Abstract
Among methanogens, only 2 genera, Methanosaeta and Methanosarcina, are known to contribute to methanogenesis from acetate, and Methanosaeta is a specialist that uses acetate specifically. However, Methanosaeta strains so far have mainly been isolated from anaerobic digesters, despite the fact that it is widespread, not only in anaerobic methanogenic reactors and freshwater environments, but also in marine environments, based upon extensive 16S rRNA gene-cloning analyses. In this study, we isolated an aceticlastic methanogen, designated strain 03d30qT, from a tidal flat sediment. Phylogenetic analyses based on 16S rRNA and mcrA genes revealed that the isolate belongs to the genus Methanosaeta. Unlike the other known Methanosaeta species, this isolate grows at Na+ concentrations of 0.20 to 0.80 M, with an optimum concentration of 0.28 M. Quantitative estimation using real-time PCR detected the 16S rRNA gene of the genus Methanosaeta in the marine sediment, and relative abundance ranged from 3.9% to 11.8% of the total archaeal 16S rRNA genes. In addition, the number of Methanosaeta organisms increased with increasing depth and was much higher than that of Methanosarcina organisms, suggesting that aceticlastic methanogens contribute to acetate metabolism to a greater extent than previously thought in marine environments, where sulfate-reducing acetate oxidation prevails. This is the first report on marine Methanosaeta species, and based on phylogenetic and characteristic studies, the name “Methanosaeta pelagica” sp. nov. is proposed for this novel species, with type strain 03d30q.
INTRODUCTION
The global budget of atmospheric CH4 is approximately 500 to 600 Tg per year (28), and approximately 74% of the emitted CH4 is derived from biological methanogenesis (6, 20, 64). Acetate is a key compound for methanogenic degradation of organic matter, and the methanogenesis accounts for two-thirds of the total CH4 generated (22). However, only 2 genera, Methanosarcina and Methanosaeta, are capable of aceticlastic methanogenesis. Methanosarcina is a versatile methanogen that prefers methanol, methylamine, and hydrogen to acetate (2), whereas Methanosaeta is a specialist that uses acetate only (4). The 2 genera use different enzymes to activate acetate (20). Complete genome sequencing of Methanosarcina acetivorans C2AT, Methanosarcina mazei Go1, Methanosaeta concilii GP6, and Methanosaeta thermophila PTT revealed that the 2 genera use different enzymes to catalyze the first step of aceticlastic methanogenesis but that the core steps of methanogenesis are similar (1, 10, 17, 54). A major difference between the genera is their affinity for acetate; the minimum threshold for acetate use is much lower in Methanosaeta (7 to 70 μM) than in Methanosarcina (0.2 to 1.2 mM) (20). Accordingly, Methanosaeta adapts to low-acetate environments and predominates over Methanosarcina in environments such as rice fields (18), landfill sites (25, 35), and anaerobic digesters (27, 52). Methanosaeta may be widely distributed in nature and may be the predominant CH4 producer on earth.
Methanogens are abundant in habitats where electron acceptors, such as oxygen, nitrate, and sulfate, are strictly limited. In anaerobic marine habitats, because of the abundance of sulfate in seawater, sulfate-reducing bacteria usually degrade organic matter, including acetate. Meanwhile, many studies on marine sediment communities have demonstrated that methanogenesis also occurs in the high-sulfate-concentration and anaerobic methane oxidation layers, as well as in the deep sulfate-depleted zone (7, 12, 44). Methylotrophic methanogens, including Methanosarcina, are regarded as major contributors to CH4 production in these areas, since methylated compounds cannot be used by sulfate-reducing bacteria and thus remain available for methanogens as noncompetitive substrates (41). In marine sediments, particularly in the deep sulfate-depleted zone, CH4 is thought to originate from H2-CO2 rather than from acetate (63). Also, various methanogens have been retrieved from brackish environments, such as estuarine and tidal flat sediments, by molecular approaches (13, 47, 48). In these environments, methanogens probably do not compete with sulfate-reducing bacteria, because organic substrates are not limiting.
As aceticlastic methanogens, several strains of Methanosarcina, but not Methanosaeta, have been isolated from marine sediments (14, 24, 56). However, as stated previously, Methanosarcina is a versatile methanogen that uses a wide range of substrates, and it may function as a methylotrophic or hydrogenotrophic methanogen. Acetate concentrations in the pore water of marine sediments are usually less than 20 μM (23, 43); thus, conditions appear to be suitable for Methanosaeta rather than for Methanosarcina. Many clones related to Methanosaeta have been detected in marine sediments (11, 24, 32, 42, 46), but their identities remain unknown. In the present study, we report on an aceticlastic methanogen belonging to the genus Methanosaeta isolated from a tidal flat sediment. In addition, based on the results of phylogenetic and ecological investigations, we discuss the contribution to CH4 production from acetate in marine ecosystems.
MATERIALS AND METHODS
Microorganisms and growth conditions.
Strain 03d30qT (= NBRC 105920T = DSM 24271T) was isolated as described below. Escherichia coli NBRC 3301 (= K-12), Methanosarcina barkeri NBRC 100474T, M. thermophila NBRC 101360T, M. concilii NBRC 103675T, and Methanosaeta harundinacea NBRC 104789T were used for analyses. The cultivation media were NBRC media no. 802, 837, 897, 994, and 925 (39), respectively, and all cultures were cultivated at a temperature of 37°C, except M. thermophila (55°C).
Study area, sample collection, and measurements.
