Abstract
The identity of metabolites encoded by the majority of nonribosomal peptide synthetases in the opportunistic pathogen, Aspergillus fumigatus, remains outstanding. We found that the nonribosomal peptide (NRP) synthetases PesL and Pes1 were essential for fumigaclavine C biosynthesis, the end product of the complex ergot alkaloid (EA) pathway in A. fumigatus. Deletion of either pesL (ΔpesL) or pes1 (Δpes1) resulted in complete loss of fumigaclavine C biosynthesis, relatively increased production of fumitremorgins such as TR-2, fumitremorgin C and verruculogen, increased sensitivity to H2O2, and increased sensitivity to the antifungals, voriconazole, and amphotericin B. Deletion of pesL resulted in severely reduced virulence in an invertebrate infection model (P < 0.001). These findings indicate that NRP synthesis plays an essential role in mediating the final prenylation step of the EA pathway, despite the apparent absence of NRP synthetases in the proposed EA biosynthetic cluster for A. fumigatus. Liquid chromatography/diode array detection/mass spectrometry analysis also revealed the presence of fumiquinazolines A to F in both A. fumigatus wild-type and ΔpesL strains. This observation suggests that alternative NRP synthetases can also function in fumiquinazoline biosynthesis, since PesL has been shown to mediate fumiquinazoline biosynthesis in vitro. Furthermore, we provide here the first direct link between EA biosynthesis and virulence, in agreement with the observed toxicity associated with EA exposure. Finally, we demonstrate a possible cluster cross-talk phenomenon, a theme which is beginning to emerge in the literature.
INTRODUCTION
Nonribosomal peptide (NRP) synthetases are multimodular enzymes in which each module is responsible for individual amino acid recognition, tethering, and subsequent incorporation into an NRP product (59). NRP synthesis (NRPS) in fungi, and in Aspergillus spp. in particular, provides a biosynthetic route for the biosynthesis of a range of bioactive metabolites, including gliotoxin, a redox active dipeptide, and siderophores such as triacetylfusarinine C (12, 47). Moreover, brevianamide F, an NRP product, has been demonstrated to be a precursor for a variety of prenylated alkaloids, including the fumitremorgins A, B, and C and tryprostatin B (28). Ergot alkaloids (EA), such as fumigaclavines A, B, and C, are also produced by A. fumigatus, whereas ergotamines, consisting of lysergic acid and a peptide component, are produced by Claviceps purpurea (11, 16). EA biosynthetic gene clusters have been identified in A. fumigatus (11, 52), C. purpurea (9, 18, 34, 51), and Neotyphodium lolli (15), and many of the steps in the EA biosynthetic pathways have been deciphered through a combination of in vitro biochemical studies and functional characterization of cluster genes (54). A gene cluster responsible for fumigaclavine C biosynthesis in A. fumigatus has been identified (11, 52); however, the requirement for NRP synthetase functionality for EA biosynthesis has not been reported. However, incomplete biosynthetic routes for fumigaclavine and ergotamine biosynthesis, based primarily on in vitro biochemical studies, have been proposed (54), and NRP synthetase involvement, in trans, cannot therefore be excluded.
Fumiquinazolines A to G are among a variety of quinazoline-containing natural products produced by fungi (1, 2). Anthranilate (Ant; a nonproteinogenic aryl β-amino acid) is a precursor for these compounds (2), whereby a biochemical approach confirmed that an NRP synthetase module (AnaPS module 1) from the known gene cluster for acetylaszonalenin from the Neosartorya fisheri NRRL 181 strain (57) activates Ant. Sequence comparison identified an A. fumigatus NRP synthetase, PesM (CADRE identifier AFUA_6G12080), with homology to AnaPS, and recombinant module 1 of PesM was also shown to preferably activate Ant (2). The authors of that study proposed that the trimodular NRP synthetase, PesM, is likely to produce fumiquinazoline F, and that an adjacent monomodular NRP synthetase, PesL (AFUA_6G12050), could function in the conversion of fumiquinazoline F to fumiquinazoline A, by activating alanine and acting with an N-acyltransferase (AFUA_6G12100), to couple alanine to both N-1′ and C-2′ in the indole ring part of fumiquinazoline F (2). Subsequently, recombinantly expressed PesL and an FAD-dependent monooxygenase (AFUA_6G12060) were shown to be required for the conversion of fumiquinazoline F into fumiquinazoline A (1). In contrast to earlier findings about the clustering of genes with pesL (33), the authors noted that pesM is part of an eight-gene cluster, along with pesL, which they predicted to be involved in the production of the Ant-containing alkaloid fumiquinazoline A (1). However, Cramer et al. (13) observed differential expression of pesL and pesM, whereby significantly higher pesL expression (3- to 4-fold higher, respectively) was observed. Moreover, pesM expression only, was detectable in A. fumigatus conidia, which suggested alternate functionality for either NRP synthetase, PesL in particular (13).
LaeA is a transcriptional regulator of secondary metabolite (SM) biosynthetic gene clusters in A. fumigatus and A. nidulans (5). pesL was proposed to be part of a five-gene SM cluster spanning the region from AFUA_6G12040 to AFUA_6G12080 (33), and transcriptional studies revealed that pesL and all other genes in this proposed cluster were found to be under LaeA regulation (35). Expression of another NRP synthetase gene pes1 (AFUA_1G10380; [38]) was downregulated in the A. fumigatus ΔlaeA strain, along with expression of the genes found in the gliotoxin, fumitremorgin B, and EA biosynthetic clusters (35). Pes1 is an orphan NRP synthetase, and although it was demonstrated to mediate A. fumigatus virulence, the identification of the NRP peptide product encoded by pes1 is outstanding (38). Analysis by Cramer et al. (13) showed that pes1 expression was only evident in liquid cultures of Sabouraud broth in the A. fumigatus Af293 strain. Concurrently, Reeves et al. (38) also confirmed differential pes1 expression. Schrettl et al. (41) used a genome-wide microarray to investigate the genes regulated by the SreA transcription factor, a repressor of siderophore production in iron-replete conditions. This profiling identified 1,147 genes that were differentially expressed in a ΔsreA mutant, which included pes1 (upregulated in ΔsreA mutants after 30 min). Since pes1 expression is upregulated in the absence of this regulator, this may indicate that pes1 plays a role in the protection against oxidative stress during sudden changes in Fe3+ levels, potentially by signaling or interacting with related biosynthetic genes. Moreover, the expression of pes1 as well as the neighboring ABC multidrug transporter was downregulated in an A. fumigatus strain lacking stuA, which is involved in the hypoxic adaptation of the fungus (43). Like the A. fumigatus Δpes1 strain, the ΔstuA strain was sensitive to hydrogen peroxide, indicating that this sensitivity exhibited by the ΔstuA strain may be due, in part, to the downregulation of pes1 expression. Consequently, targeted gene deletion and comparative metabolomic studies were undertaken to elucidate the NRP products encoded by PesL and Pes1, respectively.
MATERIALS AND METHODS
Strains, growth conditions, and general DNA manipulation.
A. fumigatus strains were grown at 37°C in Aspergillus minimal medium (AMM) or fungal minimal medium (MM), and fungal culturing was carried out as described by Schrettl et al. (40). Fungal strains used in the present study are listed in Table 1.
Table 1.
