Abstract
We have created new complementation constructs for use in Neisseria gonorrhoeae and Neisseria meningitidis. The constructs contain regions of homology with the chromosome and direct the insertion of a gene of interest into the intergenic region between the genes iga and trpB. In order to increase the available options for gene expression in Neisseria, we designed the constructs to contain one of three different promoters. One of the constructs contains the isopropyl-β-d-thiogalactopyranoside-inducible lac promoter, which has been widely used in Neisseria. We also designed a construct that contains the strong, constitutive promoter from the gonococcal opaB gene. The third construct contains a tetracycline-inducible promoter, a novel use of this promoter in Neisseria. We demonstrate that anhydrotetracycline can be used to induce gene expression in the pathogenic Neisseria at very low concentrations and without negatively affecting the growth of the organisms. We use these constructs to complement an arginine auxotrophy in N. gonorrhoeae as well as to express a translational fusion of alkaline phosphatase with TraW. TraW is a component of the gonococcal type IV secretion system, and we demonstrate that TraW localizes to the periplasm.
INTRODUCTION
Neisseria gonorrhoeae and Neisseria meningitidis are naturally transformable and do not regulate DNA uptake (2, 9, 18, 59). As a result, a variety of genetic tools for making mutations via transformation are available, and it is relatively easy to delete specific genes, construct point mutations, and perform insertional mutagenesis (17, 20, 47, 55). However, the tools that are available for performing complementation in the pathogenic Neisseria are limited. Prophages have been identified in the gonococcal and meningococcal chromosomes (14, 50), but no transducing phages have been isolated (16). Several plasmids are available for complementation in N. gonorrhoeae and N. meningitidis, including the Hermes vectors, pLES2, and pEN11 (7, 29, 62), but complementation using replicating plasmids in Neisseria can be problematic since engineered replicating plasmids are often unstable. Furthermore, since plasmids are nicked and taken up as a single strand during natural transformation, introduction of a replicating plasmid follows two-hit kinetics and thus occurs at a low frequency if the plasmid does not have homology to itself or the gonococcal chromosome (6, 18).
Instead, complementation in gonococci is usually performed by inserting the gene of interest into a site on the gonococcal chromosome. Several chromosomal loci have been characterized for use in gene complementation, including proB, iga, and the porin pseudogene (60, 61, 67). The intergenic region between lctP and aspC has also been used extensively for complementation, and a variety of constructs have been engineered to direct gene insertion at this site (19, 39, 64). Collectively, the existing complementation constructs have significant limitations. With the iga and proB constructs, gene insertion occurs within an open reading frame (61, 67), introducing the possibility that the growth or pathogenesis of the organism might be altered. Additionally, the available promoters for controlling gene expression are limited. In most cases, the gene of interest is either cloned along with its native promoter or expressed from the promoter of an upstream antibiotic resistance marker. To date, the lac promoter is the only inducible promoter that has been described for use in Neisseria (12, 54, 66). Additional tools for gene complementation in the pathogenic Neisseria are needed.
In this study, we describe a new set of complementation constructs that contain regions of homology with iga and trpB and direct gene insertion to the iga-trpB intergenic region following double-crossover recombination with the chromosome. These constructs were designed to contain one of three different promoters: the well-characterized lac promoter, a strong constitutive promoter, or a tetracycline-inducible promoter. This is the first report of tetracycline-inducible gene expression in Neisseria. We demonstrate that these constructs can be used to complement an arginine auxotrophy in gonococcal strain DGI2. We also use these constructs to express TraW, a required component of the gonococcal type IV secretion system (T4SS) (46), and investigate its localization within the cell. TraW, along with the other proteins required for type IV secretion, is encoded on the 57-kb gonococcal genetic island (GGI). In gonococci, the T4SS secretes single-stranded DNA into the extracellular environment (13, 18, 52), but little is known about the function of TraW in the process of secretion. Using alkaline phosphatase (AP) fusions in one of our new complementation constructs, we show that TraW localizes to the periplasm.
MATERIALS AND METHODS
Bacteria and growth conditions.
Bacterial strains used in this study are described in Table 1. Escherichia coli strains were grown on Luria-Bertani (LB) agar plates or in LB broth at 37°C (53). Gonococcal and meningococcal strains were grown either on gonococcal base (GCB) agar plates (Difco) containing Kellogg's supplements (25) or in GCB liquid (GCBL) medium containing Kellogg's supplements and 0.042% NaHCO3 (41) as described previously (12). Erythromycin was used at 10 μg/ml for meningococci, 2 μg/ml for gonococcal strain MR574, 10 μg/ml for all other gonococcal strains, and 500 μg/ml for E. coli. Kanamycin was used at 40 μg/ml for E. coli. Chloramphenicol was used at 10 μg/ml for gonococci and 25 μg/ml for E. coli. Gene expression in gonococci was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) or 2 ng/ml anhydrotetracycline (ATc), except where otherwise noted. To observe alkaline phosphatase activity, meningococcal strains were grown on GCB-Tris agar plates containing 5-bromo-4-chloro-3-indolylphosphate (XP) (8) with or without 1 mM IPTG or 20 ng/ml anhydrotetracycline. To measure growth of the AHU (arginine, hypoxanthine, and uracil) auxotrophic strain DGI2 and the complemented strain MR574, the strains were grown in liquid gonococcal genetic medium (GGM) (30) without soluble starch in the presence or absence of l-arginine, and the optical density at 540 nm (OD540) was recorded.
Table 1.
Bacterial strains and plasmids
| Plasmid or strain | Description | Reference or source |
|---|---|---|
| Plasmids | ||
| pCK1 | PCR of MS11 traW with traWF-XmaI and traWR-SpeI into pKH116 XmaI and SpeI sites (TraWFL::′PhoA) | This work |
| pGCC6del | Derivative of pGCC6 that underwent a spontaneous deletion of portions of lacIq and downstream DNA | 39 |
| pHSS6 | Backbone for complementation constructs; Knr | 56 |
| pIDN1 | Cloning vector, source of ermC | 20 |
| pIDN3 | Cloning vector, source of ermC | 20 |
| pKH21 | BamHI/blunt HpaI fragment from pRKO2 cloned into BamHI/blunt BstBI sites of a pGCC6del derivative modified to contain an improved polylinker | This work |
| pKH22 | BamHI/blunt HpaI fragment from pRKO2 cloned into BamHI/blunt BstBI sites of a pGCC6del derivative modified to contain an improved polylinker | This work |
| pKH35a | lctP-aspC complementation construct, source of lac regulatory elements in pMR39 | 19 |
| pKH37a | lctP-aspC complementation construct, source of lac regulatory elements in pMR33 | 27 |
| pKH39 | PCR of lacZ from JD1504 with lacZ5′-PstI and lacZ3′-MfeI into pKH35 PstI and MfeI sites | This work |
| pKH116 | ′phoA on blunt ApaI/KpnI fragment from pUI1158 cloned into pIDN1 Ecl136 and KpnI sites | This work |
| pMR18 | PCR of trpB-iga from MS11 with trpBF-NotI and igaR-NotI cloned into pHSS6 NotI site | This work |
| pMR19 | PCR around pMR18 with pMR18F and pMR18R-KpnI, digested with BsaI, and self-ligated | This work |
| pMR24 | PCR from pKH37 with pKH37F and pKH37R-KpnI cloned into pMR19 KpnI site | This work |