The Futtsu tidal flat is a foreshore sand flat located from the Koito River estuary to the sea in Tokyo Bay, Chiba Prefecture, Japan. Sediment samples were collected on 24 June 2005 using a peat sampler (model DIK-105A; Daiki Rika Kogyo). The collected samples were immediately (approximately 2 h) transported to the laboratory in a sealed nylon bag with an O2-absorbing and CO2-generating agent (Anaero-Pack; Mitsubishi Gas Chemical) and inoculated into medium as soon as possible. Samples for molecular analysis were stored in 2-ml tubes at −80°C until extraction of DNA. Sediment temperature was measured on site using a thermometer. Pore water salinity and pH were measured on site using a refractometer (ATC-S/Mill-E; Atago) and pH meter (D-13; Horiba), respectively.
Enrichment, isolation, and purity test for aceticlastic methanogens.
NBRC medium no. 1108 was used to enrich and isolate the aceticlastic methanogens. The medium was composed of the following salts and solutions (liter−1): 1.19 g KH2PO4, 0.21 g K2HPO4, 3.05 g MgCl2 · 6H2O, 0.15 g CaCl2 · 2H2O, 0.54 g NH4Cl, 20 g NaCl, 6.56 g sodium acetate, 1.5 g Bacto Yeast Extract (Difco), 0.4 g Bacto Tryptone (Difco), 0.14 g coenzyme M (2-mercaptoethanesulfonic acid sodium salt), 2.5 g NaHCO3, 1 mg resazurin, 2 ml trace element solution (37), 10 ml vitamin solution (37), and 0.36 g Na2S · 9H2O. The medium was prepared in culture vessels with butyl-rubber stoppers and aluminum seals under N2-CO2 (80/20 [vol/vol]). Enrichments and routine cultivations were conducted in 50-ml culture vessels containing 20 ml of the liquid medium.
The purity of the isolate was verified by microscopic observation, inoculation into the various media, and determination of the 16S rRNA gene sequences amplified using the following primer sets: universal primer set 530f and 1392r, bacterial primer set 27f and 1492r (26), and archaeal primer set A109f (18) and ARC915 (58) (Table 1).
Table 1.
PCR primer sets for this study
| Gene | Primers (forward/reverse) | Target group | Amplicon length (bp) | Reference |
|---|---|---|---|---|
| 16S rRNA | 530f/1392r | Universal | 900 | 26 |
| A109f/ARC915 | Archaea | 800 | 18, 58 | |
| Ar0023mLF/Ar1530R | Archaea | 1,500 | 36 | |
| 27f/1492r | Bacteria | 1,500 | 26 | |
| Bac349F/Bac806R | Bacteria | 460 | 59 | |
| A344F/ARC915 | Archaea | 570 | 5, 58 | |
| MX825cm/ARC915 | Methanosaeta | 90 | This study, 58 | |
| MS821c/ARC915 | Methanosarcina | 90 | 49, 58 | |
| mcrA | MR1mod/ME2mod | Archaea | 1,100 | This study |
Phenotypic and chemotaxonomic features.
Optimum temperatures, initial pH values, and Na+ concentrations for growth were determined by examining the time course of CH4 production. The pH of the medium was adjusted by adding Na2CO3 or HCl at room temperature. The range of Na+ concentrations for the growth of each Methanosaeta species was examined using each medium and adding various concentrations of NaCl. The gas phase of the cultures was analyzed by gas chromatography using a thermal-conductivity detector and a Molecular Sieve 60/80 column (both from Shimadzu) (34).
The G+C content of extracted genomic DNA was analyzed using high-performance liquid chromatography (HPLC) with a reverse-phase column (31). The Gram reaction and susceptibility and motility tests were performed as described by Boone and Whitman (3). To determine DNA-DNA relatedness, the fluorometric microplate hybridization method developed by Ezaki et al. (15, 16) was applied.
Microscopy.
An Olympus AX70 microscope was used for routine observation. For the observation of ultrathin sections of cells, cells were prepared by using rapid freezing and the freeze-substitution method (65). After the procedures, the cells were embedded, stained with uranyl acetate and lead citrate after prestaining with 0.2% oolong tea extract in potassium phosphate buffer (51, 65), and observed using a transmission electron microscope (H-7650; Hitachi) operating at 100 kV.
DNA extraction, PCR, sequencing, and quantitative PCR.
Extraction and purification of the genomic DNA of the microorganisms were performed as described previously (38). The PCR primers used in this study are summarized in Table 1. The 16S rRNA gene of the microorganisms was amplified and sequenced as described previously (36). The partial mcrA (alpha subunit of methyl-coenzyme M reductase) gene was amplified using primers MR1mod (5′-GAC CTS CAC TWC GTV AAC AAC-3′) and ME2mod (5′-TCA TBG CRT AGT TNG GRT AGT-3′), which were slightly modified MR1 (53) and ME2 (19) primers based on the genome sequence of M. thermophila (CP000477). PCR amplification and sequencing of the products were performed as described previously (33). The genomic DNA of the microbial cells in the sediments was extracted using a Power Soil DNA Isolation Kit (MoBio). Quantitative PCR was performed as described previously (36), except for the primer sets and standards. The following 16S rRNA gene-targeted PCR primer sets were used: Bac349F and Bac806R (59) for the domain Bacteria, A344F (5) and ARC915 (58) for the domain Archaea, MX825cm (modified MX825c; 5′-GCT AGG TGT CRG YCA CGG TGC GA-3′) (8) and ARC915 for the genus Methanosaeta, and MS821c (5′-GCT CGC TAG GTG TCA GGC ATG GCG-3′) (49) and ARC915 for the genus Methanosarcina. The template standards for each primer set were constructed using the dilution series of the 16S rRNA gene PCR products of E. coli (for Bacteria), M. thermophila (for Archaea and Methanosaeta), and M. barkeri (for Methanosarcina). These PCR products were used in each real-time PCR analysis to calculate the copy number of the 16S rRNA genes in the sediment samples. The efficiencies of quantitative PCR analyses for Bacteria, Archaea, Methanosaeta, and Methanosarcina were 1.01, 0.77, 0.77, and 0.78, respectively. Copy numbers were calculated from duplicate data in 2 independent analyses using the same DNA extract. Two tests were performed to confirm the specificity of the real-time PCR assay: a melting-curve analysis performed after the amplification and a size analysis of the PCR products confirmed by gel electrophoresis. In addition, to confirm specificity, real-time PCR products for the assays of the genera Methanosaeta and Methanosarcina were cloned using a TOPO TA Cloning Kit (Invitrogen), and several representatives were sequenced using the M13 primer set.
