, germ tube and appressorium, expose mainly chitin, while the endophytic infection structures, namely, substomatal vesicle and infection hyphae, expose mainly chitosan on the surface of their cell walls; growing tips of the infection hyphae are known to expose chitin rather than chitosan (12). Germlings were double stained using the chitin-specific lectin WGA coupled to Texas Red (4, 36) and the chitosan-specific CAP, and fluorescence was observed by confocal laser scanning fluorescence microscopy (Fig. 5). eGFP and Texas Red fluorescence was monitored using excitation/emission wavelengths of 488/595 nm and 500 to 545/608 to 700 nm, respectively. WGA bound to the germ tubes and appressoria as well as to the tip of the infection hyphae (red fluorescence), while CAP strongly stained the substomatal vesicle and weakly stained the infected hyphae (green fluorescence).
Fig 5.
Double staining of in vitro-induced infection structures of the wheat stem rust fungus Puccinia graminis f. sp. tritici. 1, spore; 2, germ tube; 3, appressorium; 4, substomatal vesicle; 5, infection hypha. Germ tube, appressorium, and the tip of the infection hypha were labeled by WGA conjugated to Texas Red (red fluorescence), indicating the presence of chitin, while CAP staining revealed the presence of chitosan in the substomatal vesicle and, less marked, infection hypha (green fluorescence).
DISCUSSION
Conversion of surface-exposed cell wall chitin into chitosan during penetration and colonization of the host tissue appears to be a widespread pathogenicity mechanism in biotrophic fungi, for both plant and human pathogens (3, 12). This conversion was often deduced from a concomitant induction of a chitin deacetylase rather than by direct histochemical proof of the presence of chitosan due to lack of a commercially available specific stain for chitosan (15). Polyclonal chitosan-specific antisera have been produced repeatedly, and recently, the generation of a monoclonal antibody specifically directed against chitosan has been described (12, 18, 20, 23, 30, 31, 40), but they have rarely been used in histochemical staining due to their typically low affinity toward the antigen (12, 38). Lectins with specificity to chitosan have been described (10, 25) but are not commercially available, and our attempts at reproducing the findings were not successful. The anionic dye eosin Y was used to stain chitosan in the human pathogen Cryptococcus neoformans (3) and in the maize pathogen Ustilago maydis (37). However, this anionic dye, which is typically used as a counterstain to hematoxylin, binds to cationic chitosan presumably through electrostatic interactions only, limiting its specificity. Clearly, alternative chitosan-specific probes are needed to test for chitosan on the surfaces of pathogenic fungi.
Even though lectins with high affinity and good specificity for chitin such as WGA are easily available, a number of alternative chitin-specific probes have been described. A wild-type chitinase was successfully used to stain chitin in pathogenic fungi and fungus-infected body fluids (6). A chitin binding domain fused to GFP has been used for staining chitin in Saccharomyces cerevisiae and in human tissue with various fungal infections (19). In spite of one report of using a gold-complexed chitosanase for the cytochemical detection of chitosan (17), this approach does not seem to have been followed up. In the present work, we have extended this enzyme-based strategy by engineering a chitosan-specific probe from a site-directed mutagenesis-inactivated chitosanase fused to eGFP and two affinity tags for purification and detection. The specificity of the chitosanase chosen for the degradation of highly deacetylated chitosans (unpublished) confers the same binding specificity to the mutein, and exchanging a catalytically active residue prevents hydrolysis of the bound substrate. Active site residues have been investigated in detail in three bacterial hydrolases belonging to the CAZY glycoside hydrolase family 8, namely, in a chitosanase from Bacillus sp. strain 17, in endoglucanase CelA from Clostridium thermocellum, and in endoglucanase K from Bacillus sp. strain KSM-330 (1, 2, 27). In all three enzymes, a glutamic acid residue acts as a proton donor and an aspartic acid residue acts as a nucleophile in catalysis. These amino acids are conserved in all GH-8 family members. Previously, both the glutamic acid and the aspartic acid residues in the Bacillus sp. strain 17 chitosanase have been substituted for with glutamine and asparagine, respectively, to show that both are important in catalysis and that the aspartic acid residue is involved in substrate binding too (1). In the present work, we therefore substituted only the glutamic acid residue for glutamine, leaving the aspartic acid residue unchanged. This abolished the hydrolytic activity completely but retained the substrate affinity.