| pMR28 | PsiI/PacI fragment of PCR from FA1090 with OpaPro-PsiI and OpaPro-PacI cloned into PacI/blunt BseRI sites of pMR24 | This work |
| pMR30 | PCR around pMR18 with pMR18F and pMR18R-SacI, digested with BsaI, and self-ligated | This work |
| pMR32b | iga-trpB complementation construct (PopaB), polylinker sites in same orientation as pKH37 | This work |
| pMR33b | iga-trpB complementation construct (lacPO), polylinker sites in same orientation as pKH37 | This work |
| pMR35 | PCR from pKH35 with pKH37F and pKH37R-SacI cloned into pMR30 SacI site | This work |
| pMR38 | PsiI/PacI fragment of PCR from FA1090 with OpaPro-PsiI and OpaPro-PacI cloned into PacI/blunt BseRI sites of pMR35 | This work |
| pMR39b | iga-trpB complementation construct (lacPO), polylinker sites in same orientation as pKH35 | This work |
| pMR41b | iga-trpB complementation construct (PopaB), polylinker sites in same orientation as pKH35 | This work |
| pMR47 | PCR around pCK1 with deltraWF and deltraWR, digested with BamHI, and self-ligated (TraWSS::′PhoA) | This work |
| pMR49 | Blunted XhoI/FspI fragment of pKH21 cloned into blunted pMR19 KpnI site | This work |
| pMR50 | Blunted SacI/FspI fragment of pKH22 cloned into blunted pMR19 KpnI site | This work |
| pMR53 | PCR of pMR47 with pMR47F-SacII and pMR47R-XhoI into pMR33 SacII and XhoI sites | This work |
| pMR57 | PCR of pMR47 with pMR47F-SacII and pMR47R-XhoI into pMR32 SacII and XhoI sites | This work |
| pMR58 | PCR of pMR47 with pMR47F-SacII and pMR47R-XhoI into pKH37 SacII and XhoI sites | This work |
| pMR61 | PCR of pCK1 with pMR47F-SacII and pMR47R-XhoI into pMR33 SacII and XhoI sites | This work |
| pMR68b | iga-trpB complementation construct (Ptet), polylinker sites in same orientation as pKH37 | This work |
| pMR69b | iga-trpB complementation construct (Ptet), polylinker sites in same orientation as pKH35 | This work |
| pMR70 | PCR of pMR47 with pMR47F-SacII and pMR47R-SpeI into pMR68 SacII and SpeI sites | This work |
| pMR71 | PCR of pMR47 with pMR47F-XhoI and pMR47R-SacII into pMR69 XhoI and SacI sites | This work |
| pMR90 | PCR of argJ from MS11 with argJF-SacI and argJR-SpeI into pMR68 SacI and SpeI sites | This work |
| pRKO2 | Original source of tetracycline repressor and p57opt promoter | 63 |
| pUI1158 | Original source of ′phoA and polylinker | 45 |
| SM44 | pWC147 containing mTnCm-3 insertion 44 downstream of cnp1 | 57 |
| N. gonorrhoeae | ||
| DGI2 | AHU N. gonorrhoeae | 44 |
| MS11 | WT N. gonorrhoeae | 65 |
| FA1090 | WT N. gonorrhoeae | H. S. Seifert |
| JD1504 | MS11 pilE2::mTnCmLac | 31 |
| ND500 | MS11 ΔGGI | 19 |
| MR544 | MS11 transformed by pMR53, lacPO TraWSS::′PhoA at iga-trpB | This work |
| MR546 | MS11 transformed by pMR57, PopaB TraWSS::′PhoA at iga-trpB | This work |
| MR547 | MS11 transformed by pMR58, lacPO TraWSS::′PhoA at lctP-aspC | This work |
| MR553 | MS11 transformed by pMR61, lacPO TraWFL::′PhoA at iga-trpB | This work |
| MR554 | MS11 transformed by pMR71, Ptet TraWSS::′PhoA at iga-trpB | This work |
| MR555 | MR544 transformed by SM44, lacPO TraWSS::′PhoA at iga-trpB, Cmr | This work |
| MR556 | MR553 transformed by SM44, lacPO TraWFL::′PhoA at iga-trpB, Cmr | This work |
| MR557 | MS11 transformed by pMR70, Ptet TraWSS::′PhoA at iga-trpB | This work |
| MR558 | PK180 transformed by pMR70 | This work |
| MR559 | ND500 transformed by pMR53, lacPO TraWSS::′PhoA at iga-trpB | This work |
| MR560 | ND500 transformed by pMR61, lacPO TraWFL::′PhoA at iga-trpB | This work |
| MR561 | MR559 transformed by SM44, lacPO TraWSS::′PhoA at iga-trpB, Cmr | This work |
| MR562 | MR560 transformed by SM44, lacPO TraWFL::′PhoA at iga-trpB, Cmr | This work |
| MR574 | DGI2 transformed by pMR90 | This work |
| PK180 | MS11 transformed by pKH39 | P.L. Kohler |
| N. meningitidis | ||
| ATCC 13102 | ATCC, 13 | |
| MR1000 | ATCC 13102 transformed by pMR57, PopaB TraWSS::′PhoA at iga-trpB | This work |
| MR1002 | ATCC 13102 transformed by pMR33, lac regulatory elements at iga-trpB | This work |
| MR1003 | ATCC 13102 transformed by pMR68, tet regulatory elements at iga-trpB | This work |
| MR1004 | MR1002 transformed by pMR53, lacPO TraWSS::′PhoA at iga-trpB | This work |
| MR1005 | MR1003 transformed by pMR70, Ptet TraWSS::′PhoA at iga-trpB | This work |
Constructs for complementation at the lctP-aspC site.
Constructs for complementation at the iga-trpB site.
DNA techniques.
Plasmid DNA was isolated using a QIAPrep miniprep kit (Qiagen). Gel purification of digested DNA was performed using a QIAQuick gel extraction kit (Qiagen). Blunting of digested DNA ends was performed using T4 DNA polymerase (New England BioLabs). After ligation, DNA was transformed into chemically competent RapidTrans TAM1 E. coli cells (Active Motif). Plasmid screening was conducted by generating whole-cell lysates using the lysis solution of Kado and Liu and analyzing the lysates by agarose gel electrophoresis (24). Further screening of possible transformants was conducted by restriction enzyme digest and PCR.
Plasmid construction.
See Table 1 for a description of the plasmids used in this study. The sequences of all PCR primers are provided in Table 2. Briefly, to make the complementation constructs for IPTG-inducible gene expression, the lac regulatory elements, chloramphenicol acetyltransferase (CAT) gene (cat), and a polylinker were PCR amplified from pKH37 (27) using the primers pKH37F and pKH37R-KpnI. The PCR product was digested with KpnI and ligated into the KpnI site of pMR19 to generate pMR24. To make the same plasmid as pMR24 but with the polylinker sites in the reverse orientation, the primers pKH37F and pKH37R-SacI were used to generate a PCR product from pKH35 (19). The PCR product was digested with SacI and ligated into the SacI site of pMR30 to make pMR35. The cat markers in these plasmids were replaced with ermC by ligating the blunt NsiI/SphI fragment from pIDN3 (20) into the blunt Bpu10I/BseRI fragment of pMR24 and pMR35 to form pMR33 and pMR39, respectively.
Table 2.
PCR primers
| Primer name | Sequence (5′ → 3′)a |
|---|---|
| argJF-SacI | GGTGGTGAGCTCCGCAAGGAGAACCGTTATGG |
| argJR-SpeI | GGTGGTACTAGTGTGGAGGTCATGGTATGTTGTC |
| deltraWF | GGTGGTGGATCCGTTCTGGAAAACCGGGCTGCTCAGG |
| deltraWR | GGTGGTGGATCCTTCCGCTATTGGGTATGTGG |
| igaR-NotI | GGTGCGGCCGCTATGTGGCCGGCGATATTGG |
| lacZ5′-PstI | GCGACTGCAGCACACAGGAAACAGCTATGA |
| lacZ3′-MfeI | GAGCCAATTGGGCCTGCCCGGTTATTATTA |
| OpaPro-PsiI | CGACGGTTATAAGGAATGACGGCGGAAAGATG |
| OpaPro-PacI | CGTTAATTAAGGGCGGATTATATCGGGTTC |
| pKH37F | CCATCCGTTCTGCTCTATAC |
| pKH37R-KpnI | GGTGGTACCTTGGTCATGGCCAGCTTATC |
| pKH37R-SacI | GGTGAGCTCTTGGTCATGGCCAGCTTATC |
| pMR18F | GCCGGGTCTCCCAGTTTATGCATCCCTTAAATTTAGTATTTTAGAAACGAATCT |
| pMR18R-KpnI | GCCGGGTCTCCACTGCATCCCTTAAGGGGGTACCTCATAGCAAAATAAAATGCCGTCT |
| pMR18R-SacI | GCCGGGTCTCCACTGCATCCCTTAAGGGGAGCTCTCATAGCAAAATAAAATGCCGTCT |
| pMR47F-SacII | GGTGGTCCGCGGGCCGGCTTAATCCATTCAGG |
| pMR47F-XhoI | GGTGGTCTCGAGGCCGGCTTAATCCATTCAGG |
| pMR47R-SacII | GGTGGTCCGCGGCCGTGATCTGCCATTAAGTC |
| pMR47R-SpeI | GGTGGTACTAGTCCGTGATCTGCCATTAAGTC |
| pMR47R-XhoI | GGTGGTCTCGAGCCGTGATCTGCCATTAAGTC |
| traWF-XmaI | GGTCCCGGGCCGGCTTAATCCATTCAGGGC |
| traWR-SpeI | GGTACTAGTCGGTTTCATGACCTCTACACG |
| trpBF-NotI | GGTGCGGCCGCAAAGCGCAGATGCAGGAAGC |
Restriction sites are underlined.
To make the complementation constructs for constitutive gene expression, the opaB promoter (PopaB) was PCR amplified from chromosomal DNA of gonococcal strain FA1090 using the primers OpaPro-PsiI and OpaPro-PacI. The PCR product was digested with PsiI and PacI and ligated into the PacI/blunted BseRI fragment of pMR24 and pMR35 to generate pMR28 and pMR38, respectively. The cat markers in these plasmids were replaced with ermC by ligating the blunted NsiI/SphI fragment from pIDN3 (20) into the blunted Bpu10I/BseRI fragment from pMR28 and pMR38 to form pMR32 and pMR41, respectively.