Fungal strains and plasmid constructs
| Strain or plasmid | Description or genotype | Source or reference |
|---|---|---|
| Strains | ||
| ATCC 46645 | Wild-type A. fumigatus strain | 19 |
| CEA17 | ΔakuB | 14 |
| ΔpesL mutant | ΔakuB pesL::ptrA | This study |
| Δpes1akuB mutant | ΔakuB pes1::ptrA | This study |
| Δpes146645 mutant | ATCC 46645; pes1::ptrA | This study |
| Plasmid | ||
| pSK275 | Plasmid containing ptrA cassette conferring resistance to pyrithiamine | 21 |
PCRs for generation of DNA manipulation constructs were performed using an Expand long- range template kit (Roche). For general cloning procedures, the bacterial strain Escherichia coli DH5α was used which was cultivated in LB (1% [wt/vol] Bacto tryptone, 0.5% [wt/vol] yeast extract, 1% [wt/vol] NaCl; pH 7.5) medium. Genomic DNA was extracted by using a Zymogen fungal DNA extraction kit (Zymo Research Corp.).
Deletion of A. fumigatus nonribosomal peptide synthetase genes.
A. fumigatus transformation was carried out according to the method of Schrettl et al. (40). To generate the ΔpesL and Δpes1 mutant strains, the bipartite marker technique was used (32), with modifications. pesL is predicted to encode an NRP synthetase of 1,161 amino acid residues. In the ΔpesL-ptrA mutant, the deleted region comprises amino acids 1 to 940 of PesL. Pes1 is predicted to encode a NRP synthetase of 6,269 amino acid residues. In the Δpes1-ptrA mutant, the deleted region comprises amino acids 1 to 904 of Pes1, and this strategy also resulted in the deletion of 570 bp upstream of the pes1 start codon.
The A. fumigatus ΔakuB and ATCC 46645 strains were each cotransformed with two DNA constructs, containing an incomplete fragment of a pyrithiamine resistance gene (ptrA) (40) fused to 1.2 kb and 1.3 kb of pes1 up- and downstream sequences, respectively, which flanked the regions to be deleted. The marker fragments shared a 557-bp overlap within ptrA, serving as a potential recombination site during transformation. Two rounds of PCR generated each fragment. Table S1 in the supplemental material provides a complete list of all of the primers used in the present study. For the disruption of pesL, each flanking region was amplified from ΔakuB genomic DNA using the primers opesL-1 and opesL-4 for flanking region A (1.2 kb), and the primers opesL-2 and opesL-3 for flanking region B (1.3 kb). After gel purification, the fragments were digested with PstI and HindIII, respectively. The ptrA selection marker was released from plasmid pSK275 (kindly provided by Sven Krappmann, Gottingen, Germany) by digestion with PstI and HindIII and ligated with the two flanking regions A and B. For transformation, two overlapping fragments were amplified from the ligation products using the primers opesL-5 and optrA-2 for fragment C (2.6 kb) and the primers opesL-6 and optrA-1 for fragment D (2.1 kb). Subsequently, A. fumigatus was transformed simultaneously with the overlapping fragments C and D. A similar approach was taken for generating pes1 disruption constructs with some modifications as follows. Flanking regions A and B and pSK275 were digested with PvuI and KpnI, respectively. Final disruption constructs were 2.9 kb (fragment C) and 2.4 kb (fragment D). Deletion strains were screened by Southern blot analysis, and digoxigenin hybridization probes were generated by using the primers opesL-3 and opesL-6 for pesL and the primers opes1-5 and opes1-3 for pes1. In order to obtain homokaryotic transformants, colonies from single homokaryotic spores were picked, and single genomic integration was confirmed by Southern blot analysis.
RNA isolation and real-time PCR.
Fungal RNA was isolated and purified from A. fumigatus hyphae crushed in liquid nitrogen using an RNeasy plant minikit (Qiagen). RNA was treated with DNase 1 (Invitrogen), and cDNA synthesis from mRNA (500 ng) was performed using a first-strand transcription cDNA synthesis kit (Roche) with oligo(dT) primers. The gene encoding calmodulin (calm) (AFUA_4G10050), which is constitutively expressed in A. fumigatus, served as a control in reverse transcription-PCR (RT-PCR) experiments (6).
Real-time PCR was performed using the LightCycler 480 Sybr green 1 master mix (Roche) on a LightCycler 480 real-time PCR system. PCRs were carried out in 96-well plates in a reaction volume of 20 μl containing 5 μl of template cDNA. Standard curves were prepared for calm, pesL, and pes1 by generating 5 orders of 10-fold serial dilutions of cDNA in H2O and performing five replicate PCRs on these dilutions. All primers used for RT-PCR experiments are labeled “RT.” Primer sequences and culture conditions used are listed in Table S1 in the supplemental material and in Table 2, respectively. Cycling conditions for PCRs were calculated according to the recommendations from Roche, and 40 cycles of PCR were performed. Standard curves with a PCR efficiency of ≥1.8 and with an error of <0.2, were accepted, and the cycling conditions were used for subsequent real-time PCR analysis by relative quantification using LightCycler 480 software. For real-time PCRs, a 1/10 dilution of cDNA from each sample was used as a template, and each reaction was performed in triplicate.
Table 2.
Summary of all the culture conditions used to examine pesL and pes1 expression
| Gene and culture conditiona | Time period (h) | NRPS expressedb |
|---|---|---|
| pesL | ||
| MEM supplemented with 5% FCS | 24 | Y |
| 48 | Y | |
| 72 | Y | |
| 96 | Y | |
| YG medium | 24 | N |
| 48 | Y | |
| 72 | Y | |
| 96 | N | |
| Czapek's medium | 24 | Y |
| 48 | Y | |
| 72 | Y | |
| 96 | Y | |
| AMM | 24 | Y |
| 48 | N | |
| pes1 | ||
| Sabouraud broth | 24 | Y |
| 48 | Y | |
| 72 | N | |
| MM | 24 | Y |
| 48 | Y | |
| 72 | N | |
| AMM | 24 | Y |
| 48 | Y | |
| 72 | Y |
MEM, minimal Eagle medium; FCS, fetal calf serum; YG, yeast-glucose; MM, minimal medium; AMM, Aspergillus MM. Shaking at 37°C was performed for all culture conditions.
Y, yes; N, no.
Metabolite extraction and analysis by high-pressure liquid chromatography/diode array detection/mass spectrometry (HPLC-DAD-MS).
Metabolic profiling was performed after culture on AMM and Czapek medium using the microextraction procedure of Smedsgaard (44), wherein plugs (0.6 cm2) were taken from petri dish cultures and extracted with 1 ml of methanol-dichloromethane-ethyl acetate (1:2:3 [vol/vol]). Extraction solutions were evaporated, and residues were redissolved in 400 μl of methanol and stored at −20°C until analysis. Ultra-HPLC-DAD analyses were performed on a Dionex RSLC Ultimate 3000 (Dionex, Sunnyvale, CA) equipped with a diode array detector. Separation was obtained on a Kinetex C18 column (150 by 2.1 mm, 2.6 μm; Phenomenex, Torrance, CA) maintained at 60°C using a linear gradient starting from 15% (vol/vol) CH3CN in water (containing 50 ppm of trifluoroacetic acid) increasing to 100% (vol/vol) CH3CN over 7 min at a flow rate of 0.8 ml/min. The injection volume was 1 μl. HPLC-DAD-MS analysis was performed on an LCT orthogonal acceleration time of flight (oaTOF) mass spectrometer (Micromass, Manchester, United Kingdom) as described by Nielsen et al. (30, 31). Chromatography was performed on a 5-cm, 3-μm Luna C18 (2) column (Phenomenex) using a water-acetonitrile gradient from 15% (vol/vol) CH3CN to 100% (vol/vol) CH3CN over 20 min, with both solvents containing 20 mM formic acid. Authentic standards for the following compounds were analyzed in sequence with extracts: fumitremorgin B and C, fumigaclavine A, B, and C, and fumiquinazoline F and G.
Phenotypic analysis of A. fumigatus NRP synthetase mutants.