Phylogenetic analysis.
Phylogenetic analyses were carried out using the 16S rRNA gene sequence and deduced amino acid sequence of the mcrA gene. The 16S rRNA gene sequences were aligned with an ARB data set using ARB software (29), and the resulting alignment was recorrected manually considering primary and secondary structures. The data set of the mcrA gene was aligned using Clustal X software (60). Phylogenetic trees were inferred using the neighbor-joining (NJ) method in the Clustal X packages, and 1,000 replicate data sets were used for the bootstrap analysis (50, 60).
Nucleotide sequence accession numbers.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA and mcrA gene sequences are AB679167 to AB679171.
RESULTS
Site description.
Core samples of sandy sediments were collected from 2 points (sample no. 031 and 032) at low tide. As some sea grass grew near sample no. 031 and the sediments were predominantly blackish, the conditions were inferred to be completely anaerobic. For sample no. 032, the sediments changed gradually from gray to black with increasing depth, and the deeper sediments probably maintained anaerobic conditions. Sediment samples were collected at 3 depths (10, 35, and 60 cm) for molecular analysis, and the sediments from sample no. 032 at depths of 35 cm and 60 cm were used for the enrichment of aceticlastic methanogens. The average temperature, pH, and salinity were 21°C, 7.3, and 32‰, respectively, in the sediments and 25°C, 7.8, and 31‰, respectively, in the surrounding seawater. The measurement values of the sediments were not much different at different depths.
Enrichment and isolation.
Primary enrichment cultures of aceticlastic methanogens were obtained by inoculating the medium with 1 g of each sediment sample and incubating it at 20°C and 30°C. After 1 month of cultivation, microbial growth and CH4 formation were observed in all cultures, and the primary enrichment cultures were transferred to fresh medium. Microscopic observation of enrichment after several passages indicated that Methanosaeta-like cells were enriched in the culture of sample no. 032 (60-cm depth) at 30°C. Therefore, the acquisition of pure culture was focused on the enrichment. Since colony formation in medium solidified with 1.5% agar was not successful, serial dilution was used for the isolation. It became clear that the Methanosaeta-like cells were dominant and that the transferred culture at higher dilution (>107) grew continuously in the medium even after removing yeast extract and tryptone, although more than 3 months was required to confirm growth. After repeating the dilution culture several times, an aceticlastic methanogen, designated strain 03d30qT, was obtained (Fig. 1A). The purity of the isolate was verified by determining the 16S rRNA gene sequence amplified from the extracted DNA using various primer sets and inoculation into test media. No growth was observed in medium under H2-CO2 (80/20 [vol/vol]; 150 kPa) as a substitute for N2-CO2; medium supplemented with 10 mM sodium lactate, 10 mM sodium sulfate, and 10 mM sodium thiosulfate; and NBRC medium no. 325 (39) for aerobic heterotrophs. These results confirmed that the culture was free of hydrogenotrophic methanogens and heterotrophs.
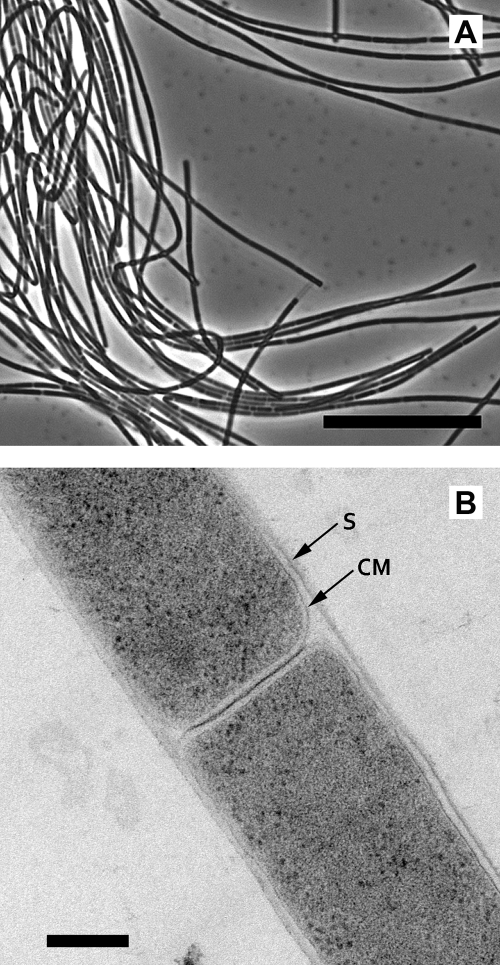
Fig 1.
(A) Phase-contrast micrograph of strain 03d30qT (bar, 10 μm). (B) Ultrathin section of strain 03d30qT observed with a transmission electron microscope (bar, 0.2 μm). CM, cytoplasmic membrane; S, sheath.
Phylogenetic analysis.