To make the complementation constructs for tetracycline-inducible gene expression, the tet regulatory elements were cloned from pRKO2 (63) into pGCC6del. This plasmid was further modified, and new polylinkers were generated to make plasmids pKH21 and pKH22. The tet regulatory elements, the polylinker, and the cat marker contained on the blunted XhoI/FspI fragment from pKH21 and the blunted SacI/FspI fragment from pKH22 were cloned into the blunted KpnI site of pMR19 to form pMR49 and pMR50, respectively. To replace the cat markers in these plasmids with ermC and to add additional DNA uptake sequences (DUS), the blunted KpnI/NheI fragment from pIDN1 (20) was cloned into the blunted AflII/SacI-HF fragments from pMR49 and pMR50 to form pMR68 and pMR69, respectively.
Transformation of gonococci and meningococci.
Spot transformations were used to transform gonococci and meningococci with complementation construct plasmid DNA. Plasmids smaller than 8,000 bp were linearized by digestion with NheI or PciI. Between 1 and 10 μg plasmid DNA in 20 μl water was spotted onto a GCB agar plate and allowed to dry. Several piliated colonies of N. gonorrhoeae strain MS11, ND500, or DGI2 or N. meningitidis strain ATCC 13102 were streaked over the spots, and the plate was incubated overnight at 37°C with 5% CO2. A Dacron swab was used to transfer colonies from the spots onto GCB agar plates containing 10 μg/ml erythromycin (2 μg/ml for DGI2) or 10 μg/ml chloramphenicol. Individual transformants were restreaked and screened by PCR.
Immunoblot analysis.
Proteins were electrophoresed on 10% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Bio-Rad) either for 1 h at 100 V or overnight at 30 V. Membranes were blocked with 5% milk in Tris-buffered saline containing 0.5% Tween 20 (TTBS) either for 1 h or overnight. Membranes were incubated with primary antibody in TTBS for 1 h, washed with TTBS for 15 min, and incubated with the secondary antibody in TTBS for 1 h. Blots were then washed with TTBS for 30 min, developed using an Immun-Star horseradish peroxidase substrate kit (Bio-Rad), and exposed to film. For the primary antibodies, anti-PhoA (Chemicon) was used at 1:1,000, anti-CAT (Sigma) was used at 1:7,000, and anti-PilQ (H. S. Seifert) was used at 1:10,000. The appropriate horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology) were used at a 1:10,000 dilution. Densitometry analysis of immunoblots was performed using ImageJ software (1).
β-Galactosidase and alkaline phosphatase assays.
Gonococcal strains were grown overnight at 37°C on GCB agar plates. Strains were inoculated into 3 ml of GCBL medium containing Kellogg's supplements and 0.042% NaHCO3 (41) at an OD540 of 0.25 and grown for 3 h. Cultures were vortexed, and 1 ml of cells was subcultured into 2 ml fresh medium at an OD540 of approximately 0.15. Cultures were induced with 0.5 mM IPTG or 2 ng/ml ATc and grown for an additional 2 h.
Assays for alkaline phosphatase activity were conducted as described previously (36). Cells from a 0.5-ml volume of culture were harvested by centrifugation at 13,600 × g for 30 s, washed once in 10 mM MgSO4 in 10 mM Tris-HCl, pH 8.0, and resuspended in 0.5 ml cold 1 M Tris-HCl, pH 8.0. To this suspension was added 0.5 ml 0.1 mM ZnCl2 in 1 M Tris-HCl, pH 8.0, followed by 50 μl chloroform and 50 μl 0.1% SDS. Cells were vortexed, incubated for 5 min at 37°C, and then incubated for 5 min on ice. A volume of 100 μl of 4 mg/ml p-nitrophenylphosphate in 1 M Tris-HCl, pH 8.0, was added, and the solution was incubated at 37°C until it turned pale yellow, usually within 25 min. The reaction was terminated by the addition of 120 μl of 83 mM EDTA, 166 mM KH2PO4. Tubes were placed on ice, and the OD550 and OD420 were measured.
β-Galactosidase assays were conducted as previously described (40). Briefly, cells from a 0.5-ml volume of the culture were harvested by centrifugation at 13,600 × g for 30 s. The pellet was washed once in 10 mM MgSO4 in 10 mM Tris-HCl, pH 8.0, and resuspended in cold 1 M Tris-HCl, pH 8.0. To this cell suspension was added a 0.5-ml volume of Z buffer (60 mM Na2HPO4 · 7H2O, 40 mM Na2HPO4 · H2O, 10 mM KCl, 50 mM β-mercaptoethanol, pH adjusted to 7.0), followed by 50 μl chloroform and 50 μl 0.1% SDS. Cells were vortexed and incubated for 5 min at 37°C, followed by 5 min on ice. A volume of 200 μl 4 mg/ml o-nitrophenyl-β-galactopyranoside in Z buffer was added, and the reaction mixture was incubated at 28°C until a pale yellow color resulted. The time was noted, and the reaction was terminated by the addition of 120 μl 1 M Na2CO3. The OD420 and OD550 were measured.
For both assays, a 1.5-ml volume of bacterial culture was harvested by centrifugation at 13,600 × g for 30 s, resuspended in 0.5 ml 1 M Tris-HCl, pH 8.0, and lysed by sonication, and a Bradford assay was performed to measure total protein. Phosphatase activity and β-galactosidase activity were calculated using the following equation: 1,000 × (OD420 − [1.75 × OD550])/(time in minutes × mg of protein × volume in ml).
Subcellular fractionation.
Gonococcal strains were grown overnight at 37°C on GCB agar plates. For each strain, two 4-ml cultures were inoculated in GCBL medium containing Kellogg's supplements and 0.042% NaHCO3 (41) at an OD540 of 0.25. The cultures were induced with 0.5 mM IPTG and grown for 3 h. The cells were harvested by centrifugation at 11,952 × g for 5 min and washed once in ice-cold 0.01 M Tris-HCl, pH 7.0. The cell pellet was resuspended in 4 ml 0.01 M Tris-HCl, pH 7.0, and sonicated for 30 s at 45% amplitude with a 1-s on-off pulse using a Branson digital sonifier. The sonicate was centrifuged at 15,557 × g in an F-20/Micro rotor for 10 min to remove any unlysed cells, and the supernatant was passed through a 0.2-μm-pore-size filter. The supernatant was centrifuged at 126,000 × g in a TLA110 rotor for 1.5 h to separate soluble from membrane fractions. The membrane fraction was washed with 750 μl 0.01 M Tris-HCl, pH 7.0, and centrifuged at 127,000 × g in a TLA120.2 rotor for 1.5 h. The pellet was resuspended in 50 μl 0.1 M Tris-HCl buffer, pH 7.0. A Bradford assay was used to normalize the amount of protein electrophoresed on SDS-polyacrylamide gels.
RESULTS
Design of complementation constructs.
In order to identify a chromosomal region for complementation, we reviewed sites that had successfully been used in the past and chose the iga locus (67). This locus encodes immunoglobulin A1 (IgA1) protease, a secreted protease produced only by the pathogenic Neisseria that cleaves the hinge region of IgA (28, 42) and also cleaves LAMP1 in epithelial cells (32). The iga locus was also attractive since it was shown that insertions within iga do not affect infection in human male volunteers (22). We improved upon the existing constructs for complementation at iga by designing our constructs so that gene insertion occurs in the intergenic region between iga and the neighboring gene, trpB, thereby leaving the iga open reading frame intact. Plasmid pHSS6 was used as the starting point to construct the complementation constructs because of its small size and that fact that it replicates in E. coli but not N. gonorrhoeae (56).
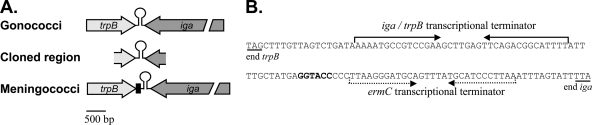
The iga and trpB genes share a transcriptional terminator (Fig. 1A) (23). A 1,134-bp region of chromosomal DNA from gonococcal strain MS11 containing portions of the iga and trpB genes as well as the intergenic region was cloned into pHSS6 (Fig. 1A). The iga-trpB region is conserved between gonococci and meningococci. The sequence of the iga-trpB locus from MS11 is between 89 and 92% identical to the sequence from several sequenced meningococcal genomes (GenBank accession no. CP000381.1, AE002098.2, AM421808.1, AL157959.1, and AM889136.1), and the 1,134-bp sequence cloned into pHSS6 is even more highly conserved (94 to 95% identical). Sequence analysis of the iga-trpB region revealed that meningococci differ from gonococci by the presence of a 103-bp Correia element inserted between the stop codon of trpB and the shared transcriptional terminator (Fig. 1A) (10, 11, 58). Given the high overall conservation within the iga and trpB open reading frames, however, we predicted that these constructs would be useful for complementation of both gonococci and meningococci.
Fig 1.