The site-directed-mutagenesis-inactivated chitosanase was fused to eGFP as well as to two affinity tags, allowing easy one-step purification via affinity chromatography and providing alternative methods of detection. eGFP can be visualized directly due to its fluorescence, and all three tags can be detected using commercially available antibodies coupled to a large variety of markers. As an example, we have used the very sensitive detection method based on horseradish peroxidase-coupled antibodies and chemiluminescence of its enzymatic product. This method is clearly more sensitive than fluorescence detection of eGFP, but the latter is the easiest way of in situ detection of chitosan, e.g., in the cell walls of pathogenic fungi, using fluorescence microscopy.
It is interesting to note that CAP gave a binding pattern that slightly differed from the one seen previously using a polyclonal antichitosan antiserum (12). In both cases, chitin was seen to be exposed in the epiphytic infection structures of the wheat stem rust fungus, namely, germ tubes and appressoria, and chitosan was detected on the surface of endophytic infection structures, namely, substomatal vesicles and infection hyphae. However, while both chitosan-specific probes stained the substomatal vesicle strongly, the antibody bound more strongly to the infection hyphae than CAP. We assume that this differential staining pattern is due to subtle differences in the specificities of the two probes. While we have shown in this work that the affinity of the chitosanase-based CAP is highest for chitosans with very low degrees of acetylation, the antiserum appears to bind best to chitosans with intermediate degrees of acetylation (unpublished). We may, thus, conclude that the chitosan in the substomatal vesicle is more strongly deacetylated than the one exposed on the surface of the infection hyphae. To date, no methods exist for the in situ analysis of the degree of acetylation of chitosans in a cell wall, and the degree of acetylation of chitosans present in fungal cell walls has not been determined accurately. The differential binding of the two chitosan-specific probes may guide the way to the development of a histochemical assay for the determination of DA.
Supplementary Material
Footnotes
Published ahead of print 24 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Adachi W, et al. 2004. Crystal structure of family GH-8 chitosanase with subclass II specificity from Bacillus sp. K17. J. Mol. Biol. 343:785–795 [DOI] [PubMed] [Google Scholar]
- 2. Alzari PM, Souchon H, Dominguez R. 1996. The crystal structure of endoglucanase CelA, a family 8 glycosyl hydrolase from Clostridium thermocellum. Structure 4:265–275 [DOI] [PubMed] [Google Scholar]
- 3. Baker LG, Specht CA, Donlin MJ, Lodge JK. 2007. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot. Cell 6:855–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baldo BA, Barnett D, Lee JW. 1984. Lectins as cytochemical probes of the developing wheat-grain.V. Demonstration of separate polysaccharides containing N-acetyl-d-glucosamine and d-galactose in nucellar epidermal-cell walls. Aust. J. Plant Physiol. 11:179–190 [Google Scholar]
- 5. Benhamou N, Asselin A. 1989. Attempted localization of a substrate for chitinases in plant-cells reveals abundant N-acetyl-d-glucosamine residues in secondary walls. Biol. Cell 67:341–350 [Google Scholar]
- 6. Benjaminson MA. 1969. Conjugates of chitinase with fluorescein isothiocyanate or lissamine rhodamine as specific stains for chitin in situ. Stain Technol. 44:27–31 [DOI] [PubMed] [Google Scholar]
