Abstract
The relative abundance of micromonosporas in the bacterial communities inhabiting cellulose baits, water columns, and sediments of two freshwater lakes was determined by quantitative PCR (qPCR) of reverse-transcribed 16S rRNA. Micromonospora spp. were shown to be significant members of the active bacterial population colonizing cellulosic substrates in the lake sediment, and their increased prevalence with greater depth was confirmed by enumeration of CFU.
TEXT
The genus Micromonospora comprises slow-growing actinomycetes found in terrestrial, aquatic, and marine environments and abundant in soils and freshwater sediments (9). Members of this genus are considered to be particularly suited to an aquatic lifestyle due to the absence of aerial hyphae and the production of hydrophilic spores, and in some early studies, micromonosporas were found to be the most common actinomycetes isolated from lake sediments (2). Micromonosporas are well known for their ability to degrade complex recalcitrant materials, such as cellulose and chitin (8, 9, 18), which confers upon these organisms a potentially important role in recycling particulate organic matter in freshwater environments.
Recently, evidence has been obtained for the presence, growth, and cellulolytic activity of micromonosporas in two lakes in the English Lake District (3). Micromonosporas were shown to grow on cellulose baits (dewaxed cotton strings) and to be able to degrade cellulose when growing in unsupplemented lake water. However, the importance of micromonosporas in the ecology of lake sediments remains unknown, particularly as lakes stratify in the summer months, leading to much lower oxygen tensions in the sediments precisely when most of the biological activity occurs. However, there is evidence that micromonosporas can grow under microaerophilic conditions (6, 17), and a recent study has described a chicken cecum isolate with 99% 16S rRNA gene sequence homology to Micromonospora that was capable of degrading cyanophycin in anoxic conditions (15).
Here, the importance of members of the genus Micromonospora in freshwater environments was addressed by determining their relative abundance in the total bacterial community using quantitative PCR (qPCR) (16) alongside enumeration of Micromonospora isolates obtained from lake water, cellulose baits, and lake sediments.
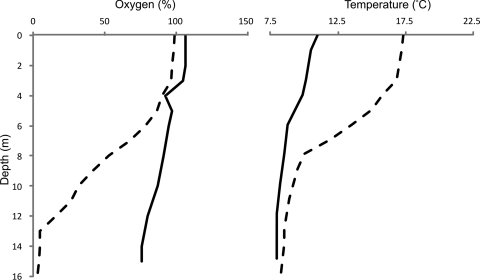
The sampling sites were Esthwaite Water and Priest Pot, two lakes of contrasting trophic statuses in the English Lake District. The former is a eutrophic lake, ca. 16 m in depth and 113.3 ha in area with an average chlorophyll a concentration of 54 μg liter−1, and is considered one of the most productive lakes in the English Lake District (5). Priest Pot is a small (1-ha) and shallow (3.4-m-deep) lake; it is considered hypereutrophic (chlorophyll a concentration of up to 800 μg liter−1) (4). Cellulose baits (cotton strings dewaxed by repeated chloroform-ethanol extraction) (19), lake water, and sediments were retrieved from the lakes as described previously (3). The baits were placed in the lakes on 28 April 2008 and recovered on 3 June 2008. At each depth, a 0.5-g nylon rope sample was included as a control for benign surface attachment of microorganisms. Temperature and oxygen concentrations were measured in Esthwaite Water using a YSI 85 dissolved oxygen, conductivity, salinity, and temperature meter (Geo Scientific Ltd., Vancouver, Canada). On introduction of the cotton baits, the water column of Esthwaite Water was mixed and the oxygen concentrations remained relatively high, even in the deeper sections of the lake (Fig. 1); when the baits were recovered 5 weeks later, the water column was distinctly stratified, with little dissolved oxygen below 13 m (Fig. 1). The temperature increased in the upper water column but changed little in the deep parts of the lake. Based on previous studies in this laboratory demonstrating that Priest Pot stratifies earlier and more persistently than Esthwaite Water (data not shown), it is assumed that this neighboring lake was also stratified at the time of sampling.
Fig 1.
Dissolved oxygen and temperature depth profiles in Esthwaite Water samples obtained from the constant monitoring station on 28 April 2008 (solid line) and 3 June 2008 (dotted line). Dissolved oxygen concentrations are expressed as percentages of air saturation.