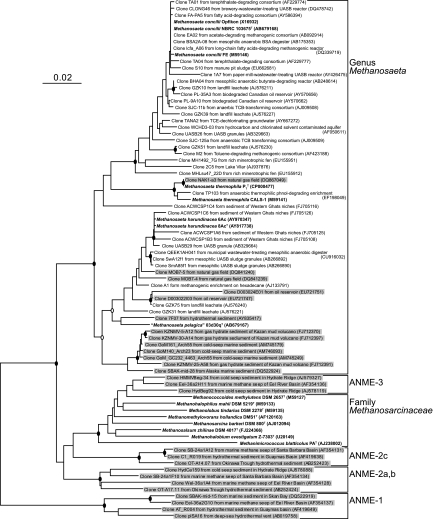
We determined nearly full-length 16S rRNA gene sequences of strain 03d30qT (1,383 bp; AB679167) and M. concilii NBRC 104675T (1,407 bp; AB679168). Phylogenetic analysis based on the 16S rRNA gene sequence (Fig. 2) indicated that strain 03d30qT should be placed in the genus Methanosaeta and that the sequence similarities between the isolate and the valid species M. concilii, M. thermophila, and M. harundinacea were 92.8%, 92.5%, and 97.0%, respectively. In the genus Methanosaeta, strain 03d30qT formed a distinct cluster with environmental clone sequences retrieved from saline environments, such as hydrothermal sediments, cold-seep sediments, oil reservoirs, marine methane hydrates, and ancient seawater (11, 24, 32, 40, 42, 46), though it is distantly related to the sequences from terrestrial environments, anaerobic methanogenic reactors, and M. harundinacea isolated from an upflow anaerobic sludge blanket (UASB) reactor treating brewery wastewater (30). The cluster was clearly separated from the anaerobic methane-oxidizing archaeal (ANME) clusters that have often been retrieved from marine sediments. In addition to the 16S rRNA gene sequence, the mcrA gene sequences of strain 03d30qT (1,150 bp; AB679169), M. concilii NBRC 103675T (1,150 bp; AB679170), and M. harundinacea NBRC 104789T (1,150 bp; AB679171) were also determined. The NJ tree for McrA (Fig. 3) demonstrated that strain 03d30qT was also associated with the genus Methanosaeta, and the similarities between the isolate and M. concilii, M. thermophila, and M. harundinacea were 82.2%, 85.6%, and 90.9%, respectively. DNA-DNA hybridization studies between strain 03d30qT and M. harundinacea NBRC 104789T showed relatedness values of less than 1%.
Fig 2.
Neighbor-joining tree based on 16S rRNA gene sequences of strain 03d30qT and relatives. The probability scores obtained at branch points are indicated by solid circles (above 95%) and open circles (above 90%). The sequences obtained from isolates are in boldface. Environmental clonal sequences retrieved from saline environments are shaded. GenBank/EMBL/DDBJ accession numbers are shown in parentheses. Bar, 0.02 substitutions per compared nucleotide site.
Fig 3.
Neighbor-joining tree showing the relationships between strain 03d30qT and relatives based on deduced McrA sequences. The probability scores obtained at branch points are indicated by solid circles (above 95%) and an open circle (above 90%). GenBank/EMBL/DDBJ accession numbers are shown in parentheses. Bar, 0.02 substitutions per compared position.
Physiology of the isolate and Na+ requirement.
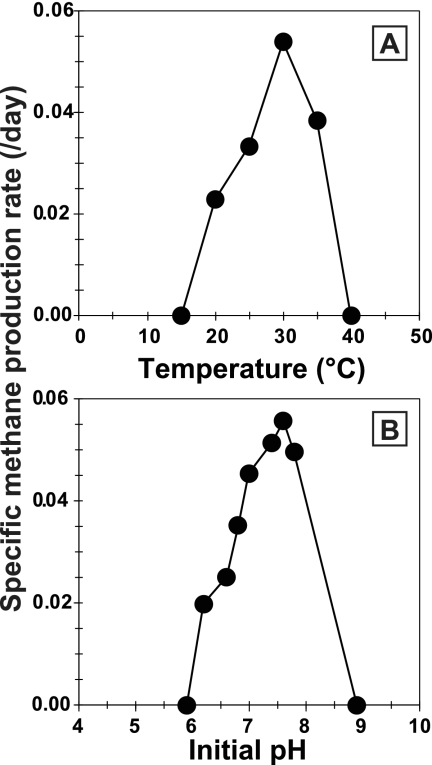
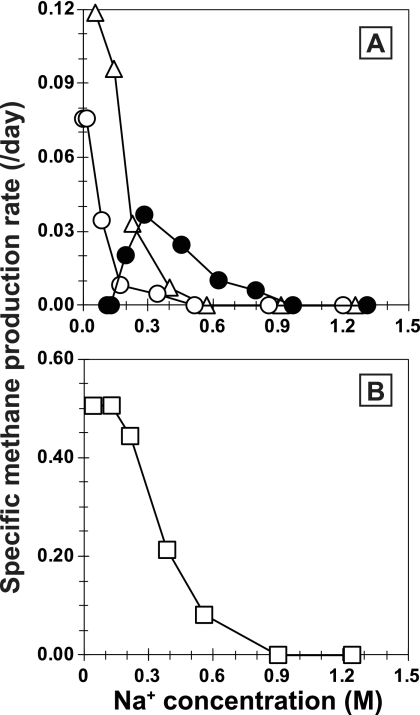
Morphologically, the cells of strain 03d30qT were weakly autofluorescent nonmotile rods, and many multicellular filaments were observed microscopically (Fig. 1A). A tubular sheath enclosed the cells, and the individual cells within the sheath had a cytoplasmic membrane (Fig. 1B). Gram staining of the cells was negative, and susceptibility to lysis by detergent (SDS; 0.1 g liter−1) or hypotonic conditions was not observed. The G+C content of the genomic DNA of strain 03d30qT was 45.4 mol%. Strain 03d30qT used acetate as the sole energy and carbon source, and yeast extract stimulated its growth. Growth and methane formation were not observed on H2-CO2 (80/20; 125 kPa), formate (40 mM), methanol (20 mM), ethanol (20 mM), methylamine (20 mM), dimethylamine (20 mM), trimethylamine (20 mM), or dimethyl sulfide (4 mM). Strain 03d30qT was able to grow at temperatures between 20°C and 35°C and initial pH values between 6.0 and 7.8, with optimum growth at 30°C and initial pH 7.5 (Fig. 4). The effect of the Na+ concentration in the medium on growth, determined by CH4 production of strain 03d30qT and other Methanosaeta species, is shown in Fig. 5. The Na+ concentrations for growth of strain 03d30qT ranged from 0.20 to 0.80 M, with optimum growth at 0.28 M. The other Methanosaeta species grew optimally in each medium containing the lowest Na+ concentration. The upper Na+ concentrations for growth of M. concilii, M. thermophila, and M. harundinacea were 0.40 M, 0.56 M, and 0.34 M, respectively. Strain 03d30qT had a doubling time of 12.4 days under optimum growth conditions (30°C, pH 7.5, and 0.28 M Na+).