Analysis of the intergenic region between the genes iga and trpB. (A) Schematic of iga-trpB locus in gonococci and meningococci. The 1,134-bp region of genomic DNA from N. gonorrhoeae strain MS11 that was cloned into the complementation constructs is also represented. The transcriptional terminator shared by iga and trpB is indicated by a stem-loop structure. Meningococcal strains contain a 103-bp Correia element (black rectangle) inserted between the stop codon of trpB and the transcriptional terminator. (B) Changes made to the iga-trpB intergenic region in the complementation constructs. The stop codons of trpB and iga are underlined. A second transcriptional terminator from the erythromycin resistance gene ermC was inserted downstream of iga, and a restriction site was added between the two terminators for cloning purposes (bold type). All gene insertion is directed into this restriction site, such that both iga and trpB maintain transcriptional terminators.
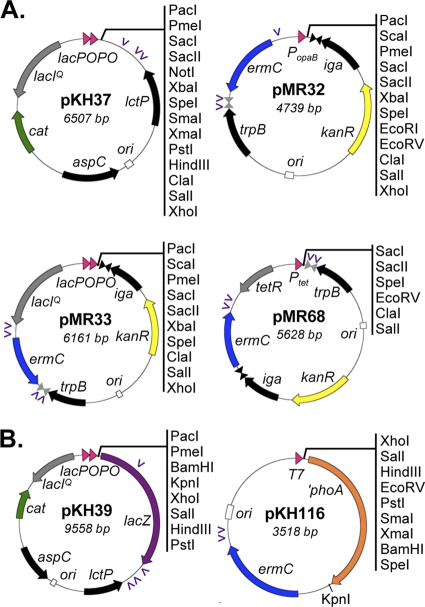
To ensure that both trpB and iga would maintain transcriptional terminators following gene insertion in the intergenic region, we cloned the transcriptional terminator from the erythromycin resistance gene (ermC) downstream of iga in the complementation constructs (Fig. 1B). We also added a restriction site between the two terminators, and this site served as the insertion site for the promoters, polylinkers, and selectable markers (Fig. 1B). In order to make the iga-trpB complementation constructs useful in combination with the existing constructs that direct gene insertion at the lctP-aspC site (pKH35 and pKH37) (19, 27), we designed the iga-trpB constructs to contain a different selectable marker (erythromycin resistance, ermC) (Fig. 2).
Fig 2.
Description of complementation constructs and plasmids for constructing lacZ and phoA fusions. (A) Plasmid maps of complementation constructs. Regions of homology with the gonococcal and meningococcal chromosomes are shown in black. Plasmid pKH37 directs gene insertion between lctP and aspC and contains tandem copies of the lac promoter and operator (lacPOPO), the lac repressor (lacIq), and the selectable marker chloramphenicol acetyltransferase (cat). Plasmids pMR32, pMR33, and pMR68 all direct gene insertion between trpB and iga and encode an erythromycin resistance marker (ermC). Plasmid pMR33 contains lacPOPO and lacIq, pMR32 contains the constitutive promoter from the opaB gene of N. gonorrhoeae strain FA1090 (PopaB), and pMR68 contains the tetracycline-inducible promoter (Ptet) and tet repressor (tetR). The transcriptional terminator found in the chromosome between iga and trpB is indicated by gray inverted triangles. The added transcriptional terminator from the ermC gene is indicated by black inverted triangles. Unique restriction sites in the polylinkers are shown. DNA uptake sequences are indicated by the purple arrowheads. Plasmids pKH35, pMR41, pMR39, and pMR69 (data not shown) are identical to pKH37, pMR32, pMR33, and pMR68, respectively, except that the orientation of the sites in the polylinker is reversed to facilitate cloning. (B) Plasmids for creating lacZ and phoA fusions in N. gonorrhoeae. Plasmid pKH39 contains lacZ with a ribosome binding site in pKH35, while pKH116 encodes phoA without its signal sequence (′phoA).
We designed the iga-trpB constructs to contain one of three different promoters (Fig. 2A). Plasmid pMR32 contains the strong constitutive opaB promoter from N. gonorrhoeae strain FA1090 (PopaB). PopaB differs by only 1 bp in the −35 hexamer (TTGAAA) from the sigma-70 consensus promoter sequence (5). Plasmid pMR33 contains the tandem lac promoter/operator (lacPOPO) and lac repressor (lacIq) from pKH37 (27). Plasmid pMR68 contains the tetracycline-inducible P57opt promoter/operator (Ptet) and tetracycline repressor (tetR) from pRKO2 (63). The P57opt promoter has been optimized so that the −35 hexamer matches the sigma-70 consensus sequence, resulting in increased promoter activity (63). The tetracycline-regulated promoter has been used in a variety of prokaryotic and eukaryotic expression systems (4, 63), but this is the first description of its use in Neisseria. We also constructed variants of pMR32, pMR33, and pMR68 in which the orientation of the polylinker sites is reversed to facilitate cloning (pMR41, pMR39, and pMR69, respectively) (Fig. 2A).
Functional complementation of an arginine auxotrophy.
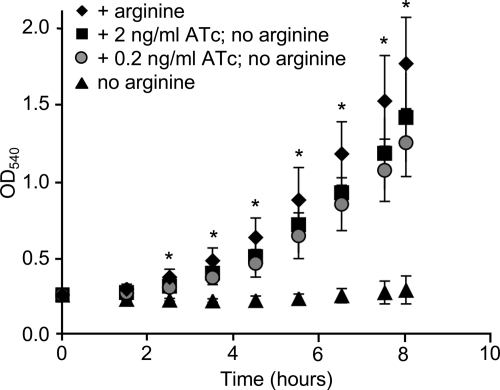
To demonstrate that the constructs that we describe can be used for functional complementation, we used pMR68 to complement an arginine auxotrophy in gonococcal strain DGI2, which is part of a group of strains requiring arginine, hypoxanthine, and uracil for growth (AHU auxotrophs) (44). The requirement for arginine in AHU strains is due to a mutation in the argJ gene, encoding ornithine acetyltransferase (37, 38, 49). Strain DGI2 is an argJ mutant (GenBank accession no. ACIG00000000.1) and does not grow in GGM lacking arginine (data not shown). We transformed DGI2 with pMR90, which contains the wild-type (WT) argJ gene from strain MS11, and measured growth of the resulting strain, MR574, in the presence or absence of arginine and the anhydrotetracycline (ATc) inducer. Strain MR574 grew in the presence but not the absence of arginine, suggesting that appreciable amounts of ArgJ are not produced from Ptet in the absence of inducer (Fig. 3). The addition of 0.2 or 2 ng/ml ATc enabled growth of MR574 in the absence of arginine (Fig. 3).
Fig 3.
Complementation of argJ in strain DGI2, which requires arginine, hypoxanthine, and uracil for growth. DGI2 was transformed with pMR90 to generate strain MR574, which expresses the wild-type argJ under the control of Ptet at the iga-trpB locus. Strain MR574 was grown in liquid gonococcal genetic medium (GGM) containing arginine or in medium without added arginine. ArgJ production was induced during growth in GGM lacking arginine by the addition of 0.2 ng/ml or 2 ng/ml ATc. The mean ± standard deviation of three independent experiments is shown. *, growth of MR574 in the presence of ATc inducer was not significantly different from growth in the presence of arginine (Student's two-tailed t test, P > 0.05), but growth under both conditions was significantly different from growth in the absence of both arginine and ATc (Student's two-tailed t test, P < 0.05).
Characterization of gene expression from complementation constructs.
In order to better quantify expression from the different complementation constructs, we made use of two plasmids that allow the construction of lacZ or phoA fusions. Plasmid pKH39 is an lctP-aspC complementation construct containing lacZ, while pKH116 encodes phoA without its signal sequence (′phoA) (Fig. 2B). We decided to characterize the relative expression from the lacPO, PopaB, and Ptet constructs by measuring the activity of an alkaline phosphatase (PhoA) fusion to TraW. TraW is a component of the gonococcal type IV secretion system and contains a predicted signal sequence (SignalP, version 4.0, server) (48), suggesting that TraW may localize to the periplasm. We constructed two different PhoA fusions containing either the full-length TraW open reading frame (TraWFL::′PhoA) or the predicted TraW signal sequence (TraWSS::′PhoA). Expression of both PhoA fusions in E. coli resulted in blue colonies when the bacteria were grown on XP indicator plates, indicating that both fusions localize to the periplasm in E. coli (data not shown).