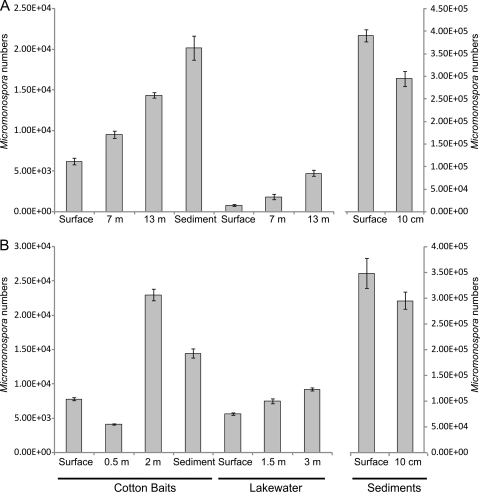
Micromonospora strains were identified on the basis of their characteristic morphology (12), i.e., slow-growing small orange, red, brown, or black colonies with little or no aerial mycelium and finely branching substrate hyphae that are <0.5 μm in diameter, ultimately differentiating into large numbers of single spores within the agar. They were isolated from cotton baits, sediment, and lake water using the methods described previously (3) with 3 replicate cotton baits per depth. The numbers of micromonosporas counted on isolation plates increased with depth in both the cotton baits and the lake water itself in Esthwaite Water (Fig. 2A). In Priest Pot, the numbers of micromonosporas in cotton baits were greater at 2 m (Fig. 2B), whereas in the water column, they increased with depth as in Esthwaite Water. The greatest numbers of isolates overall in both lakes were obtained from the sediments. One-way analysis of variance (ANOVA) performed on CFU numbers from cotton baits and lake water showed that the depth profile differences observed were significant (P < 0.005). For effective selective isolation of Micromonospora strains, a heat treatment step was used (10 min at 65°C, followed by plating on M3 agar) (3, 14) such that the CFU counts (Fig. 2) represent Micromonospora spores. Therefore, for an accurate quantification of micromonosporas, including active hyphae, their 16S rRNA was assessed by quantitative PCR of reverse-transcribed RNA and compared to that from total bacterial 16S rRNA to provide relative abundance determinations.
Fig 2.
Micromonospora numbers in cotton baits (CFU g−1), lake water (CFU liter−1), and sediments (CFU g−1 of wet sediment) obtained from Esthwaite Water (A) and Priest Pot (B) (n = 3) on 3 June 2008. Error bars represent standard errors.
Specific primers for Micromonospora were designed using taxon-specific sequences identified in the ARB databases (11), which contain sequences downloaded from SILVA. The primers were named M1C (5′-TTACCTGGGTTTGACATGG-3′) and M2D (5′-ACATCGAACGAGGGTTGC-3′). The specificity analysis on ProbeMatch (1) and ProbeCheck (10) demonstrated that this primer pair could also amplify some members of other genera within the Micromonosporaceae, i.e., a limited number of Actinoplanes and Dactylosporangium spp. This primer pair is therefore considered a semispecific Micromonospora primer set that may amplify DNA from other members of the Micromonosporaceae. Ten 142- to 149-bp clones were obtained from environmental DNA amplified with M1C and M2D primers. All clones matched Micromonospora strains from the public databases at >83% identity, with 5 having >90% identity, including one clone that had a 100% match to Micromonospora chalcea. Micromonospora spp., or closely related strains from the Micromonosporaceae family, were the top BLASTn hits for 7 of the clones when uncultured environmental sequences were excluded from the analysis. When mismatches in the qPCR primer sequences were taken into account, all clones were clearly identified as Micromonosporaceae, providing evidence that this primer set will only amplify members of the target group for which they were designed. RNA was extracted using the phenol-chloroform nucleic acid extraction method of Griffiths et al. (7). Cellulose baits, sediment (0.3 g), or the 0.2-μm-pore-size polycarbonate membranes (Millipore) used to filter lake water (10 to 200 ml) were added to lysing matrix E tubes (Qbiogene, Montreal, Canada), and the DNA/RNA was extracted. All glassware was washed with chloroform to remove RNases, and reagents were produced with distilled water treated with 0.1% (vol/vol) diethylpyrocarbonate (Sigma, Gillingham, United Kingdom). DNA was removed from nucleic acid extracts used in cDNA synthesis for the qPCR assays using the Turbo DNA-free kit (Applied Biosystems, Carlsbad, CA). The absence of residual DNA was confirmed by qPCR. RNA was reverse transcribed using Bioscript Moloney murine leukemia virus (MMLV) reverse transcriptase (Bioline) according to the manufacturer's protocol.