Fig 4.
Effect of temperature (A) and initial pH (B) on CH4 production of strain 03d30qT.
Fig 5.
Effect of Na+ concentration on CH4 production of strain 03d30qT (solid circles), M. concilii (triangles), and M. harundinacea (open circles) (A) and M. thermophila (B). The Na+ concentration was adjusted by the addition of NaCl to each growth medium.
Environmental quantification.
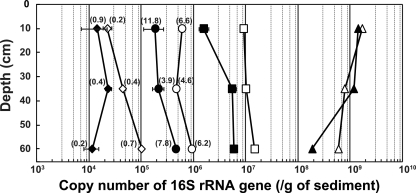
Field estimation of aceticlastic methanogens in tidal flat sediments was performed with quantitative real-time PCR assays using the 16S rRNA gene copy number (Fig. 6). The relative abundances of Methanosaeta and Methanosarcina were calculated as the percentages of the total archaeal 16S rRNA gene copy number in the DNA extracts (Fig. 6, in parentheses). The number of Bacteria tended to decrease and the number of Archaea tended to increase with increasing depth. The number of Methanosaeta organisms increased slightly with increasing depth, and the proportion of total Archaea ranged from 3.9 to 11.8%. The number of Methanosarcina organisms was less than 1/10 the number of Methanosaeta organisms, and the maximum proportion was 0.9% of total Archaea.
Fig 6.
Copy numbers of 16S rRNA genes of Bacteria (triangles), Archaea (squares), Methanosaeta (circles), and Methanosarcina (diamonds) detected by real-time PCR analyses. The open and solid symbols represent samples 031 and 032, respectively. The percentages of Methanosaeta and Methanosarcina among the total Archaea are shown in parentheses.
DISCUSSION
Ecology and physiology of a marine Methanosaeta strain.
Previous studies predicted the occurrence of Methanosaeta-type organisms in marine environments based on molecular analyses. They indicated that Methanosaeta species inhabit saline environments, such as marine sediments, gas hydrate sediments, hydrothermal sediments, oil reservoirs, and natural gas fields (11, 24, 32, 40, 42, 46) (Fig. 2), but the identities of the organisms remained unknown. Sowers reported that an acetate-utilizing rod-shaped methanogen was obtained from marine sediments (55), but it is impossible to confirm it as a marine Methanosaeta species. In the present study, we successfully isolated an aceticlastic methanogen belonging to the genus Methanosaeta from a marine environment. Unlike the other Methanosaeta species isolated so far, our isolate is obviously “moderately halophilic” and adapts to saline environments (Fig. 5). The results directly show that Methanosaeta thrives and contributes to anaerobic acetate degradation, not only in terrestrial environments, but also in saline environments. This information indicates that Methanosaeta-type organisms in marine environments may have evolved specifically by adapting to a saline environment, in which all previously known Methanosaeta species would not survive.
In marine ecosystems, hydrogen and acetate, the major substrates for methanogens, are consumed mainly by sulfate reducers in the sulfate-rich surface layer of sediments. However, sulfate is generally depleted at a depth of tens of centimeters to several meters, and methanogenesis potentially becomes the dominant terminal oxidation process in the sulfate-depleted zone. In addition, methane production has been detected and methanogens have been retrieved even from sediments with high sulfate concentrations (7, 12, 23, 24, 44). In the present study, the 16S rRNA genes of Methanosaeta were detected in tidal flat sediments by real-time PCR analysis, and the amount (number of copies per gram sample) increased slightly with increasing depth (Fig. 6). Interestingly, the number of Methanosaeta genes was 10 times higher than that of Methanosarcina genes. The results may indicate that the methane produced from acetate originates from Methanosaeta rather than from Methanosarcina and that Methanosaeta contributes significantly to the anaerobic degradation of organic matter in tidal flat sediments. Acetate concentrations in the pore water of marine sediments are usually less than 20 μM (24, 43). Therefore, in marine sediments, Methanosaeta has an advantage over Methanosarcina in acetate use based on affinity (20).