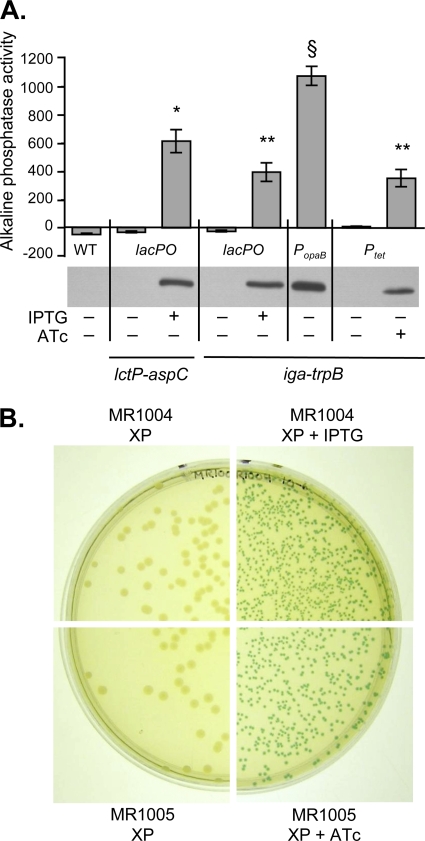
The TraWSS::′PhoA fusion was cloned into pKH37, pMR32 (PopaB), pMR33 (lacPO), and pMR68 (Ptet). The resulting plasmids were introduced into gonococci by natural transformation to generate strains MR547, MR546, MR544, and MR557, respectively. We measured the growth of these strains in GCBL medium and confirmed that gene insertion at the iga-trpB site did not alter their growth rate (data not shown). We then conducted alkaline phosphatase assays to assess production of TraWSS::′PhoA in these strains (Fig. 4A). Wild-type N. gonorrhoeae does not express AP activity (Fig. 4A) (8). AP activity was similar in strains expressing TraWSS::′PhoA under the control of lacPO at either the lctP-aspC site (MR547) or the iga-trpB site (MR544), although AP activity was slightly lower in the strain transformed with the iga-trpB construct (Fig. 4A). In both cases, AP activity was observed only in the presence of IPTG, and growth in the absence of the inducer for 2 h resulted in background levels of AP activity (Fig. 4A). Compared to MR544, a strain expressing TraWSS::′PhoA from PopaB at the iga-trpB site (MR546) produced almost three times as much AP activity, confirming that the PopaB construct provides strong, constitutive gene expression (Fig. 4A). In a strain expressing TraWSS::′PhoA from the tetracycline-inducible promoter (MR557), the levels of AP activity in the presence of 2 ng/ml ATc were similar to the levels of AP activity produced by MR544. AP activity was at background levels in the absence of the ATc after 2 h of growth (Fig. 4A). An immunoblot detected levels of PhoA correlating with the activity measurements. These results demonstrate that the new complementation constructs can be used to produce substantial amounts of a protein of interest from the very strong constitutive opaB promoter or from the regulatable lac or tet promoters. Furthermore, they establish Ptet as the second inducible promoter available for gene expression in N. gonorrhoeae.
Fig 4.
Characterization of gene expression from the complementation constructs. (A) Alkaline phosphatase activity and immunoblot analysis of TraWSS::′PhoA expression in gonococcal strain MS11 (WT) and in gonococcal strains transformed with the complementation constructs. Expression of TraWSS::′PhoA from the lctP-aspC site was driven by the lac promoter (lacPO) in strain MR547. Expression of TraWSS::′PhoA from the iga-trpB site was driven by one of three promoters: lacPO in strain MR544, the constitutive opaB promoter (PopaB) in strain MR546, or the tetracycline-inducible promoter (Ptet) in strain MR557. Production of TraWSS::′PhoA was induced with either 0.5 mM IPTG or 2 ng/ml ATc. Alkaline phosphatase activity was normalized to mg of protein. The amount of TraWSS::′PhoA fusion protein was detected by immunoblotting with anti-PhoA (α-PhoA) antibodies. The mean ± standard deviation of at least three independent experiments is shown. *, alkaline phosphatase activity from the strain with lacPO at the lctP-aspC site in the presence of inducer was significantly different from that of the uninduced control (P < 0.005) and also different from the activities of strains with lacPO, PopaB, and Ptet at the iga-trpB site (P ≤ 0.01) by Student's two-tailed t test; **, alkaline phosphatase activity from the strains with lacPO or Ptet at the iga-trpB site in the presence of inducer was significantly different from that of the respective uninduced controls (P ≤ 0.001) but was not significantly different between strains (P > 0.05) by Student's two-tailed t test; §, alkaline phosphatase activity from the strain with PopaB at the iga-trpB site was significantly different from the activities of all other inducible strains (P < 0.001) by Student's two-tailed t test. (B) Use of the iga-trpB complementation constructs in N. meningitidis. Strains MR1004 (producing TraWSS::′PhoA under the control of lacPO) and MR1005 (producing TraWSS::′PhoA under the control of Ptet) were grown on GCB-Tris-XP agar plates containing either the XP substrate alone, XP with 1 mM IPTG, or XP with 20 ng/ml ATc.
Complementation in N. meningitidis.
Due to the similarity of the iga-trpB region in meningococci and gonococci, we expected that these constructs would also be useful for complementation in meningococci. We transformed meningococcal strain ATCC 13102 with pMR53 (lacPO), pMR57 (PopaB), and pMR70 (Ptet), which all encode traWSS::′phoA. We were able to transform strain ATCC 13102 with pMR57, and the resulting strain, MR1000, produced blue colonies when grown on XP indicator plates (data not shown). We were unable to transform ATCC 13102 with pMR53 or pMR70 after several attempts, likely because the size of the nonhomologous insert in these plasmids was significantly larger than that in pMR57. However, we were able to transform these constructs into ATCC 13102 in two steps. We first transformed ATCC 13102 with the empty complementation constructs pMR33 and pMR68. The size of the nonhomologous insert is smaller in these constructs since they do not contain the traWSS::′phoA fusion, and we were able to obtain erythromycin-resistant colonies that carried the lac or tet regulatory elements on the chromosome. We then transformed these strains with pMR53 and pMR70 to introduce traWSS::′phoA and screened for blue PhoA+ colonies on XP indicator plates, generating strains MR1004 and MR1005, respectively.
We grew strains MR1004 (producing TraWSS::′PhoA under the control of lacPO) and MR1005 (producing TraWSS::′PhoA under the control of Ptet) on XP indicator plates with or without the appropriate inducer. Both strains produced white colonies on plates containing the XP substrate without inducer (Fig. 4B). On plates that contained both XP and either the IPTG or ATc inducer, both strains produced colonies that appeared light blue by 24 h of growth and were dark blue after 40 h of growth (Fig. 4B). The lac promoter has been used before in meningococci (66), but this is the first report of tetracycline-inducible gene expression in meningococci.
Optimal induction of Ptet promoter.
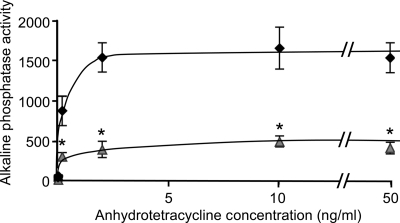
Since this is the first description of tetracycline-inducible gene expression in Neisseria, we sought to better characterize the induction of the Ptet promoter with ATc. In our initial induction experiments in gonococci, ATc concentrations of both 2 ng/ml and 20 ng/ml were sufficient to generate high levels of AP activity from TraWSS::′PhoA. To determine optimal induction conditions, we then tested a range of ATc concentrations. Maximum levels of AP activity were detected with ATc concentrations as low as 2 ng/ml, and increasing the ATc concentration to as high as 50 ng/ml did not result in any additional increases in AP activity (Fig. 5). Importantly, ATc did not negatively affect cell growth, even at relatively high concentrations. Concentrations of ATc lower than 2 ng/ml (for example, 0.2 and 0.02 ng/ml) produced intermediate levels of AP activity, making it possible to modulate promoter strength by changing the concentration of the inducer (Fig. 5).
Fig 5.
Alkaline phosphatase activity of gonococcal strains expressing TraWSS::′PhoA under the control of the tetracycline-inducible promoter in the presence of different concentrations of ATc. Black diamonds, MR554 (MS11 transformed with pMR71); gray triangles, MR557 (MS11 transformed with pMR70). Alkaline phosphatase activity was normalized to mg of protein. The mean ± standard deviation of three independent experiments is shown. *, P ≤ 0.002 compared to MR554, Student's two-tailed t test.
While performing these experiments, we noticed that a strain of gonococci that had been transformed with pMR71 (strain MR554) consistently produced higher levels of AP activity than a strain of gonococci that had been transformed with pMR70 (strain MR557) (Fig. 5). Plasmids pMR70 and pMR71 both contain TraWSS::′PhoA under the control of Ptet and differ only in the orientation of the restriction sites in the polylinker. The reason for the observed difference in AP activity between the two strains is unknown, but DNA sequencing confirmed that it is not due to any sequence mutations in the region containing the Ptet promoter or the promoter for the tet repressor (data not shown).
Simultaneous use of two inducible promoters.
Since the pKH35 and pKH37 complementation constructs contain a cat marker and recombine at lctP-aspC and all of the new constructs described in this work contain an ermC marker and recombine at iga-trpB, it should be possible to express two different gene products from the two chromosomal sites and to induce the expression of these genes either independently or simultaneously. To investigate this possibility, the WT N. gonorrhoeae strain MS11 was transformed with pKH39 to generate a strain that expresses lacZ under the control of lacPO at the lctP-aspC site (strain PK180). We then transformed this strain with pMR70, which produces TraWSS::′PhoA under the control of Ptet to generate strain MR558.