Quantitative PCR sample analysis was performed in a 7500 Fast real-time PCR system (Applied Biosystems) using a SensiMixPlus SYBR kit (Bioline). The amplification cycle comprised a 10-min activation step at 95°C, followed by 35 cycles starting at 95°C for 15 s, 30 s at 60°C, and 5 to 10 s at 72°C for extension. Each well on the multiwell tray or strip contained 12.5 μl of 2× Sensimix qPCR reagent mix, 1 μl of cDNA template (35 ng), 400 nM (final concentration) of each primer, and distilled water to a final volume of 25 μl. The products of qPCR amplification from natural samples were also run on 1 or 2% agarose gels to confirm the absence of erroneous amplification products. The 16S rRNA gene of Micromonospora lake strain 13 (3) was used to produce calibration curves; for the quantification of total bacterial 16S rRNA, the primers 1369F and Prok 1429R (16) were used, all as described in McDonald et al. (13). Three replicate cotton baits, sediments, or water samples were analyzed per depth. All of the calibration curves had R2 values of ≥0.99, and the primer pair efficiencies were >97%.
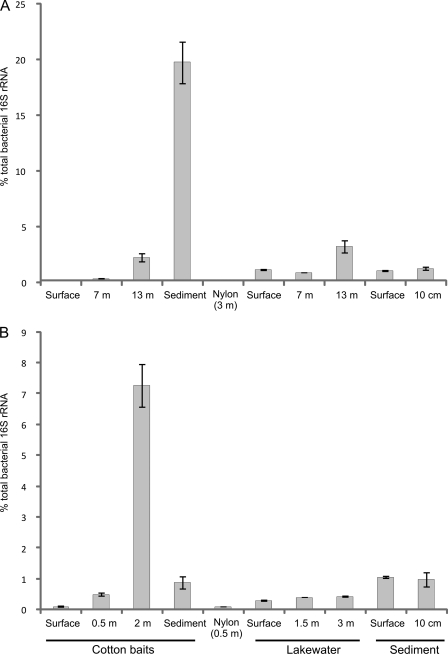
There was a similar trend in the depth profiles of Micromonospora numbers obtained by colony enumeration and those obtained by relative 16S rRNA abundance determination (Fig. 2 and 3). The relative abundance of Micromonospora 16S rRNA in the cotton baits increased with depth, reaching 20% of the total bacterial community in the cotton bait placed just above the sediment in Esthwaite Water and 7% in the 2-m-deep bait in Priest Pot. In the surface cotton baits and in the nylon control sample, they formed ≤0.1% of the bacterial community in both lakes (Fig. 3). A one-way ANOVA showed that in the cotton bait and lake water samples of both lakes, the differences in Micromonospora relative abundances according to depth were significant (P < 0.05). The increase in relative Micromonospora abundance with greater depth might be explained by the lack of competition from faster-growing organisms that produced thick biofilms on the cotton baits placed in shallow water. Although oxygen concentrations are lower toward the bottom of the lake (Fig. 1), there may be sufficient oxygen to sustain microaerophilic growth. In addition, periodic storms and strong winds can disrupt the oxycline and replenish the oxygen in the deep layers of the water column. There are suggestions that Micromonospora spp. can grow under microaerophilic conditions (6, 15, 17), a theory which the environmental data presented here now support. The low oxygen concentrations at greater depth may therefore restrict the growth of other microorganisms more so than that of micromonosporas but remain sufficient to prevent significant growth of strict anaerobes. In addition, the Micromonospora species abundances in baits close to the sediment will be promoted by the large number of spores serving as growth centers and the higher concentration of nutrients present in the sediment. Micromonospora abundance in the water column itself was ca. 1% or less except in the 13-m-deep sample from Esthwaite Water, in which it was approximately 3% (Fig. 3A).
Fig 3.
Relative abundances of Micromonospora spp. as determined by qPCR of cDNA prepared from RNA extracts from Esthwaite Water (A) and Priest Pot (B) on 3 June 2008 (n = 3). Error bars represent standard errors.