Although several strains of aceticlastic methanogens belonging to the genus Methanosarcina have been isolated from marine sediments (14, 24, 56, 61), isotopic evidence suggests that acetate is a minor precursor for methanogenesis and that the rate of acetate oxidation to CO2 is higher than the rate of CH4 production in the surface layer (43, 63). On the other hand, sometimes Methanosaeta growth was not observed although the organisms were detected by molecular analysis (25). Certainly, Methanosaeta is one of the most recalcitrant methanogens and is difficult to enrich and isolate, primarily because of slow growth. In addition, Wellsbury et al. reported difficulty distinguishing between abiological thermogenic and biological aceticlastic methanogenesis in marine sediments (9, 62). These facts suggest that aceticlastic methane generation in marine sediments has been underestimated. Parkes et al. showed that traces of Methanosaeta were detected and acetate concentrations in marine sediments remained low, although the rate of CH4 production from acetate was equivalent to one-quarter of the rate from H2-CO2 (43), indicating the presence of sufficient acetate-using populations to maintain the final steps of in situ degradation of organic matter. Aceticlastic methanogens may be widespread and perhaps dominant in contributing to acetate degradation in marine sediments, in particular the tidal flat sediments, which have an abundant supply of organic matter.
In conclusion, we have isolated marine Methanosaeta species from tidal flat sediments and suggested a possible contribution to acetate metabolism in saline environments. Further analyses for acetate degradation in tidal flat sediments, such as competition between methanogens and sulfate-reducing bacteria, are now under way.
Taxonomy.
The characteristics of strain 03d30qT and Methanosaeta species are summarized in Table 2. The phylogenetic tree based on the 16S rRNA gene sequence (Fig. 2) revealed that strain 03d30qT belongs to the genus Methanosaeta and that M. harundinacea is a related cultured relative (97.0% sequence similarity). Phylogenetic analysis based on McrA showed a similar trend (Fig. 3). The results from a DNA-DNA hybridization study indicated that the degree of DNA relatedness between strain 03d30qT and M. harundinacea was <1%. These results suggest that a new species should be created for the isolate (57). In addition, the G+C content of genomic DNA, sensitivity to the Na+ concentration, and temperature for growth differentiated strain 03d30qT from the other Methanosaeta species. In particular, the difference in Na+ sensitivity was a criterion for discriminating strain 03d30qT from the other Methanosaeta species, because strain 03d30qT was the only moderately halophilic species. For these reasons, strain 03d30qT represents a novel species within the genus Methanosaeta, for which the name “Methanosaeta pelagica” sp. nov. is proposed.
Table 2.
Comparison of properties of Methanosaeta species
| Property | Value |
|||
|---|---|---|---|---|
| “M. pelagica” | M. conciliia | M. thermophilaa | M. harundinaceaa | |
| Type strain | 03d30q | GP6 | PT | 8Ac |
| Habitat | Tidal flat sediment | Anaerobic digester | Anaerobic digester | Anaerobic digester |
| Optimum growth conditions | ||||
| Temp (°C) | 30 | 35–40 | 55–60 | 34—37 |
| pH | 7.5 | 7.1–7.5 | 6.5–6.7 | 7.2–7.6 |
| Na+ concn (M) | 0.28 | <0.06 | <0.13 | <0.02 |
| Effect on yeast extract for growth | Stimulate | Inhibit | Inhibitb | Required |
| G+C content (mol%) | 45.4 (HPLC) | 50.3 (Tm)c | 52.7 (HPLC) | 55.7 (Tm) |
Description of “Methanosaeta pelagica” sp. nov.
“Methanosaeta pelagica” (pe.la′gi.ca. L. fem. adj. pelagica, pertaining to the sea). Single cells are 0.5 μm wide and 2.5 to 11.0 μm long, and long sheathed chains of cells form. It shows weakly blue-green autofluorescence by epifluorescence microscopy. Nonmotile. Gram negative. Susceptibility to lysis by SDS or hypotonic conditions is not observed. Strictly anaerobic. Acetate is the only substrate for growth and CH4 production: H2-CO2, formate, methanol, ethanol, methylamine, dimethylamine, trimethylamine, or dimethyl sulfide does not support growth. Yeast extract stimulates growth. The optimum growth conditions are 30°C, initial pH 7.5, and 0.28 M Na+ concentration. The DNA G+C content of the type strain is 45.4 mol%.
The type strain, 03d30q (= NBRC 105920 = DSM 24271) was isolated from a tidal flat sediment in Tokyo Bay, Chiba Prefecture, Japan.
ACKNOWLEDGMENTS
We thank the staff of the Futtsu fishermen's union for collecting sediment samples. We also thank Kuniko Shimamura and Shinobu Iwasaki for technical support.