A. fumigatus wild-type and mutant strains were grown on either AMM or MEA agar plates for 1 week at 37°C. Conidia were harvested aseptically in PBST (phosphate-buffered saline, 0.1% [vol/vol] Tween 80) and filtered through sterile Miracloth to remove mycelial matter. Conidia, at 102 or 104 per spot as indicated, were spotted onto a variety of test plates as described in Table 3. The plates were incubated at 37°C unless otherwise stated. The colony diameter was measured periodically, and statistical analysis was carried out using two-way analysis of variance.
Table 3.
Phenotypic plate assays
| Phenotypic test | Reagent(s) used | Concn tested | Phenotypic observations |
|---|---|---|---|
| Role of pesL in siderophore biosynthesis | Iron stresses (high, low, and none) | 10 μM, 1.5 mM, and 200 μM BPSb | NDa |
| Oxidative stress | Menadione | 20, 30, and 40 μM | The A. fumigatus ΔpesL mutant is more resistant to menadione at all of the concentrations tested |
| Diamide | 0.1, 0.2, 0.4, 1, and 2 mM | ND | |
| Hydrogen peroxide | 1, 2, and 3 mM | The ΔpesL and Δpes1 mutants display increased sensitivity to H2O2 | |
| Antifungal susceptibility | Voriconazole | 0.05-0.25, 0.5, 0.75, and 1.0 μg/ml | The ΔpesL and Δpes1 mutants display increased sensitivity to voriconazole |
| Amphotericin B | 0.125, 0.25, 0.5, and 1.0 μg/ml | The ΔpesL and Δpes1 mutants display increased sensitivity to amphotericin B | |
| Caspofungin | 0.2, 0.5, and 1.0 μg/ml | ND | |
| Heavy metal stress | Cobalt chloride | 0.1, 0.5, and 1 mM | ND |
| Cell wall stress | Caffeine | 2 and 5 mM | ND |
| Congo Red | 5, 10, and 15 μg/ml | ND | |
| Calcofluor White | 100 and 200 μg/ml | ND | |
| High temp (48°C) | NA | ND | |
| Membrane stress | SDS | 0.01 and 0.02% (wt/vol) | ND |
ND, no difference between wild-type and NRPS mutants.
BPS, bathophenanthroline disulfonate.
Galleria mellonella infection experiments.
G. mellonella virulence testing was carried out according to the method of Reeves et al. (37). Sixth-instar larvae of G. mellonella (Lepidoptera: Pyralidae, the greater wax moth) (Mealworm Company, Sheffield, England) were stored in wood shavings in the dark at 15°C prior to use. Only larvae weighing between 0.2 and 0.4 g were used during the present study. Larvae (n = 20 or 30) were inoculated into the hind pro-leg with a 20-μl inoculum volume of either 106 or 107 conidia as indicated for each experiment. Mortality, defined by the lack of movement in response to stimulation, and discoloration (melanization) rates were recorded at 24-h intervals for up to 96 h after injection. Kaplan-Meier survival curves were analyzed using the Mantel-Cox log-rank test for significance.
RESULTS
Disruption of pesL and pes1 in A. fumigatus.
To identify the NRPs produced by PesL and Pes1, A. fumigatus ΔpesL and A. fumigatus Δpes1 mutants were generated. The A. fumigatus ΔpesL mutant was generated in a ΔakuB mutant background, while the A. fumigatus Δpes1 mutant was generated in ΔakuB mutant and ATCC 46645 backgrounds. Potential mutants were initially identified by resistance to pyrithiamine after transformation. Southern blot analysis was used to identify pesL (negative) and ptrA (positive) colonies by probing for a 3,271-bp EcoRI restriction fragment in the ΔpesL deletion and a 6,641-bp fragment in the ΔakuB deletion (see Fig. S1 in the supplemental material). Southern analysis of 14 isolates confirmed the disruption of pesL. Similarly, pyrithiamine-resistant transformants (n = 36 in a ΔakuB background and n = 27 in a ATCC 46645 background) were screened by Southern blot for pes1 disruption by the presence of a 1,922-bp PvuII restriction fragment in the Δpes1 mutant and a 4,234-bp fragment in the wild-type strain (see Fig. S1 in the supplemental material), leading to identification of A. fumigatus Δpes1ΔakuB and Δpes146645 mutants, respectively. A representative isolate of each mutant was selected for further analysis.
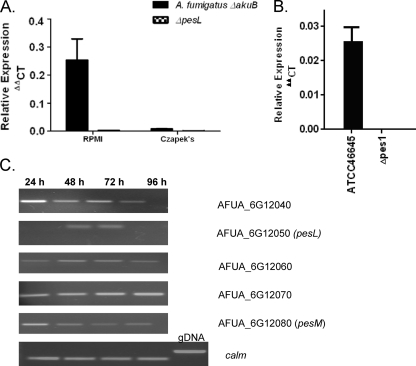
RT-PCR and real-time PCR confirmed that pesL transcripts were absent in the ΔpesL mutant but present in the A. fumigatus ΔakuB strain after 48 h growth in RPMI and Czapek broth (Fig. 1 and Table 2). Similarly, pes1 transcripts were absent in the Δpes1 mutant but present in A. fumigatus ATCC 46645 after 24 h growth in AMM (Fig. 1 and Table 2). Expression analysis (Fig. 1) also demonstrated differential expression of pesL compared to adjacent genes, including pesM, after between 24 and 96 h of culture. This finding strongly suggests that pesL plays a distinct role compared to adjacent genes and is not a component of this putative gene cluster.
Fig 1.
Real-time PCR analysis of NRPS gene expression and differential expression analysis of putative pesL cluster genes. (A) pesL expression was monitored after the growth of A. fumigatus ΔakuB and ΔpesL mutants in RPMI or Czapek broth for 48 h. Real-time PCR analysis was undertaken on cDNA samples taken from these cultures. The relative transcript abundances of both pesL and a housekeeping gene, calm, are shown. pesL expression is evident in A. fumigatus wild-type cultures in both RPMI and Czapek media but absent in the ΔpesL mutant under these conditions. pesL is expressed at a low level compared to the housekeeping gene calm (0.25 and <0.1 relative abundances in RPMI and Czapek media, respectively). (B) Real-time PCR analysis of pes1 expression in A. fumigatus ATCC 46645 and Δpes146645 strains after 24 h growth in AMM. Analysis confirmed the disruption of pes1 in ATCC 46645 since expression in the Δpes1 strain was completely absent. The data presented represent the means ± the standard errors of three replicates for each strain. (C) RT-PCR analysis of genes proposed to be in pesL cluster according to Nierman et al. (33) indicated that these five genes do not exhibit coregulated gene expression after growth of the A. fumigatus wild-type strain in yeast-glucose broth over a 96-h time period, whereas calm expression was observed at constant levels throughout the experiment. Genomic DNA (gDNA) contamination is excluded since the calm cDNA amplicon is evident at 314 bp and not at 617 bp, in accordance with the calm gDNA amplicon (6).
PesL and Pes1 are essential for fumigaclavine C biosynthesis.
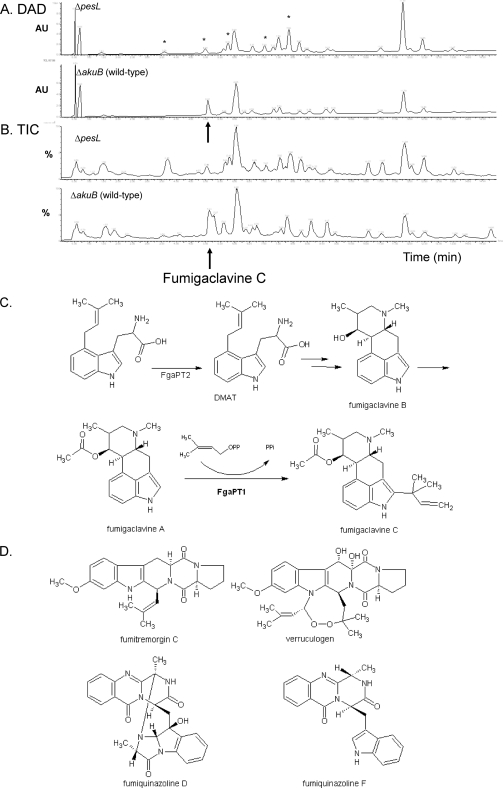
Comparative metabolite profiling by HPLC-DAD-MS analyses of metabolite extracts of A. fumigatus ΔakuB and ΔpesL strains after growth on Czapek agar for 6 days indicated significant differences in the appearance of several metabolites (Fig. 2). Based on a detailed analysis of secondary metabolites from A. fumigatus using UV spectroscopy and MS (24), it was clear that the biosynthesis of both fumigaclavines and fumitremorgins had been affected in the mutant strains (Fig. 2).