Overall, the results presented here offer strong evidence for active Micromonospora populations in the lakes studied and for their importance under microaerophilic conditions as a significant component of the active bacterial community attached to and degrading cellulosic substrates. This is demonstrated by the fact that their relative abundance was highest in baits placed just above the sediment and relatively low in the sediments themselves and in the water column. The quantification of Micromonospora rRNA transcripts by qPCR was essential to differentiate between spores and the active community and demonstrates that this group can be among the most abundant of those bacterial species that colonize and degrade cellulosic substrates in freshwater habitats.
Nucleotide sequence accession numbers.
The ten cloned DNA sequences obtained from environmental DNA amplified with the M1C and M2D primers were deposited in GenBank under accession numbers HQ644027 to HQ644036.
Footnotes
Published ahead of print 2 March 2012
REFERENCES
- 1. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cross T. 1981. Aquatic actinomycetes—a critical survey of the occurrence, growth and role of actinomycetes in aquatic habitats. J. Appl. Bacteriol. 50:397–423 [DOI] [PubMed] [Google Scholar]
- 3. de Menezes AB, Lockhart RJ, Cox MJ, Allison HE, McCarthy AJ. 2008. Cellulose degradation by micromonosporas recovered from freshwater lakes and classification of these actinomycetes by DNA gyrase B gene sequencing. Appl. Environ. Microbiol. 74:7080–7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Finlay BJ, Maberly SC. 2000. Microbial diversity in Priest Pot: a productive pond in the English Lake District. Freshwater Biological Association special publication 9. Freshwater Biological Association, Ambleside, Cumbria, United Kingdom [Google Scholar]
- 5. George DG, Talling JF, Rigg E. 2000. Factors influencing the temporal coherence of five lakes in the English Lake District. Freshwater Biol. 43:449–461 [Google Scholar]
- 6. Goodfellow M, Williams ST. 1983. Ecology of actinomycetes. Annu. Rev. Microbiol. 37:189–216 [DOI] [PubMed] [Google Scholar]
- 7. Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jendrossek D, Tomasi G, Kroppenstedt RM. 1997. Bacterial degradation of natural rubber: a privilege of actinomycetes? FEMS Microbiol. Lett. 150:179–188 [DOI] [PubMed] [Google Scholar]
- 9. Kawamoto I. 1984. Genus Micromonospora, p 2442–2450 In Williams ST, Sharpe ME, Holt JG. (ed), Bergey's manual of systematic bacteriology, vol 4 Williams & Wilkins, Baltimore, MD [Google Scholar]
- 10. Loy A, et al. 2008. probeCheck—a central resource for evaluating oligonucleotide probe coverage and specificity. Environ. Microbiol. 10:2894–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ludwig W, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCarthy AJ, Williams ST. 1990. Methods for studying the ecology of actinomycetes. Methods Microbiol. 22:533–563 [Google Scholar]
- 13. McDonald JE, de Menezes AB, Allison HE, McCarthy AJ. 2009. Molecular biological detection and quantification of novel Fibrobacter populations in freshwater lakes. Appl. Environ. Microbiol. 75:5148–5152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rowbotham TJ, Cross T. 1977. Ecology of Rhodococcus coprophilus and associated actinomycetes in freshwater and agricultural habitats. J. Gen. Microbiol. 100:231–240 [Google Scholar]
- 15. Sallam A, Steinbuchel A. 2009. Cyanophycin-degrading bacteria in digestive tracts of mammals, birds and fish and consequences for possible applications of cyanophycin and its dipeptides in nutrition and therapy. J. Appl. Microbiol. 107:474–484 [DOI] [PubMed] [Google Scholar]
- 16. Suzuki MT, Taylor LT, DeLong EF. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vobis G. 2006. The genus Actinoplanes and related genera, p 623–653 In Dworkin M. (ed), Prokaryotes, vol 3 Springer-Verlag, New York, NY [Google Scholar]
- 18. Wohl DL, McArthur JV. 2001. Aquatic actinomycete-fungal interactions and their effects on organic matter decomposition: a microcosm study. Microb. Ecol. 42:446–457 [DOI] [PubMed] [Google Scholar]
- 19. Wood TM. 1988. Preparation of crystalline, amorphous, and dyed cellulase substrates. Methods Enzymol. 160:19–25 [Google Scholar]