Footnotes
Published ahead of print 17 February 2012
REFERENCES
- 1. Barber RD, et al. 2011. Complete genome sequence of Methanosaeta concilii, a specialist in aceticlastic methanogenesis. J. Bacteriol. 193:3668–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boone DR, Mah RA. 2001. Genus I. Methanosarcina Kluyver and van Niel 1936, 400AL, emend. Mah and Kuhn 1984a, 266 (Nom. Cos., Opinion 63 of the Jud. Comm. 1986b, 492), p 269–276 In Boone DR, Castenholz RW. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 1 The Archaea and the deeply branching and phototrophic Bacteria Springer, New York, NY [Google Scholar]
- 3. Boone DR, Whitman WB. 1988. Proposal of minimal standards for describing new taxa of methanogenic bacteria. Int. J. Syst. Bacteriol. 38:212–219 [Google Scholar]
- 4. Boone DR, Whitman WB, Koga Y. 2001. Family II. Methanosaetaceae fam. nov., p 289–294 In Boone DR, Castenholz RW. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 1 The Archaea and the deeply branching and phototrophic Bacteria Springer, New York, NY [Google Scholar]
- 5. Casamayor EO, et al. 2002. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 4:338–348 [DOI] [PubMed] [Google Scholar]
- 6. Conrad R. 2009. The global methane cycle: recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 1:285–292 [DOI] [PubMed] [Google Scholar]
- 7. Cragg BA, et al. 1996. Bacterial populations and processes in sediments containing gas hydrates (ODP Leg 146: Cascadia Margin). Earth Planet. Sci. Lett. 139:497–507 [Google Scholar]
- 8. Crocetti G, Murto M, Bjönsson L. 2006. An update and optimisation of oligonucleotide probes targeting methanogenic Archaea for use in fluorescence in situ hybridisation (FISH). J. Microbiol. Methods 65:194–201 [DOI] [PubMed] [Google Scholar]
- 9. de Graaf W, Wellsbury P, Parkes RJ, Cappenberg TE. 1996. Comparison of acetate turnover in methanogenic and sulfate-reducing sediments by radiolabeling and stable isotope labeling and by use of specific inhibitors: evidence for isotopic exchange. Appl. Environ. Microbiol. 62:772–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deppenmeier U, et al. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453–461 [PubMed] [Google Scholar]
- 11. Dhillon A, et al. 2005. Methanogen diversity evidenced by molecular characterization of methyl coenzyme M reductase A (mcrA) genes in hydrothermal sediments of the Guaymas Basin. Appl. Environ. Microbiol. 71:4592–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. D'Hondt S, et al. 2004. Distributions of microbial activities in deep subseafloor sediments. Science 306:2216–2221 [DOI] [PubMed] [Google Scholar]
- 13. Edmonds JW, Weston NB, Joye SB, Moran MA. 2008. Variation in prokaryotic community composition as a function of resource availability in tidal creek sediments. Appl. Environ. Microbiol. 74:1836–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elberson MA, Sowers KR. 1997. Isolation of an aceticlastic strain of Methanosarcina siciliae from marine canyon sediments and emendation of the species description for Methanosarcina siciliae. Int. J. Syst. Bacteriol. 47:1258–1261 [DOI] [PubMed] [Google Scholar]
- 15. Ezaki T, et al. 1988. Simple genetic method to identify viridans group streptococci by colorimetric dot hybridization and fluorometric hybridization in microdilution wells. J. Clin. Microbiol. 26:1708–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ezaki T, Hashimoto Y, Yabuuchi E. 1989. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39:224–229 [Google Scholar]
- 17. Galagan JE, et al. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grosskopf R, Janssen PH, Liesack W. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hales BA, et al. 1996. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62:668–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jetten MSM, Stams AJM, Zehnder AJB. 1992. Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol. Lett. 88:181–197 [Google Scholar]
- 21. Reference deleted.
- 22. Jones WJ. 1991. Diversity and physiology of methanogens, p 219–235 In Rogers JE, Whitman WB. (ed), Microbial production and consumption of greenhouse gasses: methane, nitrogen, oxides, and halomethanes. ASM Press, Washington, DC [Google Scholar]
- 23. Kendall MM, Boone DR. 2006. Cultivation of methanogens from shallow marine sediments at Hydrate Ridge, Oregon. Archaea 2:31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kendall MM, et al. 2007. Diversity of Archaea in marine sediments from Skan Bay, Alaska, including cultivated methanogens, and description of Methanogenium boonei sp. nov. Appl. Environ. Microbiol. 73:407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laloui-Carpentier W, Li T, Vigneron V, Mazeas L, Bouchez T. 2006. Methanogenic diversity and activity in municipal solid waste landfill leachates. Antonie Van Leeuwenhoek 89:423–434 [DOI] [PubMed] [Google Scholar]
- 26. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–176 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. Jon Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 27. Leclerc M, Delgenes JP, Godon JJ. 2004. Diversity of the archaeal community in 44 anaerobic digesters as determined by single strand conformation polymorphism analysis and 16S rDNA sequencing. Environ. Microbiol. 6:809–819 [DOI] [PubMed] [Google Scholar]
- 28. Lelieveld J, Crutzen PJ, Dentener FJ. 1998. Changing concentration, lifetime and climate forcing of atmospheric methane. Tellus B 50:128–150 [Google Scholar]
- 29. Ludwig W, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma K, Liu X, Dong X. 2006. Methanosaeta harundinacea sp. nov., a novel acetate-scavenging methanogen isolated from a UASB reactor. Int. J. Syst. Evol. Microbiol. 56:127–131 [DOI] [PubMed] [Google Scholar]
- 31. Mesbah M, Premachandran U, Whitman WB. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159–167 [Google Scholar]
- 32. Mochimaru H, et al. 2007. Methanogen diversity in deep subsurface gas-associated water at the Minami-Kanto gas field in Japan. Geomicrobiol. J. 24:93–100 [Google Scholar]
- 33. Mori K, Harayama S. 2011. Methanobacterium petrolearium sp. nov. and Methanobacterium ferruginis sp. nov., novel mesophilic methanogens isolated from salty environments. Int. J. Syst. Evol. Microbiol. 61:138–143 [DOI] [PubMed] [Google Scholar]