Fig 2.
PesL is essential for fumigaclavine C biosynthesis. Chromatograms from diode array detection (DAD) and a total ion chromatogram (TIC) of the A. fumigatus ΔakuB and ΔpesL conidial extracts from growth on Czapek solid medium. (A) DAD-based chromatogram of A. fumigatus ΔpesL (upper curve). Notice the absence of the peak for fumigaclavine C (Rt = 5.07 min). A DAD-based chromatogram of the ΔakuB (lower curve) fumigaclavine C (Rt = 5.07 min) is present. Note also the apparent increase in several other compounds in the A. fumigatus ΔpesL profile (Rt = 1.43, 3.58, 4.99, 5.76, 7.02, 7.86, and 8.22 min), as indicated by asterisks. (B) TIC of the ΔpesL mutant. Notice the absence of the peak for fumigaclavine C (Rt = 5.13 min). A TIC of ΔakuB confirms the presence of fumigaclavine C (Rt = 5.13 min) (lower TIC). (C) Final steps in the biosynthesis of fumigaclavine C (53). (D) Structures of fumitremorgin C, verruculogen, and fumiquinazolines D and F detected in the present study.
At first glance, the peak corresponding to fumigaclavine C (retention time [Rt] = 5.07 min, Fig. 2A, lower trace) present in the ΔakuB extract seemed to have been produced in lower amounts by the ΔpesL mutant (Rt = 4.99, Fig. 2B, upper trace). However, careful analysis of both UV and MS data for these two peaks (see Fig. S2 in the supplemental material), revealed that fumigaclavine C was completely absent in the ΔpesL mutant and indicated that the peak at 4.99 min could be tentatively assigned to 12,13-dihydroxy fumitremorgin C based on both UV spectroscopic and MS analysis.
The absence of fumigaclavine C in A. fumigatus ΔpesL was further confirmed by ion trace analysis searching for the specific mass of protonated fumigaclavine C (Fig. 2C), m/z 367 ([M+H]+), along with analysis of an authentic standard of fumigaclavine C (see the data in Fig. S3 in the supplemental material). Similarly, analysis of authentic standards of fumigaclavine A and B (data not shown) confirmed that the peak eluting at an Rt of 1.43 min corresponded to fumigaclavine A, which was present in relatively large amounts (m/z 299, [M+H]+) in both A. fumigatus ΔakuB and A. fumigatus ΔpesL extracts, whereas only traces of fumigaclavine B (m/z 257, [M+H]+) could be detected (see Fig. S3 in the supplemental material; see also Fig. 2C).
Two slightly later-eluting compounds (Rt = 5.29 min and 5.63 min) with UV spectra similar to that of fumigaclavine C (see Fig. S3H in the supplemental material), both with the same base peak (m/z 309), which likely represent [M+H]+ of two isomeric forms of 9-deacetoxy fumigaclavine C previously reported from A. fumigatus according to Antibase (23), were also absent in extracts of A. fumigatus ΔpesL. This further supports our observation of the absence of fumigaclavine C following pesL deletion.
Increase in fumitremorgin production in mutant strains.
MS analysis of the possible fumitremorgins mentioned above indeed confirmed the presence of fumitremorgin C (Rt = 8.23 min; Fig. 2D) based on a comparison with the authentic standard. In addition, TR-2 (Rt = 7.83 min) and verruculogen (Rt = 10.95 min) could be tentatively identified according to Larsen et al. (24; data not shown). Finally, the earlier-eluting compounds could be tentatively assigned as isomers of 12,13-dihydroxy fumitremorgin C (Rt = 4.99 and 5.76 min; Fig. 2) and a monohydroxy fumitremorgin C (Rt = 7.02 min) (see Fig. S4 in the supplemental material; also, data not shown). The most polar eluting fumitremorgin-like compound at 3.58 min could not be assigned to any known compound. With the exception of this compound, all other mentioned fumitremorgins were detected in both A. fumigatus ΔakuB and ΔpesL metabolite extracts, although in increased amounts in the ΔpesL extract, as seen in Fig. 2.
Independently, fumigaclavine C was detected in extracts of A. fumigatus ATCC 46645, and analysis of A. fumigatus Δpes1 strains revealed that this metabolite was absent in both the A. fumigatus Δpes146645 and the A. fumigatus Δpes1ΔakuB mutants (see Fig. S5 in the supplemental material). Identically to what was observed during the ΔpesL strain analysis, fumigaclavine A was confirmed to be present in all wild-type and mutant strains analyzed (data not shown).
PesL is not essential for fumiquinazoline production in A. fumigatus.
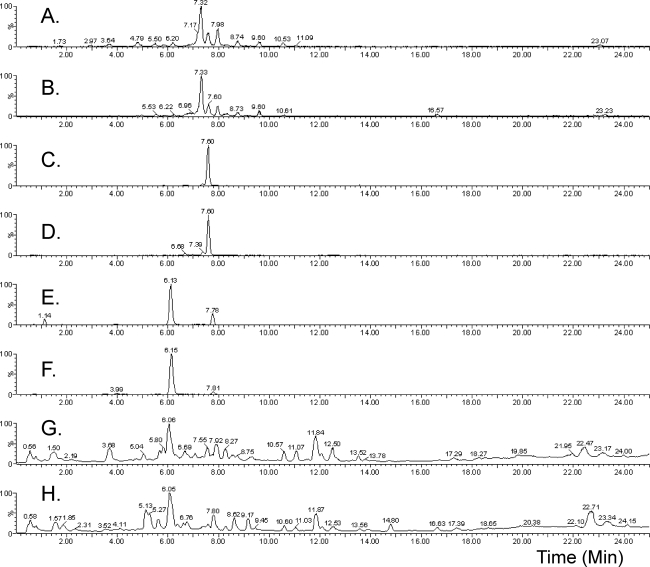
HPLC-DAD-MS analysis of A. fumigatus wild-type and ΔpesL extracts revealed the presence of fumiquinazolines A to F in both strains (Fig. 3). These nonribosomal peptide compounds could easily be detected using positive electrospray ionization as their protonated species [M+H]+ (Fig. 3), along with their characteristic UV spectra (see Fig. S6 in the supplemental material) (24). Table S2 in the supplemental material shows a list of the compounds in the fumiquinazoline family, along with their molecular weights and molecular formulas.
Fig 3.