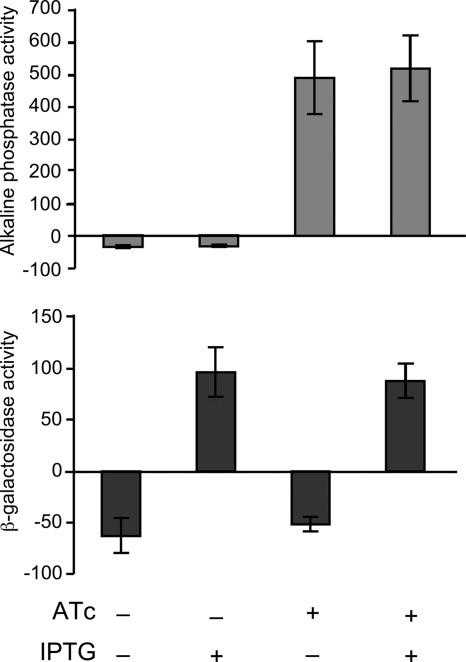
We grew MR558 either without inducer, with IPTG or ATc alone, or in the presence of both inducers. We then conducted β-galactosidase and alkaline phosphatase assays on bacteria grown under the different conditions (Fig. 6). In the absence of both inducers, we did not detect activity in either assay. In the presence of IPTG, we detected only β-galactosidase activity, and in the presence of ATc, we detected only AP activity. In the presence of both inducers, we detected activity in both assays (Fig. 6). There was no noticeable growth defect when cells were grown in the presence of both IPTG and ATc, and activity levels for alkaline phosphatase and β-galactosidase were comparable whether both inducers were present or whether a single inducer was present. Thus, it is possible to use both lacPO and Ptet in the same cell and to turn them on and off independently of one another.
Fig 6.
Independent and simultaneous induction of the lac promoter and tetracycline-inducible promoter (Ptet) within the same gonococcal cell. We constructed a strain (MR558) that expresses lacZ under the control of the lac promoter at the lctP-aspC site and traWSS::′phoA under the control of Ptet at the iga-trpB site. We then measured alkaline phosphatase activity and β-galactosidase activity in the presence or absence of 0.5 mM IPTG and/or 2 ng/ml ATc. Alkaline phosphatase activity and β-galactosidase activity were normalized to mg protein. The mean ± standard deviation of three independent experiments is shown.
Subcellular localization of TraW::PhoA fusions.
We used expression of the TraW::PhoA translational fusions from the new complementation constructs to examine subcellular localization of TraW. Expression of both TraWSS::′PhoA and TraWFL::′PhoA from the iga-trpB complementation site in N. gonorrhoeae resulted in AP activity (Fig. 7A). Since alkaline phosphatase must be transported to the periplasm in order to be active, these results confirm our hypothesis that TraW localizes to the periplasm in Neisseria and demonstrate that the predicted TraW signal sequence functions for protein export. The TraW homolog in F plasmid (TraWF) is involved in F-pilus assembly and is part of an interaction group of six proteins (21, 35). TraWF was shown to localize to the periplasm when expressed in maxicells (35) but localized to the outer membrane when expressed in cells carrying the entire F transfer region (3).
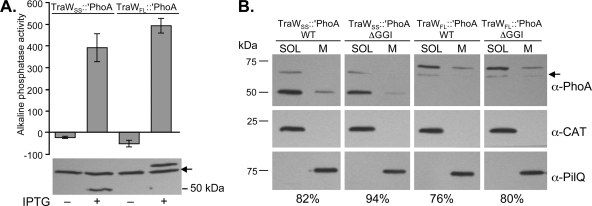
Fig 7.
Activity and localization of TraW::PhoA fusions in N. gonorrhoeae. (A) Alkaline phosphatase activity and immunoblot analysis of N. gonorrhoeae strains producing TraWSS::′PhoA (MR544) and TraWFL::′PhoA (MR553). Alkaline phosphatase units were normalized to total mg of protein. The mean ± standard deviation of three independent experiments is shown. Arrow, a band that cross-reacts with the anti-PhoA antibody. (B) Subcellular localization of TraW::PhoA fusions in gonococcal cells. Four chloramphenicol-resistant strains were constructed: MR555 (expressing traWSS::′phoA in a WT background), MR561 (expressing traWSS::′phoA in a ΔGGI background), MR556 (expressing traWFL::′phoA in a WT background), and MR562 (expressing traWFL::′phoA in a ΔGGI background). Cells were fractionated into a soluble fraction (SOL) containing the cytoplasm and periplasm as well as a membrane fraction (M) containing the inner and outer membranes. Subcellular fractions were probed with antibodies against alkaline phosphatase (PhoA), the cytoplasmic CAT, and the outer membrane protein PilQ. Densitometry analysis was performed using ImageJ software. The percentage of total TraW::PhoA fusion protein associated with the soluble fraction for each strain is reported. Arrow, a band that cross-reacts with the anti-PhoA (α-PhoA) antibody.
We asked whether the TraW::PhoA fusions that we had constructed would localize differently in N. gonorrhoeae if expressed in the presence or absence of other T4SS proteins. We expressed TraWSS::′PhoA and TraWFL::′PhoA under the control of lacPO at the iga-trpB site in the WT gonococcal strain MS11 and in an isogenic strain carrying a deletion of the entire GGI (ΔGGI). The resulting strains were fractionated into a soluble fraction containing cytoplasmic and periplasmic material and a membrane fraction containing both inner and outer membranes. The proteins present in each fraction were then analyzed by immunoblotting (Fig. 7B). CAT was detected only in the soluble fraction, and the gonococcal outer membrane protein PilQ was detected only in the membrane fraction, indicating that we were able to achieve clean fractionation of soluble and membrane material. The localization of the TraWSS::′PhoA fusion would not be expected to vary in the presence or absence of other T4SS proteins, since it contains only the signal sequence portion of TraW. Densitometry analysis indicated that 82% of the total TraWSS::′PhoA fusion localized to the soluble fraction when expressed in a WT background, while 94% localized to the soluble fraction when expressed in a ΔGGI background (Fig. 7B). For the TraWFL::′PhoA fusion, 76% of the protein localized to the soluble fraction when expressed in a WT background, and 80% localized to the soluble fraction when expressed in a ΔGGI background (Fig. 7B). These results are representative of several independent experiments and indicate that TraWSS::′PhoA and TraWFL::′PhoA primarily localize to the soluble fraction (presumably the periplasm) and that a small amount of each fusion protein is associated with the membranes. The pattern of localization for both TraW::′PhoA fusion proteins is similar regardless of whether they are produced in the presence or absence of other T4SS proteins.
DISCUSSION
Despite the overall genetic tractability of the pathogenic Neisseria, the tools available for gene complementation are limited. Due to the problems associated with the use of replicating plasmids in Neisseria, complementation is usually achieved by expressing the gene of interest from a chromosomal locus. Relatively few loci have been developed for complementation, however, and the options for promoters to drive gene expression are similarly limited. We describe a set of new complementation constructs to address these problems. The constructs undergo double-crossover recombination with the iga and trpB genes following transformation into Neisseria. Gene insertion occurs in the intergenic region between the genes, ensuring that both genes are left intact. Additionally, to minimize the possibility that gene insertion might affect neighboring genes, we engineered an additional transcriptional terminator in the constructs such that both trpB and iga maintain transcriptional terminators following gene insertion.
In order to increase the number of available promoters for gene expression in Neisseria, we designed the iga-trpB complementation constructs to contain three different promoters: lacPO, PopaB, and Ptet. We characterized gene expression from these constructs by measuring the activity of an alkaline phosphatase fusion. The tandem lac promoter/operator (lacPOPO) provides strong gene expression in Neisseria. Induction of lacPOPO inserted upstream of the pilE gene resulted in approximately 87% of wild-type pilE mRNA levels (34), while expression of the traD gene from the lctP-aspC site under the control of lacPOPO resulted in a 123-fold increase in transcript levels compared to wild-type levels (51). We show that the iga-trpB constructs containing lacPOPO generate levels of AP activity similar to the levels of these previously characterized lctP-aspC constructs (19, 27, 54). We also designed a construct containing the strong, constitutive promoter from the opaB gene. As expected on the basis of the large amounts of opacity protein produced by gonococcal strains, the PopaB construct was the strongest of those that we characterized in this study, providing three times as much AP activity as the lacPO construct. Because PopaB provides strong constitutive expression, the usefulness of these constructs largely depends on whether the gene of interest is toxic to the cell when expressed at high levels. Finally, we designed a construct containing a tetracycline-inducible promoter. The tetracycline-inducible promoter has not been described for use in Neisseria, although it has been used in a variety of prokaryotic and eukaryotic expression systems. We demonstrated that gene expression from this promoter can be induced specifically in the presence of anhydrotetracycline. Various levels of expression from the Ptet construct can be achieved by adding different amounts of inducer, and maximum expression levels are similar to those achieved with the lacPOPO constructs.