- 7. Biondo C, et al. 2003. Induction of T helper type 1 responses by a polysaccharide deacetylase from Cryptococcus neoformans. Infect. Immun. 71:5412–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Briza P, Ellinger A, Winkler G, Breitenbach M. 1988. Chemical composition of the yeast ascospore wall—the 2nd outer layer consists of chitosan. J. Biol. Chem. 263:11569–11574 [PubMed] [Google Scholar]
- 9. Brosson D, Kuhn L, Prensier G, Vivares CP, Texier C. 2005. The putative chitin deacetylase of Encephalitozoon cuniculi: a surface protein implicated in microsporidian spore-wall formation. FEMS Microbiol. Lett. 247:81–90 [DOI] [PubMed] [Google Scholar]
- 10. Chen HP, Xu LL. 2005. Isolation and characterization of a novel chitosan-binding protein from non-heading Chinese cabbage leaves. J. Integr. Plant Biol. 47:452–456 [Google Scholar]
- 11. Das S, et al. 2006. The cyst wall of Entamoeba invadens contains chitosan (deacetylated chitin). Mol. Biochem. Parasitol. 148:86–92 [DOI] [PubMed] [Google Scholar]
- 12. El Gueddari NE, Rauchhaus U, Moerschbacher BM, Deising HB. 2002. Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol. 156:103–112 [Google Scholar]
- 13. Felipe MSS, et al. 2005. Transcriptional profiles of the human pathogenic fungus Paracoccidioides brasiliensis in mycelium and yeast cells. J. Biol. Chem. 280:24706–24714 [DOI] [PubMed] [Google Scholar]
- 14. Ferreira RB, et al. 2007. The role of plant defence proteins in fungal pathogenesis. Mol. Plant Pathol. 8:677–700 [DOI] [PubMed] [Google Scholar]
- 15. Ghormade V, Kulkarni S, Doiphode N, Rajamohanan PR, Deshpande MV. 2010. Chitin deacetylase: a comprehensive account on its role in nature and its biotechnological applications, p 1054–1066 In Mendez-Vilas A. (ed), Current research, technology and education topics in applied microbiology and microbial biotechnology. Formatex Research Center, Badajoz, Spain [Google Scholar]
- 16. Gould GW, Georgala DL, Hitchins AD. 1963. Fluorochrome-labelled lysozyme—reagent for detection of lysozyme substrate in cells. Nature 200:385–386 [DOI] [PubMed] [Google Scholar]
- 17. Grenier J, Benhamou N, Asselin A. 1991. Colloidal gold-complexed chitosanase—a new probe for ultrastructural-localization of chitosan in fungi. J. Gen. Microbiol. 137:2007–2015 [Google Scholar]
- 18. Hadwiger LA, Line RF. 1981. Hexosamine accumulations are associated with the terminated growth of Puccinia striiformis on wheat isolines. Physiol. Plant Pathol. 19:249–255 [Google Scholar]
- 19. Hardt M, Laine RA. 2004. Mutation of active site residues in the chitin-binding domain ChBD(ChiAl) from chitinase A1 of Bacillus circulans alters substrate specificity: use of a green fluorescent protein binding assay. Arch. Biochem. Biophys. 426:286–297 [DOI] [PubMed] [Google Scholar]
- 20. Hartmann DJ, Sorlier P, Denuziere A, Viton C, Domard A. 2003. Preparation and development of anti-chitosan antibodies. J. Biomed. Mater. Res. A 67A:766–774 [DOI] [PubMed] [Google Scholar]
- 21. Hirai A, Odani H, Nakajima A. 1991. Determination of degree of deacetylation of chitosan by H-1-NMR spectroscopy. Polymer Bull. 26:87–94 [Google Scholar]
- 22. Kafetzopoulos D, Martinou A, Bouriotis V. 1993. Bioconversion of chitin to chitosan—purification and characterization of chitin deacetylase from Mucor rouxii. Proc. Natl. Acad. Sci. U. S. A. 90:2564–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim SY, Shon DH, Lee KH. 2000. Enzyme-linked immunosorbent assay for detection of chitooligosaccharides. Biosci. Biotechnol. Biochem. 64:696–701 [DOI] [PubMed] [Google Scholar]