PesL is not essential for fumiquinazoline production in A. fumigatus. Ion traces illustrating the presence of fumiquinazolines in both A. fumigatus ΔakuB and ΔpesL mutants are shown. (A) Ion trace (m/z 446.1 Da, [M+H]+) illustrating the presence of fumiquinazolines A and B in the A. fumigatus ΔpesL mutant. (B) Ion trace (m/z 446.1 Da, [M+H]+) illustrating the presence of fumiquinazolines A and B in the A. fumigatus ΔakuB mutant. (C) Ion trace (m/z 444.1 Da, [M+H]+) illustrating the presence of fumiquinazolines C and D in the A. fumigatus ΔpesL mutant. (D) Ion trace (m/z 444.1 Da, [M+H]+) illustrating the presence of fumiquinazolines C and D in the A. fumigatus ΔakuB mutant. (E) Ion trace (m/z 359.1 Da, [M+H]+) illustrating the presence of fumiquinazolines E and F in the A. fumigatus ΔpesL mutant. (F) Ion trace (m/z 359.1 Da, [M+H]+) illustrating the presence of fumiquinazolines E and F in the A. fumigatus ΔakuB mutant. (G) TIC of conidial extracts of the Aspergillus fumigatus ΔpesL mutant after growth on Czapek medium. (H) TIC of metabolite extracts of the Aspergillus fumigatus ΔakuB mutant after growth on Czapek medium.
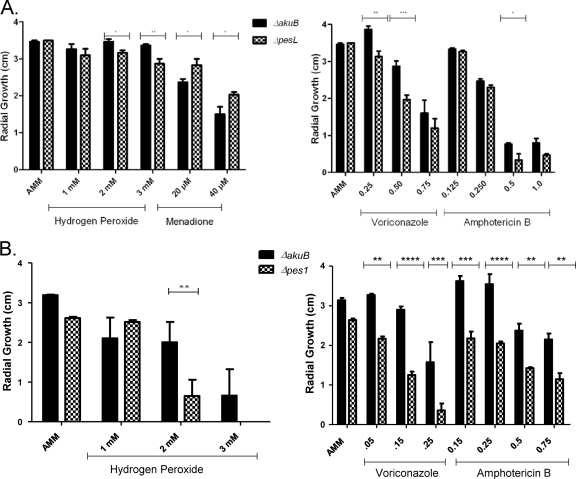
Phenotypic analysis of A. fumigatus ΔpesL and Δpes1 mutants reveals hydrogen peroxide and antifungal sensitivity.
Exposure of wild-type and NRP mutant strains to a range of agents (Table 3) indicated several mutant phenotypes. The A. fumigatus ΔpesL mutant was more sensitive to hydrogen peroxide than was the wild type (>2 mM H2O2, P < 0.01) but more resistant to menadione than was the wild type (20 μM, P < 0.05; 40 μM, P < 0.01) (Fig. 4). The A. fumigatus ΔpesL and ΔakuB mutants grew at identical rates upon exposure to diamide; the growth rates were compared in order to determine whether altered glutathione levels were evident (data not shown). Antifungal susceptibility testing indicated that the ΔpesL strain exhibited increased susceptibility to the azole voriconazole. The addition of voriconazole (0.25 to 0.75 μg/ml) led to a significantly reduced growth phenotype of the ΔpesL strain compared to the ΔakuB strain at all of the concentrations tested (Fig. 4). This increased sensitivity of the ΔpesL strain to voriconazole was most significant at 0.5 μg of voriconazole/ml at 72 h growth (P < 0.001) (Fig. 4). Similarly, the ΔpesL strain exhibited moderately increased sensitivity to the polyene antifungal amphotericin B (0.5 μg/ml; P < 0.05) (Fig. 4). A. fumigatus ΔpesL grew at a rate identical to that of the ΔakuB strain upon exposure to caspofungin (inhibitor of cell wall biosynthesis).
Fig 4.
Phenotypic analysis of NRP synthetase mutants. (A) A significant reduction in growth of the A. fumigatus ΔpesL mutant is observed compared to the ΔakuB mutant upon exposure to H2O2 (2 mM, P < 0.05; 3 mM, P < 0.01); however, the A. fumigatus ΔpesL mutant exhibited increased growth compared to the ΔakuB mutant upon exposure to menadione (20 μM, P < 0.05; 40 μM P < 0.05). Significant growth inhibition of the A. fumigatus ΔpesL mutant was also observed in the presence of voriconazole (0.25 μg/ml, P < 0.01; 0.5 μg/ml, P < 0.001) and amphotericin B (0.5 μg/ml, P < 0.05) compared to the A. fumigatus ΔakuB mutant. (B) Significant growth inhibition of the A. fumigatus Δpes1ΔakuB mutant compared to the ΔakuB mutant was observed upon exposure to H2O2 (2 mM, P < 0.01), voriconazole (0.015 μg/ml, P < 0.001), and amphotericin B (0.15 to 0.75 μg/ml, P ≤ 0.01) compared to the A. fumigatus ΔakuB mutant.
Strikingly, similar phenotypes were observed for the A. fumigatus ΔpesL and Δpes1 strains. Upon exposure to H2O2, A. fumigatus Δpes1ΔakuB displayed reduced growth compared to the ΔakuB strain (2 mM, P < 0.01). A. fumigatus Δpes1ΔakuB failed to grow upon exposure to 3 mM H2O2. The A. fumigatus Δpes1ΔakuB strain exhibited a significant decrease in radial growth compared to the ΔakuB strain upon increasing concentrations of voriconazole (0.05-0.25 μg/ml), with the most significant reduction in growth observed at 72 h (0.15 μg/ml; P < 0.0001) (Fig. 4). Furthermore, A. fumigatus Δpes1ΔakuB exhibited increased sensitivity to amphotericin B, with a reduction in radial growth compared to the ΔakuB strain at increasing concentrations (0.25 μg/ml; P < 0.0001).
pesL contributes to the virulence of A. fumigatus.
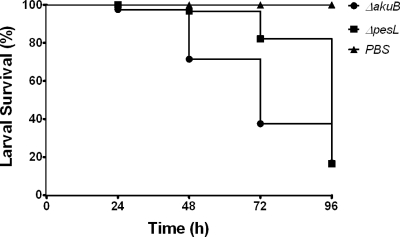
The A. fumigatus ΔpesL mutant exhibited attenuated virulence in the G. mellonella infection model, wherein larvae infected with the ΔpesL strain displayed increased survival compared to larvae infected with the ΔakuB strain (P < 0.001). The conidial inoculum for A. fumigatus ΔpesL testing was 107 conidia/larva. Larval survival (%) is shown in Fig. 5. At 24 h after infection, 97% of the larvae infected with the ΔakuB strain remained alive, whereas a 100% survival rate was observed for those infected with the ΔpesL strain. The reduction in virulence associated with the ΔpesL mutation was more pronounced at 48 h and 72 h after initial infection. At 48 h postinfection, 96% of the larvae infected with the ΔpesL mutant remained alive, in contrast to only 71% of those infected with the ΔakuB mutant. By 72 h, 82% of the larvae infected with the ΔpesL strain survived versus 37% infected with the ΔakuB mutant. The overall survival proportions between larvae infected with the ΔakuB strain or the ΔpesL strain is highly significant (P < 0.001), indicating that the loss of pesL and its encoded peptide leads to reduced virulence in the G. mellonella infection model. The virulence of A. fumigatus Δpes146645 was assessed by using a conidial inoculum of 106 and, under these conditions, no significant difference in survival (25% versus 30%) was observed between the A. fumigatus ATCC 46645 and Δpes146645 strains.
Fig 5.
Disruption of A. fumigatus pesL leads to attenuated virulence in the Galleria mellonella infection model. (A) Comparative virulence of A. fumigatus wild-type and ΔpesL strains in the G. mellonella infection model. The A. fumigatus ΔpesL strain exhibited significantly attenuated virulence in this model (P < 0.001), as indicated by increased larval survival (%) associated with ΔpesL mutant infection compared to the wild type over 96 h. Phosphate-buffered saline (PBS) was used as an injection control, and all larvae in this group remained viable for the entire experiment.