- 34. Mori K, Maruyama A, Urabe T, Suzuki K, Hanada S. 2008. Archaeoglobus infectus sp. nov., a novel thermophilic, chemolithoheterotrophic archaeon isolated from a deep-sea rock collected at Suiyo Seamount, Izu-Bonin Arc, western Pacific Ocean. Int. J. Syst. Evol. Microbiol. 58:810–816 [DOI] [PubMed] [Google Scholar]
- 35. Mori K, Sparling R, Hatsu M, Takamizawa K. 2003. Quantification and diversity of the archaeal community in a landfill site. Can. J. Microbiol. 49:28–36 [DOI] [PubMed] [Google Scholar]
- 36. Mori K, et al. 2008. First cultivation and ecological investigation of a bacterium affiliated with the candidate phylum OP5 from hot springs. Appl. Environ. Microbiol. 74:6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mori K, Suzuki K. 2008. Thiofaba tepidiphila gen. nov., sp. nov., a novel obligately chemolithoautotrophic, sulfur-oxidizing bacterium of the Gammaproteobacteria isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 58:1885–1891 [DOI] [PubMed] [Google Scholar]
- 38. Mori K, Yamamoto H, Kamagata Y, Hatsu M, Takamizawa K. 2000. Methanocalculus pumilus sp. nov., a heavy-metal-tolerant methanogen isolated from a waste-disposal site. Int. J. Syst. Evol. Microbiol. 50:1723–1729 [DOI] [PubMed] [Google Scholar]
- 39. NITE Biological Resource Center 2010. NBRC catalogue of biological resources: microorganisms, microorganism-related DNA resources, and human-related DNA resources, 2nd ed National Institute of Technology and Evaluation, Chiba, Japan [Google Scholar]
- 40. Orcutt B, et al. 2010. Impact of natural oil and higher hydrocarbons on microbial diversity, distribution, and activity in Gulf of Mexico cold-seep sediments. Deep-Sea Res. II Top. Stud. Oceanogr. 57:2008–2021 [Google Scholar]
- 41. Oremland RS, Polcin S. 1982. Methanogenesis and sulfate reduction: competitive and noncompetitive substrates in estuarine sediments. Appl. Environ. Microbiol. 44:1270–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pachiadaki MG, Lykousis V, Stefanou EG, Kormas KA. 2010. Prokaryotic community structure and diversity in the sediments of an active submarine mud volcano (Kazan mud volcano, East Mediterranean Sea). FEMS Microbiol. Ecol. 72:429–444 [DOI] [PubMed] [Google Scholar]
- 43. Parkes RJ, et al. 2007. Biogeochemistry and biodiversity of methane cycling in subsurface marine sediments (Skagerrak, Denmark). Environ. Microbiol. 9:1146–1161 [DOI] [PubMed] [Google Scholar]
- 44. Parkes RJ, Cragg BA, Wellsbury P. 2000. Recent studies on bacterial populations and processes in subseafloor sediments: a review. Hydrogeol. J. 8:11–28 [Google Scholar]
- 45. Patel GB. 2000. Genus I. Methanosaeta Patel and Sprott 1990, 80VP, p 289–294 In Boone DR, Castenholz RW. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 1 Springer, New York, NY [Google Scholar]
- 46. Pham VD, et al. 2009. Characterizing microbial diversity in production water from an Alaskan mesothermic petroleum reservoir with two independent molecular methods. Environ. Microbiol. 11:176–187 [DOI] [PubMed] [Google Scholar]
- 47. Purdy KJ, Munson MA, Cresswell-Maynard T, Nedwell DB, Embley TM. 2003. Use of 16S rRNA-targeted oligonucleotide probes to investigate function and phylogeny of sulphate-reducing bacteria and methanogenic archaea in a UK estuary. FEMS Microbiol. Ecol. 44:361–371 [DOI] [PubMed] [Google Scholar]
- 48. Purdy KJ, Munson MA, Nedwell DB, Martin Embley T. 2002. Comparison of the molecular diversity of the methanogenic community at the brackish and marine ends of a UK estuary. FEMS Microbiol. Ecol. 39:17–21 [DOI] [PubMed] [Google Scholar]
- 49. Raskin L, Stromley JM, Rittmann BE, Stahl DA. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 51. Sato S, et al. 2003. Use of oolong tea extract (OTE) for elastin staining and enhancement in ultrathin sections. Med. Electron Microsc. 36:179–182 [DOI] [PubMed] [Google Scholar]
- 52. Sekiguchi Y, et al. 1998. Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. Microbiology 144:2655–2665 [DOI] [PubMed] [Google Scholar]
- 53. Simankova MV, et al. 2003. Isolation and characterization of new strains of methanogens from cold terrestrial habitats. Syst. Appl. Microbiol. 26:312–318 [DOI] [PubMed] [Google Scholar]
- 54. Smith KS, Ingram-Smith C. 2007. Methanosaeta, the forgotten methanogen? Trends Microbiol. 15:150–155 [DOI] [PubMed] [Google Scholar]
- 55. Sowers KR. 2002. Methanogenesis in the marine environment, p 1913–1923 In Bitton G. (ed), Encyclopedia of environmental microbiology, vol 4 John Wiley & Sons, Inc, Hoboken, NJ [Google Scholar]
- 56. Sowers KR, Baron SF, Ferry JG. 1984. Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl. Environ. Microbiol. 47:971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stackebrandt E, Goebel BM. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846–849 [Google Scholar]
- 58. Stahl DA, Amann R. 1991. Development and application of nucleic acid probes, p 205–248 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. Jon Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 59. Takai K, Horikoshi K. 2000. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 66:5066–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. von Klein D, Arab H, Völker H, Thomm M. 2002. Methanosarcina baltica, sp. nov., a novel methanogen isolated from the Gotland Deep of the Baltic Sea. Extremophiles 6:103–110 [DOI] [PubMed] [Google Scholar]
- 62. Wellsbury P, et al. 1997. Deep marine biosphere fuelled by increasing organic matter availability during burial and heating. Nature 388:573–576 [Google Scholar]
- 63. Whiticar MJ, Faber E, Schoell M. 1986. Biogenic methane formation in marine and freshwater environments: CO2 reduction vs. acetate fermentation-isotope evidence. Geochim. Cosmochim. Acta 50:693–709 [Google Scholar]
- 64. Whitman WB, Bowen TL, Boon DR. 2006. The methanogenic bacteria, p 165–207 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The Prokaryotes. Springer, New York NY [Google Scholar]
- 65. Yamaguchi K, Suzuki K-I, Tanaka K. 2010. Examination of electron stains as a substitute for uranyl acetate for the ultrathin sections of bacterial cells. J. Electron Microsc. 59:113–118 [DOI] [PubMed] [Google Scholar]