We used a Ptet construct to functionally complement an arginine auxotrophy in gonococcal strain DGI2. Strain DGI2 was isolated from a patient with disseminated gonococcal infection in the 1980s and is part of a group of strains that require arginine, hypoxanthine, and uracil for growth (AHU auxotrophs) (44). These strains have been extensively studied and share a number of features in common (26), including the production of type 1 IgA1 protease (43). Most other gonococcal isolates, including MS11, produce type 2 IgA1 protease. The portion of iga cloned into the complementation constructs is conserved between type 1 and type 2 IgA1 proteases, however, and we were able to transform strain DGI2 with a Ptet construct expressing wild-type argJ. The complemented strain did not grow in the absence of arginine if the inducer was not present, but the addition of anhydrotetracycline allowed growth in the absence of arginine, demonstrating that the new constructs can be used to functionally complement a gonococcal mutant.
The iga-trpB region is also conserved between N. gonorrhoeae and N. meningitidis, and we demonstrated that the constructs that we describe function for complementation in meningococci as well as gonococci. It should be noted that the sequence of the iga-trpB intergenic region in meningococci differs from that in gonococci by the presence of a 103-bp Correia element between the stop codon of trpB and the shared transcriptional terminator. Correia elements are common repetitive elements in the Neisseria genomes (33). Previous work has indicated that these elements may function as promoters for transcription, transcriptional terminators, targets for RNA processing, or sites for DNA recombination (58). We were able to transform meningococci with the PopaB construct containing TraWSS::′PhoA and obtained blue colonies on XP indicator plates. We found that it was necessary to transform the lac- and tet-inducible constructs containing TraWSS::′PhoA into meningococci in two steps, likely because of the relatively large nonhomologous inserts in these constructs. We used the resulting strains to demonstrate tetracycline-inducible gene expression, increasing the number of inducible promoters available for gene expression in meningococci.
We designed the iga-trpB constructs to be used in combination with previously characterized complementation constructs that direct gene insertion at the lctP-aspC site. Two different gene products were expressed from these two chromosomal loci either independently or simultaneously, one under the control of the IPTG-inducible lacPO and one under the control of the tetracycline-inducible Ptet. While we experienced no difficulty constructing a strain that contained both Ptet and lacPO, we had some difficulty transforming a pMR33-derived construct into a strain that already contained the lacPO/lacIq elements at the lctP-aspC site. In this case, the incoming complementation construct has homology with the lac regulatory elements present at the lctP-aspC site, in addition to homology with the iga-trpB locus. We were able to obtain transformants carrying the expected inserts at both complementation sites only if we carried out selection in the presence of both chloramphenicol and erythromycin. In the presence of erythromycin alone, the resulting transformants did not carry the expected insert at the iga-trpB site and instead appeared to have undergone recombination between the lacPO/lacIq elements, resulting in the loss of the cat marker from the lctP-aspC site.
We constructed two different alkaline phosphatase fusions, one containing the full-length TraW open reading frame and one containing only the predicted TraW signal sequence. TraW is a component of the gonococcal type IV secretion system (T4SS), which secretes single-stranded chromosomal DNA into the extracellular environment, where it is functional for the natural transformation of other gonococci in the population (19, 52). TraW is encoded in the gonococcal genetic island (GGI) along with the other genes required for type IV secretion, but little is known about its function in DNA secretion (13, 19, 46). In the E. coli F plasmid, a homolog of TraW (TraWF) is required for F pilus assembly (15) and is likely part of the secretion apparatus. Previous studies have shown that TraWF localizes to the periplasm (35) but is associated with the membrane when expressed in the presence of the full F transfer region (3). We hypothesized that gonococcal TraW would also localize to the periplasm in gonococci and chose it as a fusion partner for ′PhoA.
In agreement with our hypothesis, production of both TraWSS::′PhoA and TraWFL::′PhoA in E. coli and in N. gonorrhoeae resulted in AP activity, indicating that TraW localizes to the periplasm. We hypothesized that TraW might be membrane associated in gonococci and asked whether the subcellular localization of TraWSS::′PhoA or TraWFL::′PhoA fusion was altered if expressed in a strain carrying a deletion of the GGI (ΔGGI). We fractionated cells into a soluble fraction and a membrane fraction. The fractionation results indicated that both TraWSS::′PhoA and TraWFL::′PhoA localize primarily in the soluble fraction, regardless of whether they were produced in a WT or ΔGGI background. These results indicate that TraW is a soluble, periplasmic protein in N. gonorrhoeae. The presence of T4SS proteins in N. gonorrhoeae did not cause the overexpressed TraW to associate with membrane material, which may indicate that it does not associate tightly with the T4SS apparatus or that there were insufficient amounts of other T4SS proteins to influence the localization of significant amounts of the expressed TraW fusion protein.
The complementation constructs developed in this study are a significant addition to the genetic tools available for N. gonorrhoeae and N. meningitidis. In combination with previously characterized constructs, these new constructs make it possible to complement multiple mutations in a single cell and assess the relative contributions of each gene to an overall phenotype. Additionally, these constructs allow the expression of new genes in strains that already contain the lac promoter or make use of existing complementation sites, allowing increasingly more complex questions to be asked regarding the basic biology, natural transformation, and pathogenesis of these significant human pathogens.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01 AI047958 awarded to J.P.D., and M.E.R. was supported by NIH grant T32 GM07215.
We thank H. S. Seifert for the PilQ antibody and Petra L. Kohler for the construction of the strain PK180.
Footnotes
Published ahead of print 10 February 2012
REFERENCES
- 1. Abramoff MD, Magalhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int. 11:36–42 [Google Scholar]
- 2. Alexander HE, Redman W. 1953. Transformation of type specificity of meningococci: change in heritable type induced by type-specific extracts containing desoxyribonucleic acid. J. Exp. Med. 97:797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arutyunov D, Arenson BMJ, Frost LS. 2010. F plasmid TraF and TraH are components of an outer membrane complex involved in conjugation. J. Bacteriol. 192:1730–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beck CF, Mutzel R, Barbe J, Mueller W. 1982. A multifunctional gene (tetR) controls Tn10-encoded tetracycline resistance. J. Bacteriol. 150:633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belland RJ, Morrison SG, Carlson JH, Hogan DM. 1997. Promoter strength influences phase variation of neisserial opa genes. Mol. Microbiol. 23:123–135 [DOI] [PubMed] [Google Scholar]
- 6. Biswas GD, Graves JF, Sox TE, Tenover FC, Sparling PF. 1982. Marker rescue by a homologous plasmid during transformation of gonococci by a hybrid Pcr plasmid. J. Bacteriol. 151:77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bos MP, Tefsen B, Geurtsen J, Tommassen J. 2004. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc. Natl. Acad. Sci. U. S. A. 101:9417–9422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boyle-Vavra S, Seifert HS. 1995. Shuttle mutagenesis: a mini-transposon for producing PhoA fusions with exported proteins in Neisseria gonorrhoeae. Gene 155:101–106 [DOI] [PubMed] [Google Scholar]
- 9. Catlin BW. 1960. Transformation of Neisseria meningitidis by deoxyribonucleates from cells and from culture slime. J. Bacteriol. 79:579–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Correia FF, Inouye S, Inouye M. 1986. A 26-base-pair repetitive sequence specific for Neisseria gonorrhoeae and Neisseria meningitidis genomic DNA. J. Bacteriol. 167:1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Correia FF, Inouye S, Inouye M. 1988. A family of small repeated elements with some transposon-like properties in the genome of Neisseria gonorrhoeae. J. Biol. Chem. 263:12194–12198 [PubMed] [Google Scholar]
- 12. Dillard JP. 2011. Genetic manipulation of Neisseria gonorrhoeae. Curr. Protoc. Microbiol. 23:4A.1.1-4A.1.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dillard JP, Seifert HS. 2001. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol. Microbiol. 41:263–278 [DOI] [PubMed] [Google Scholar]
- 14. Dunning Hotopp JC, et al. 2006. Comparative genomics of Neisseria meningitidis: core genome, islands of horizontal gene transfer and pathogen-specific genes. Microbiology 152(Pt 12):3733–3749 [DOI] [PubMed] [Google Scholar]
- 15. Frost LS, Ippen-Ihler K, Skurray RA. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goldberg ID, Steinberg VI, Siddiqui A, Hart EJ, Schaper D. 1978. Attempts to isolate bacteriophage specific for Neisseria gonorrhoeae. In Immunobiology of Neisseria gonorrhoeae. American Society for Microbiology, Washington, DC [Google Scholar]