DISCUSSION
We have shown previously that Pes1 plays a role in resistance to H2O2-mediated oxidative stress and in the virulence of A. fumigatus (38). PesL has been the subject of recent work and, along with another NRP synthetase, PesM, has been implicated in fumiquinazoline biosynthesis (1, 2). The A. fumigatus pesL and pes1 genes were disrupted as described previously (32), and a corresponding abolition of gene expression was subsequently confirmed by RT-PCR and real-time PCR. Cramer et al. reported that pesL expression was observed after 48 h of growth in RPMI and Czapek broth (13). In the work presented here, pesL expression was also found to be more abundant after growth in RPMI, whereas pesL expression was negligible after growth in Czapek broth. Cramer et al. (13) also reported that pes1 (AFUA_1G10380) was minimally expressed in Sabouraud medium and not expressed in any of the other conditions tested, whereas, in the findings presented here, pes1 expression was detected in AMM and MM, as well as Sabouraud medium. It should be noted, however, that the A. fumigatus strains used here differed from that of Cramer et al. (13), who used the reference strain A. fumigatus Af293. Furthermore, the housekeeping genes used varied in these studies (e.g., calmodulin [6] versus the actin used by Cramer et al. [13]).
Initial comparisons of metabolites derived from liquid cultures of the A. fumigatus wild type versus the ΔpesL and Δpes1 mutants revealed no differences in metabolite profiles (data not shown). It was then considered that the NRP synthetases of interest might synthesize peptides associated with conidia rather than vegetative growth. Interestingly, disruption of an NRP synthetase, MaNPS1, in the insect pathogenic fungus Metarhizium anisopliae revealed that conidium-associated serinocyclins are nonribosomally synthesized (22, 29). Trichothecenes produced by Stachybotrys chartarum are also spore associated (45). Furthermore, the ergot alkaloids (EA) of A. fumigatus (fumigaclavines A, B, and C and festuclavine) have a confirmed association with conidia (10). Fumigaclavine C, the end product of the complex EA biosynthetic pathway (Fig. 2C), was present in extracts of the A. fumigatus ΔakuB strain and completely absent in the A. fumigatus ΔpesL strain. A complete loss of fumigaclavine C in the A. fumigatus ΔpesL strain was observed wherein conidia, in part, comprised the specimens under examination, in agreement with the known EA association with A. fumigatus conidia (10). Furthermore, fumigaclavine C was also completely absent in metabolite extracts of A. fumigatus Δpes1, suggesting redundancy among these NRP synthetases. All other known A. fumigatus EA, and in particular fumigaclavine A, were detected in extracts of the A. fumigatus ΔpesL and Δpes1 strains, indicating that the biosynthetic role of PesL and Pes1 likely occurs at the final step of the EA pathway, perhaps by aiding FgaPT1 in the reverse prenylation of fumigaclavine A in the C2′ position of the indole ring of fumigaclavine A to yield fumigaclavine C (53).
Comparative phenotypic analyses revealed that both the A. fumigatus ΔpesL and the A. fumigatus Δpes1ΔakuB strains exhibited increased sensitivity to voriconazole and amphotericin B compared to wild-type strains. In contrast, sensitivity testing with caspofungin revealed no difference between the A. fumigatus ΔpesL and Δpes1 strains and their respective wild types. Furthermore, A. fumigatus ΔpesL and A. fumigatus Δpes1ΔakuB exhibited increased sensitivity to H2O2, implying roles for PesL and Pes1 in protection against the effects of voriconazole-, amphotericin B-, and H2O2-mediated oxidative stress. A. fumigatus ΔpesL exhibited increased growth compared to the wild type upon exposure to menadione in the present study, whereas all strains grew at equal rates on diamide. A range of oxidizing agents was chosen for analysis since no single agent can fully represent the conditions of oxidative stress (49, 58). The increased resistance of the A. fumigatus ΔpesL strain to menadione may be due to an oxidant defense response that was elevated in the ΔpesL strain as a protective mechanism. The transcriptional responses to various oxidizing agents, including the ones used here, was shown to differ substantially in S. cerevisiae, with respiratory gene expression influenced by hydrogen peroxide, whereas menadione influenced the NADPH-producing pentose phosphate pathway (50). Furthermore, a genome-wide comparison of gene expression profiles upon exposure to menadione, hydrogen peroxide, and diamide in A. nidulans revealed that separate response gene groups existed for the different agents (36). Importantly, the similar phenotypes observed for the A. fumigatus ΔpesL and A. fumigatus Δpes1ΔakuB strains strongly suggest redundancy among these NRP synthetases; however, it is clear that both are simultaneously required for fumigaclavine C biosynthesis. Increased production of several fumitremorgins, such as TR-2, fumitremorgin C, and verruculogen, could be seen in the extracts of the ΔpesL strain (and, to a minor extent, in the Δpes1 strain), suggesting that the increased pool of isoprene available due to less prenylation of fumigaclavine A may instead be incorporated into the fumitremorgins.
The observed phenotypic and comparative metabolite data suggest a link between oxidative stress resistance and fumigaclavine C. Interestingly, the role of NRPS in the protection of fungal species against oxidative stress has been previously reported. NPS6, which is responsible for siderophore biosynthesis in the plant pathogen Cochliobolus heterostrophus, was found to be involved in both virulence and resistance to oxidative stress (26). These findings together are in agreement with previously established links between fungal secondary metabolism and oxidative stress (39). Furthermore, since both the A. fumigatus ΔpesL and the A. fumigatus Δpes1 strains are more sensitive to voriconazole and amphotericin B, it appears that fumigaclavine C may also play a role in resistance to antifungals. The specific mechanism underlying the apparent antifungal resistance was not investigated further in the present study, although emerging hypotheses suggest that secondary metabolite production may represent a component of the oxidative stress response in fungi (39).
The A. fumigatus fumigaclavine biosynthetic cluster has been extensively studied (20, 27, 48, 53, 56); however, a number of undefined reactions persist, as do cluster-encoded genes with unknown functions. The A. fumigatus EA cluster is not reported to contain NRP synthetase genes, in contrast to other EA producing fungi (11, 52). No orthologs for the C. purpurea NRP synthetase encoding genes cpps1 or cpps2 have been found in the vicinity of fgaPT2 in the A. fumigatus EA gene cluster, which is thought to be consistent with the absence of a peptide moiety in fumigaclavines (52). Heterologously expressed A. fumigatus FgaPT1 was shown to have strict specific substrate specificity and, in vitro, to convert fumigaclavine A to fumigaclavine C (53). However, in vivo, the direct interaction of substrate and prenyl transferase may not occur as easily and may require the tethering of the substrate to PesL and Pes1 for the reaction to occur. Since PesL is a monomodular NRP synthetase, it may have a common origin with the monomodular NRP synthetase also found in C. purpurea (lpsB), and since Pes1 is a multimodular nonlinear NRP synthetase and the biosynthesis of ergotamine in C. purpurea involves a trimodular NRP synthetase (lpsA), it is possible that pes1 shares a common ancestor with the NRP synthetase gene, lpsA. The requirement for PesL and Pes1 in fumigaclavine C biosynthesis could suggest that these genes were once in a cluster with the other EA genes and have been translocated to their current location; indeed, transposable elements have been reported to be associated with other biosynthetic gene clusters in Epichloe spp. (15).
Secondary metabolite gene cluster rearrangements mediated by transposable elements might not be restricted to the EA clusters. This could allow for NRP synthetases to be used by more than one biosynthetic pathway, thereby increasing the diversity of secondary metabolites that can be produced by an organism. This hypothesis could explain how such a large repertoire of secondary metabolites can arise from 14 NRP synthetases in A. fumigatus (13). The current understanding of NRP synthetases and secondary metabolite gene clusters might need to be reconsidered in light of the findings and ideas presented here, and the current paradigm may not be as straightforward as “one NRP synthetase, one peptide” as has previously been found for NRPS in other pathways (e.g., gliotoxin biosynthesis) (3, 12). Moreover, the notion of cluster cross talk is beginning to emerge with the confirmation of interactions between two separate NRP synthetases involved in siderophore biosynthesis in a bacterial species (25). More recently, cross talk was identified between two SM clusters on different chromosomes in A. nidulans (4). Indeed, both cluster-encoded and non-cluster-encoded enzymes were required for the biosynthesis of the laspartomycin peptide antibiotics in Streptomyces viridochromogenes (55).