- 24. Levitz SM, Nong SH, Mansour MK, Huang C, Specht CA. 2001. Molecular characterization of a mannoprotein with homology to chitin deacetylases that stimulates T cell responses to Cryptococcus neoformans. Proc. Natl. Acad. Sci. U. S. A. 98:10422–10427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lienart Y, Gautier C, Domard A. 1991. Isolation from Rubus cell-suspension cultures of a lectin specific for glucosamine oligomers. Planta 184:8–13 [DOI] [PubMed] [Google Scholar]
- 26. Manocha MS, Zhonghua Z. 1997. Immunocytochemical and cytochemical localization of chitinase and chitin in the infected hosts of a biotrophic mycoparasite, Piptocephalis virginiana. Mycologia 89:185–194 [Google Scholar]
- 27. Ozaki K, Sumitomo N, Hayashi Y, Kawai S, Ito S. 1994. Site-directed mutagenesis of the putative active site of endoglucanase K from Bacillus sp. KSM-330. Biochim. Biophys. Acta 1207:159–164 [DOI] [PubMed] [Google Scholar]
- 28. Pital A, Janowitz SL, Hudak CE, Lewis EE. 1967. Fluorescein-labeled beta-glucosidase as a bacterial stain. Appl. Microbiol. 15:1165–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rajulu MBG, et al. 2011. Chitinolytic enzymes from endophytic fungi. Fungal Divers. 47:43–53 [Google Scholar]
- 30. Ryan GB, Jones WT, Mitchell RE, Mett V. 2001. Polyclonal antibody production against chito-oligosaccharides. Food Agric. Immunol. 13:127–130 [Google Scholar]
- 31. Schubert M, et al. 2010. A monoclonal antibody that specifically binds chitosan in vitro and in situ on fungal cell walls. J. Microbiol. Biotechnol. 20:1179–1184 [DOI] [PubMed] [Google Scholar]
- 32. Seibert GR, Benjaminson MA, Hoffman H. 1978. Conjugate of cellulase with fluorescein isothiocyanate-specific stain for cellulose. Stain Technol. 53:103–106 [DOI] [PubMed] [Google Scholar]
- 33. Sorlier P, Hartmann DJ, Denuziere A, Viton C, Domard A. 2003. Preparation and development of anti-chitosan antibodies. J. Biomed. Mater. Res. A 67:766–774 [DOI] [PubMed] [Google Scholar]
- 34. Studier FW. 2005. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41:207–234 [DOI] [PubMed] [Google Scholar]
- 35. Taylor MJ, et al. 2002. Detection of fungal organisms in eosinophilic mucin using a fluorescein-labeled chitin-specific binding protein. Otolaryngol. Head Neck Surg. 127:377–383 [DOI] [PubMed] [Google Scholar]
- 36. Titus JA, Haugland R, Sharrow SO, Segal DM. 1982. Texas Red, a hydrophilic, red-emitting fluorophore for use with fluorescein in dual parameter flow micro-fluorometric and fluorescence microscopic studies. J. Immunol. Methods 50:193–204 [DOI] [PubMed] [Google Scholar]
- 37. Treitschke S, Doehlemann G, Schuster M, Steinberg G. 2010. The myosin motor domain of fungal chitin synthase V is dispensable for vesicle motility but required for virulence of the maize pathogen Ustilago maydis. Plant Cell 22:2476–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tucker SL, et al. 2010. Common genetic pathways regulate organ-specific infection-related development in the rice blast fungus. Plant Cell 22:953–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vachoud L, Zydowicz N, Domard A. 1997. Formation and characterisation of a physical chitin gel. Carbohydr. Res. 302:169–177 [Google Scholar]
- 40. Walker AN, Garner RE, Horst MN. 1990. Immunocytochemical detection of chitin in Pneumocystis carinii. Infect. Immun. 58:412–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wan J, Zhang XC, Stacey G. 2008. Chitin signaling and plant disease resistance. Plant Signal Behav. 3:831–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.