- 17. Gunn JS, Stein DC. 1996. Use of a nonselective transformation technique to construct a multiply restriction/modification-deficient mutant of Neisseria gonorrhoeae. Mol. Gen. Genet. 251:509–517 [DOI] [PubMed] [Google Scholar]
- 18. Hamilton HL, Dillard JP. 2006. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol. Microbiol. 59:376–385 [DOI] [PubMed] [Google Scholar]
- 19. Hamilton HL, Domínguez NM, Schwartz KJ, Hackett KT, Dillard JP. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 55:1704–1721 [DOI] [PubMed] [Google Scholar]
- 20. Hamilton HL, Schwartz KJ, Dillard JP. 2001. Insertion-duplication mutagenesis of Neisseria: use in characterization of DNA transfer genes in the gonococcal genetic island. J. Bacteriol. 183:4718–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harris RL, Silverman PM. 2004. Tra proteins characteristic of F-like type IV secretion systems constitute an interaction group by yeast two-hybrid analysis. J. Bacteriol. 186:5480–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johannsen DB, Johnston DM, Koymen HO, Cohen MS, Cannon JG. 1999. A Neisseria gonorrhoeae immunoglobulin A1 protease mutant is infectious in the human challenge model of urethral infection. Infect. Immun. 67:3009–3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jose J, Otto GW, Meyer TF. 2003. The integration site of the iga gene in commensal Neisseria spp. Mol. Gen. Genomics 269:197–204 [DOI] [PubMed] [Google Scholar]
- 24. Kado CI, Liu ST. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kellogg DS, Jr, Peacock WL, Jr, Deacon WE, Brown L, Pirkle CL. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 85:1274–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Knapp JS, et al. 1978. Phenotypic and epidemiologic correlates of auxotype in Neisseria gonorrhoeae. J. Infect. Dis. 138:160–165 [DOI] [PubMed] [Google Scholar]
- 27. Kohler PL, Hamilton HL, Cloud-Hansen K, Dillard JP. 2007. AtlA functions as a peptidoglycan lytic transglycosylase in the Neisseria gonorrhoeae type IV secretion system. J. Bacteriol. 189:5421–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koomey JM, Gill RE, Falkow S. 1982. Genetic and biochemical analysis of gonococcal IgA1 protease: cloning in Escherichia coli and construction of mutants of gonococci that fail to produce the activity. Proc. Natl. Acad. Sci. U. S. A. 79:7881–7885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kupsch EM, et al. 1996. Construction of Hermes shuttle vectors: a versatile system useful for genetic complementation of transformable and non-transformable Neisseria mutants. Mol. Gen. Genet. 250:558–569 [DOI] [PubMed] [Google Scholar]
- 30. La Scolea LJ, Jr, Young FE. 1974. Development of a defined minimal medium for the growth of Neisseria gonorrhoeae. Appl. Microbiol. 28:70–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laskos L, Dillard JP, Seifert HS, Fyfe JAM, Davies JK. 1998. The pathogenic neisseriae contain an inactive rpoN gene and do not utilize the pilE σ54 promoter. Gene 208:95–102 [DOI] [PubMed] [Google Scholar]
- 32. Lin L, et al. 1997. The Neisseria type 2 IgA1 protease cleaves LAMP1 and promotes survival of bacteria within epithelial cells. Mol. Microbiol. 24:1083–1094 [DOI] [PubMed] [Google Scholar]
- 33. Liu SV, Saunders NJ, Jeffries A, Rest RF. 2002. Genome analysis and strain comparison of Correia repeats and Correia repeat-enclosed elements in pathogenic Neisseria. J. Bacteriol. 184:6163–6173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Long CD, et al. 2001. Modulation of gonococcal piliation by regulatable transcription of pilE. J. Bacteriol. 183:1600–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maneewannakul S, Maneewannakul K, Ippen-Ihler K. 1992. Characterization, localization, and sequence of F transfer region products: the pilus assembly gene product TraW and a new product, TrbI. J. Bacteriol. 174:5567–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manoil C. 1991. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 34:61–75 [DOI] [PubMed] [Google Scholar]
- 37. Martin PR, Mulks MH. 1992. Molecular characterization of the argJ mutation in Neisseria gonorrhoeae strains with requirements for arginine, hypoxanthine, and uracil. Infect. Immun. 60:970–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martin PR, Mulks MH. 1992. Sequence analysis and complementation studies of the argJ gene encoding ornithine acetyltransferase from Neisseria gonorrhoeae. J. Bacteriol. 174:2694–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mehr IJ, Long CD, Serkin CD, Seifert HS. 2000. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154:523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 41. Morse SA, Bartenstein L. 1974. Factors affecting autolysis of Neisseria gonorrhoeae. Proc. Soc. Exp. Biol. Med. 145:1418–1421 [DOI] [PubMed] [Google Scholar]
- 42. Mulks MH, AG Plaut. 1978. IgA protease production as a characteristic distinguishing pathogenic from harmless neisseriaceae. N. Engl. J. Med. 299:973–976 [DOI] [PubMed] [Google Scholar]
- 43. Mulks MH, Knapp JS. 1987. Immunoglobulin A1 protease types of Neisseria gonorrhoeae and their relationship to auxotype and serovar. Infect. Immun. 55:931–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Brien JP, Goldenberg DL, Rice PA. 1983. Disseminated gonococcal infection: a prospective analysis of 49 patients and a review of pathophysiology and immune mechanisms. Medicine (Baltimore) 62:395–406 [PubMed] [Google Scholar]
- 45. Ouchane S, Kaplan S. 1999. Topological analysis of the membrane-localized redox-responsive sensor kinase PrrB from Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 274:17290–17296 [DOI] [PubMed] [Google Scholar]
- 46. Pachulec E. 2010. The type IV secretion systems of Neisseria gonorrhoeae. Ph.D. thesis University of Groningen, Groningen, Netherlands [Google Scholar]
- 47. Pelicic V, Morelle S, Lampe D, Nassif X. 2000. Mutagenesis of Neisseria meningitidis by in vitro transposition of Himar1 mariner. J. Bacteriol. 182:5391–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 49. Picard FJ, Dillon JR. 1989. Biochemical and genetic studies with arginine and proline auxotrophs of Neisseria gonorrhoeae. Can. J. Microbiol. 35:1069–1075 [DOI] [PubMed] [Google Scholar]
- 50. Piekarowicz A, et al. 2007. Characterization of the dsDNA prophage sequences in the genome of Neisseria gonorrhoeae and visualization of productive bacteriophage. BMC Microbiol. 7:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Salgado-Pabón W, et al. 2010. Increased expression of the type IV secretion system in piliated Neisseria gonorrhoeae. J. Bacteriol. 192:1912–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Salgado-Pabón W, Jain S, Turner N, van der Does C, Dillard JP. 2007. A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae. Mol. Microbiol. 66:930–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 54. Seifert HS. 1997. Insertionally inactivated and inducible recA alleles for use in Neisseria. Gene 188:215–220 [DOI] [PubMed] [Google Scholar]
- 55. Seifert HS, Ajioka RS, Paruchuri D, Heffron F, So M. 1990. Shuttle mutagenesis of Neisseria gonorrhoeae: pilin null mutations lower DNA transformation competence. J. Bacteriol. 172:40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Seifert HS, Chen EY, So M, Heffron F. 1986. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 83:735–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Seifert HS, Wilson D. 1992. Characterization of a cryptic gene pair from Neisseria gonorrhoeae that is common to pathogenic Neisseria species. Infect. Immun. 60:1232–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Siddique A, Buisine N, Chalmers R. 2011. The transposon-like Correia elements encode numerous strong promoters and provide a potential new mechanism for phase variation in the meningococcus. PLoS Genet. 7:e1001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sparling PF. 1966. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J. Bacteriol. 92:1364–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Steichen CT, Cho C, Shao JQ, Apicella MA. 2011. The Neisseria gonorrhoeae biofilm matrix contains DNA, and an endogenous nuclease controls its incorporation. Infect. Immun. 79:1504–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Steichen CT, Shao JQ, Ketterer MR, Apicella MA. 2008. Gonococcal cervicitis: a role for biofilm in pathogenesis. J. Infect. Dis. 198:1856–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stein DC, Silver LE, Clark VL, Young FE. 1983. Construction and characterization of a new shuttle vector, pLES2, capable of functioning in Escherichia coli and Neisseria gonorrhoeae. Gene 25:241–247 [DOI] [PubMed] [Google Scholar]
- 63. Stieger M, Wohlgensinger B, Kamber M, Lutz R, Keck W. 1999. Integrational plasmids for the tetracycline-regulated expression of genes in Streptococcus pneumoniae. Gene 226:243–251 [DOI] [PubMed] [Google Scholar]
- 64. Stohl EA, et al. 2003. Escherichia coli RecX inhibits RecA recombinase and coprotease activities in vitro and in vivo. J. Biol. Chem. 278:2278–2285 [DOI] [PubMed] [Google Scholar]
- 65. Swanson J, Kraus SJ, Gotschlich EC. 1971. Studies on gonococcus infection. I. Pili and zones of adhesion: their relation to gonococcal growth patterns. J. Exp. Med. 134:886–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. van Dam V, Bos MP. 2012. Generating knock-out and complementation strains of Neisseria meningitidis. Methods Mol. Biol. 799:55–72 [DOI] [PubMed] [Google Scholar]
- 67. Wolfgang M, van Putten JP, Hayes SF, Dorward D, Koomey JM. 2000. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 19:6408–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]