Importantly, we showed here that gene knockout and in vitro biochemical analyses are the most appropriate means to unambiguously show an essential biosynthetic function for any given gene. PesL has recently been implicated in fumiquinazoline biosynthesis (1, 2), and the observed role for PesL in fumigaclavine C biosynthesis reported here suggests redundant roles for PesL. It remains to be seen whether this is a feature of the remainder of the uncharacterized NRP synthetases within A. fumigatus and other fungi. Support for redundancy among NRP synthetases may come from observations that NRP synthetases show less strict substrate selection and incorporation than other adenylating enzymes (8, 46).
We also observed the presence of a catalase-encoding gene (AFUA_2G18030) in the EA cluster vicinity (http://www.cadre-genomes.org.uk), which has recently been included in the A. fumigatus cluster and was shown to be necessary for EA biosynthesis in A. fumigatus (17). A putative catalase gene (cpcat2) has also been identified in the EA cluster of C. purpurea, although no function has yet been assigned (9). The appearance of catalases in the EA clusters may also suggest a link between the production of EA and oxidative stress, since catalases are known antioxidant enzymes (7). This inclusion of a catalase in the A. fumigatus EA cluster suggests that the core EA cluster in general is still undergoing refinement.
Initially, pesL was proposed to be part of a putative five-gene SM cluster (33) and, more recently, part of an eight-gene cluster proposed to be responsible for the biosynthesis of the fumiquinazoline family of secondary metabolites in A. fumigatus (1). However, coregulated expression of the cluster genes, with or without simultaneous secondary metabolite production, a feature that is a hallmark of SM biosynthetic gene clusters (13, 40, 47), has not been demonstrated. All genes in the proposed pesL cluster (AFUA_6G12040 to AFUA_6G12080), according to Nierman et al. (33), were found to be expressed here in YG medium over a 96-h time period. This observation was important since secondary metabolite gene clusters have been found to be transcriptionally silent under standard laboratory conditions (42). However, the proposed cluster genes did not all exhibit the same pattern of expression, suggesting that they are not coregulated in the production of a particular secondary metabolite. Furthermore, gene clusters encoding secondary metabolites are usually coregulated with the production of the specific metabolite(s) (4, 13), and this has also been observed for the C. purpurea EA biosynthetic cluster (9, 51). Such a study had not been reported for the genes involved in either EA or fumiquinazoline biosynthesis in A. fumigatus. The presence of several fumiquinazolines in the A. fumigatus ΔpesL strain indicates that fumiquinazoline biosynthesis may be more complex than is currently thought and also actually suggests an alternative route for fumiquinazoline A production in A. fumigatus ΔpesL.
Despite advances in the field of secondary metabolite biosynthesis, there still remains a large deficit relating NRP synthetases to peptide products in the important human pathogen A. fumigatus, an observation previously noted by others (13, 47). As more NRP synthetases are functionally characterized, one can predict that the potential and complexity of these remarkable enzymes will become even more apparent.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a Health Research Board project grant (RP/2006/043) and an Irish Research Council for Science, Engineering, and Technology (IRCSET) Embark Ph.D. fellowship to K.A.O. The quantitative PCR facilities were funded by Science Foundation Ireland (SFI/07/RFP/GEN/F571/ECO7). T.O.L. was funded by the Danish Research Agency for Technology and Production (grant 09-064967).
Footnotes
Published ahead of print 17 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ames BD, Liu X, Walsh CT. 2010. Enzymatic processing of fumiquinazoline F: a tandem oxidative-acylation strategy for the generation of multicyclic scaffolds in fungal indole alkaloid biosynthesis. Biochemistry 49:8564–8576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ames BD, Walsh CT. 2010. Anthranilate-activating modules from fungal nonribosomal peptide assembly lines. Biochemistry 49:3351–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balibar CJ, Walsh CT. 2006. GliP, a multimodular nonribosomal peptide synthetase in Aspergillus fumigatus, makes the diketopiperazine scaffold of gliotoxin. Biochemistry 45:15029–15038 [DOI] [PubMed] [Google Scholar]
- 4. Bergmann S, et al. 2010. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl. Environ. Microbiol. 76:8143–8149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bok JW, et al. 2005. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell 4:1574–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burns C, et al. 2005. Identification, cloning, and functional expression of three glutathione transferase genes from Aspergillus fumigatus. Fungal Genet. Biol. 42:319–327 [DOI] [PubMed] [Google Scholar]
- 7. Chauhan N, Latge JP, Calderone R. 2006. Signalling and oxidant adaptation in Candida albicans and Aspergillus fumigatus. Nat. Rev. Microbiol. 4:435–444 [DOI] [PubMed] [Google Scholar]
- 8. Christiansen G, Philmus B, Hemscheidt T, Kurmayer R. 2011. Genetic variation of adenylation domains of the anabaenopeptin synthesis operon and evolution of substrate promiscuity. J. Bacteriol. 193:3822–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Correia T, Grammel N, Ortel I, Keller U, Tudzynski P. 2003. Molecular cloning and analysis of the ergopeptine assembly system in the ergot fungus Claviceps purpurea. Chem. Biol. 10:1281–1292 [DOI] [PubMed] [Google Scholar]
- 10. Coyle CM, Kenaley SC, Rittenour WR, Panaccione DG. 2007. Association of ergot alkaloids with conidiation in Aspergillus fumigatus. Mycologia 99:804–811 [DOI] [PubMed] [Google Scholar]
- 11. Coyle CM, Panaccione DG. 2005. An ergot alkaloid biosynthesis gene and clustered hypothetical genes from Aspergillus fumigatus. Appl. Environ. Microbiol. 71:3112–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cramer RA, et al. 2006. Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot. Cell 5:972–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cramer RA, et al. 2006. Phylogenomic analysis of non-ribosomal peptide synthetases in the genus Aspergillus. Gene 383:24–32 [DOI] [PubMed] [Google Scholar]
- 14. Ferreira MED, et al. 2006. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5:207–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fleetwood DJ, Scott B, Lane GA, Tanaka A, Johnson RD. 2007. A complex ergovaline gene cluster in Epichloe endophytes of grasses. Appl. Environ. Microbiol. 73:2571–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flieger M, Wurst M, Shelby R. 1997. Ergot alkaloids: Sources, structures, and analytical methods. Folia Microbiol. 42:3–29 [DOI] [PubMed] [Google Scholar]
- 17. Goetz KE, Coyle CM, Cheng JZ, O'Connor SE, Panaccione DG. 2011. Ergot cluster-encoded catalase is required for synthesis of chanoclavine-I in Aspergillus fumigatus. Curr. Genet. 57:201–211 [DOI] [PubMed] [Google Scholar]
- 18. Haarmann T, et al. 2005. The ergot alkaloid gene cluster in Claviceps purpurea: extension of the cluster sequence and intra species evolution. Phytochemistry 66:1312–1320 [DOI] [PubMed] [Google Scholar]
- 19. Hearn VM, Mackenzie DWR. 1980. Mycelial antigens from 2 strains of Aspergillus fumigatus: an analysis by two-dimensional immunoelectrophoresis. Mykosen 23:549–562 [PubMed] [Google Scholar]
- 20. Kato N, et al. 2009. Identification of cytochrome P450s required for fumitremorgin biosynthesis in Aspergillus fumigatus. Chembiochem 10:920–928 [DOI] [PubMed] [Google Scholar]
- 21. Krappmann S, et al. 2006. The Aspergillus nidulans F-box protein GrrA links SCF activity to meiosis. Mol. Microbiol. 61:76–88 [DOI] [PubMed] [Google Scholar]
- 22. Krasnoff SB, et al. 2007. Serinocyclins A and B, cyclic heptapeptides from Metarhizium anisopliae. J. Nat. Prod. 70:1919–1924 [DOI] [PubMed] [Google Scholar]
- 23. Laatsch H. 2011. AntiBase 2011: the natural compound identifier. Wiley-VCH, Weinheim, Germany [Google Scholar]
- 24. Larsen TO, et al. 2007. Production of mycotoxins by Aspergillus lentulus and other medically important and closely related species in section Fumigati. Med. Mycol. 45:225–232 [DOI] [PubMed] [Google Scholar]
- 25. Lazos O, et al. 2010. Biosynthesis of the putative siderophore erythrochelin requires unprecedented crosstalk between separate nonribosomal peptide gene clusters. Chem. Biol. 17:160–173 [DOI] [PubMed] [Google Scholar]
- 26. Lee BN, et al. 2005. Functional analysis of all nonribosomal peptide synthetases in Cochliobolus heterostrophus reveals a factor, NPS6, involved in virulence and resistance to oxidative stress. Eukaryot. Cell 4:545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maiya S, Grundmann A, Li SM, Turner G. 2009. Improved tryprostatin B production by heterologous gene expression in Aspergillus nidulans. Fungal Genet. Biol. 46:436–440 [DOI] [PubMed] [Google Scholar]
- 28. Maiya S, Grundmann A, Li SM, Turner G. 2006. The fumitremorgin gene cluster of Aspergillus fumigatus: identification of a gene encoding brevianamide F synthetase. Chembiochem 7:1062–1069 [DOI] [PubMed] [Google Scholar]
- 29. Moon YS, et al. 2008. Agrobacterium-mediated disruption of a nonribosomal peptide synthetase gene in the invertebrate pathogen Metarhizium anisopliae reveals a peptide spore factor. Appl. Environ. Microbiol. 74:4366–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nielsen KF, Månsson M, Rank J, Frisvad JC, Larsen TO. 2011. Dereplication of microbial natural products by LC-DAD-TOFMS: experiences gained from an in-house database of 719 mycotoxins and fungal metabolites. J. Nat. Prod. 74:2338–2348 [DOI] [PubMed] [Google Scholar]
- 31. Nielsen KF, Smedsgaard J. 2003. Fungal metabolite screening: database of 474 mycotoxins and fungal metabolites for dereplication by standardized liquid chromatography-UV-mass spectrometry methodology. J. Chromatogr. A 1002:111–136 [DOI] [PubMed] [Google Scholar]
- 32. Nielsen ML, Albertsen L, Lettier G, Nielsen JB, Mortensen UH. 2006. Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet. Biol. 43:54–64 [DOI] [PubMed] [Google Scholar]
- 33. Nierman WC, et al. 2006. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156 [DOI] [PubMed] [Google Scholar]
- 34. Panaccione DG, et al. 2001. Elimination of ergovaline from a grass-Neotyphodium endophyte symbiosis by genetic modification of the endophyte. Proc. Natl. Acad. Sci. U. S. A. 98:12820–12825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Perrin RM, et al. 2007. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 3:508–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pocsi I, et al. 2005. Comparison of gene expression signatures of diamide, H2O2 and menadione exposed Aspergillus nidulans cultures: linking genome-wide transcriptional changes to cellular physiology. BMC Genomics 6:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reeves EP, Messina CGM, Doyle S, Kavanagh K. 2004. Correlation between gliotoxin production and virulence of Aspergillus fumigatus in Galleria mellonella. Mycopathologia 158:73–79 [DOI] [PubMed] [Google Scholar]
- 38. Reeves EP, et al. 2006. A nonribosomal peptide synthetase (Pes1) confers protection against oxidative stress in Aspergillus fumigatus. FEBS J. 273:3038–3053 [DOI] [PubMed] [Google Scholar]
- 39. Reverberi M, Ricelli A, Zjalic S, Fabbri AA, Fanelli C. 2010. Natural functions of mycotoxins and control of their biosynthesis in fungi. Appl. Microbiol. Biotechnol. 87:899–911 [DOI] [PubMed] [Google Scholar]
- 40. Schrettl M, et al. 2010. Self-protection against gliotoxin: a component of the gliotoxin biosynthetic cluster, GliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathog. 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schrettl M, et al. 2008. SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 70:27–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schroeckh V, et al. 2009. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. U. S. A. 106:14558–14563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sheppard DC, et al. 2005. The Aspergillus fumigatus StuA protein governs the up-regulation of a discrete transcriptional program during the acquisition of developmental competence. Mol. Cell. Biol. 16:5866–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smedsgaard J. 1997. Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. J. Chromatogr. A 760:264–270 [DOI] [PubMed] [Google Scholar]
- 45. Sorenson WG, Frazer DG, Jarvis BB, Simpson J, Robinson VA. 1987. Trichothecene mycotoxins in aerosolized conidia of Stachybotrys atra. Appl. Environ. Microbiol. 53:1370–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stachelhaus T, Mootz HD, Marahiel MA. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493–505 [DOI] [PubMed] [Google Scholar]
- 47. Stack D, Neville C, Doyle S. 2007. Nonribosomal peptide synthesis in Aspergillus fumigatus and other fungi. Microbiology 153:1297–1306 [DOI] [PubMed] [Google Scholar]
- 48. Steffan N, Grundmann A, Yin WB, Kremer A, Li SM. 2009. Indole prenyltransferases from fungi: a new enzyme group with high potential for the production of prenylated indole derivatives. Curr. Med. Chem. 16:218–231 [DOI] [PubMed] [Google Scholar]
- 49. Temple MD, Perrone GG, Dawes IW. 2005. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 15:319–326 [DOI] [PubMed] [Google Scholar]
- 50. Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW. 2004. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc. Natl. Acad. Sci. U. S. A. 101:6564–6569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tudzynski P, et al. 1999. Evidence for an ergot alkaloid gene cluster in Claviceps purpurea. Mol. Gen. Genet. 261:133–141 [DOI] [PubMed] [Google Scholar]
- 52. Unsold IA, Li SM. 2005. Overproduction, purification and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiology 151:1499–1505 [DOI] [PubMed] [Google Scholar]
- 53. Unsold IA, Li SM. 2006. Reverse prenyltransferase in the biosynthesis of fumigaclavine C in Aspergillus fumigatus: gene expression, purification, and characterization of fumigaclavine C synthase FGAPT1. Chembiochem 7:158–164 [DOI] [PubMed] [Google Scholar]
- 54. Wallwey C, Li SM. 2011. Ergot alkaloids: structure diversity, biosynthetic gene clusters and functional proof of biosynthetic genes. Nat. Prod. Rep. 28:496–510 [DOI] [PubMed] [Google Scholar]
- 55. Wang Y, Chen Y, Shen Q, Yin X. 2011. Molecular cloning and identification of the laspartomycin biosynthetic gene cluster from Streptomyces viridochromogenes. Gene 483:11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Willingale J, Perera KP, Mantle PG. 1983. An intermediary role for the tremorgenic mycotoxin TR-2 in the biosynthesis of verruculogen. Biochem. J. 214:991–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yin WB, Grundmann A, Cheng J, Li SM. 2009. Acetylaszonalenin biosynthesis in Neosartorya fischeri identification of the biosynthetic gene cluster by genomic mining and functional proof of the genes by biochemical investigation. J. Biol. Chem. 284:100–109 [DOI] [PubMed] [Google Scholar]
- 58. Zhao W, et al. 2006. Deletion of the regulatory subunit of protein kinase a in Aspergillus fumigatus alters morphology, sensitivity to oxidative damage, and virulence. Infect. Immun. 74:4865–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.