Abstract
We isolated and characterized a novel endophyte from hybrid poplar. This unique endophyte, identified as Enterobacter sp. strain PDN3, showed high tolerance to trichloroethylene (TCE). Without the addition of inducers, such as toluene or phenol, PDN3 rapidly reduced TCE levels in medium from 72.4 μM to 30.1 μM in 24 h with a concurrent release of 127 μM chloride ion, and nearly 80% of TCE (55.3 μM) was dechlorinated by PDN3 in 5 days with 166 μM chloride ion production, suggesting TCE degradation.
TEXT
Trichloroethylene (TCE) is a common environmental contaminant (16). High levels of TCE have the potential to cause liver damage and malfunctions in the central nervous system, and it is considered a likely human carcinogen (13). More than 54% of Superfund sites in the United States are contaminated with TCE (19), and hence, its removal has become a priority for many contaminated sites all over the industrialized world. With the discovery of a number of soil microorganisms that are capable of degrading organic pollutants, microbial bioremediation is increasingly being considered a reasonable and effective method for removing environmental contaminants (11, 17).
Several classes of aerobic microorganisms use a process called cometabolic oxidation to degrade chlorinated solvents. Since there is no gain of energy from this process, field application of cometabolism for TCE degradation can be expensive. Another limitation is that substrates such as phenol or toluene are toxic, and introducing them into the soil can result in secondary contamination problems. Researchers have tried to engineer microbes to degrade TCE constitutively without addition of cosubstrates such as toluene and phenol (21, 23, 30). However, engineered bacteria may have limitations to field applications. More recently, microbes such as Rhodococcus sp. strain L4 (31, 32) and Bacillus sp. strain 2479 (5) were found to degrade TCE in the absence of toxic substrates.
Many recent studies have shown that endophytes have a natural capacity for xenobiotic degradation (27) in addition to providing other benefits to the plants, such as nitrogen fixation, phosphate solubilization, and stress tolerance (26). The addition of nonnative or genetically modified endophytes to assist the phytoremediation of contaminated soil and ground water has been explored (2, 6, 35–38). In this study, we report that a novel isolate, termed Enterobacter sp. strain PDN3, which was isolated from a poplar tree growing on a TCE-contaminated site, showed high tolerance to TCE and degraded TCE to chloride without the addition of any commercial inducing phenolic compounds. To our knowledge, this is the first report on a natural TCE-degrading bacterium isolated from a poplar tree.
Endophyte isolation.
Dormant cuttings of several varieties of young poplars were collected from TCE-contaminated sites. Surface-sterile cuttings (7) were suspended in sterile distilled water and incubated at 30°C for 24 h with shaking. To enrich for potential TCE degraders, 1 milliliter of this sample was added into 25 ml of synthetic medium containing 5.5 mM TCE (5). Cultures that grew well at this high level of TCE were screened for degradation activity by the Fujiwara test (20, 25). One culture, strain PDN3, from hybrid poplar DN177 (Populus deltoides × Populus nigra) demonstrated TCE degradation (data not shown).
Growth measurements.
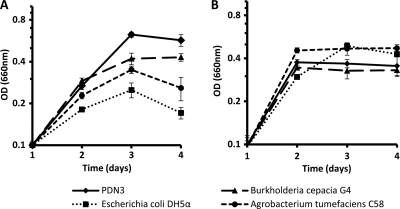
Cultures were grown in 20 ml of M9 minimal medium with 0.2% peptone instead of glucose and with 5.5 mM TCE in 125-ml flasks and shaken for 3 days at 200 rpm at 30°C. Growth was monitored every 24 h using a spectrophotometer. Burkholderia cepacia G4 (18) was included as a positive control, and Agrobacterium tumefaciens C58 (12) and Escherichia coli DH5α (10) were used as negative controls. As seen in Fig. 1A, PDN3 grew better than the other cultures in the presence of 5.5 mM TCE. Escherichia coli DH5α and Agrobacterium tumefaciens strain C58 did grow, but they were slightly inhibited in the presence of TCE (Fig. 1A), and all of the cultures grew well in the absence of TCE, reaching saturation within 24 h in M9 minimal medium with 0.2% peptone instead of glucose (Fig. 1B). Since PDN3 grew to a higher optical density in medium containing TCE than in the same medium without TCE, these results suggest that PDN3 used TCE as an additional carbon source.
Fig 1.
Growth curves of PDN3 in the presence of TCE (containing 5.5 mM TCE) (A) and absence of TCE (B) in M9 minimal medium with 0.2% peptone instead of glucose. Burkholderia cepacia G4 was included as a positive control, and Agrobacterium tumefaciens C58 and Escherichia coli DH5α served as negative controls. The experiments were performed in triplicate; error bars indicate standard deviations.
TCE degradation and chloride production.
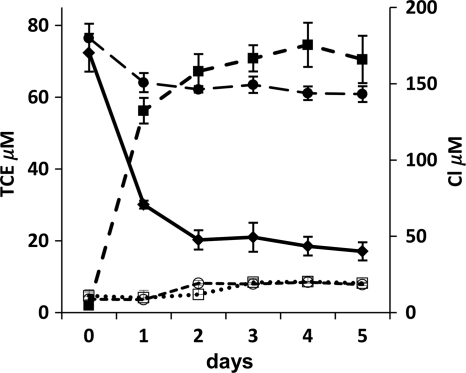
PDN3 was grown overnight in M9 medium with 0.2% peptone, harvested, and washed three times with chloride-free medium and then adjusted to an optical density (OD) at 660 nm of 1.0 using a spectrophotometer. Trichloroethylene was then added at a concentration of 76.3 μM to 20 ml of chloride-free medium in 125-ml Erlenmeyer flasks with Teflon Mininert valves, and the cultures were shaken at 150 rpm at 30°C for 5 days. At 24-h intervals, TCE was extracted into hexane and sodium chloride and analyzed by a gas chromatograph-electron capture detector (GC-ECD) (9). To confirm PDN3 degradation activity, we monitored the chloride release in the medium that was coincident with TCE removal by ion chromatography (IC) (15). As seen in Fig. 2, PDN3 rapidly reduced TCE levels in the medium from 72.4 μM to 30.1 μM in 24 h with a concurrent release of 127 μM chloride ion and nearly 80% of TCE (55.3 μM) was dechlorinated by PDN3 in 5 days with 166 μM chloride ion production. Neither chloride release nor TCE removal was observed in samples without PDN3. Also, intermediates of TCE degradation were not detected by a gas chromatograph–time-of-flight (GC-TOF) mass spectrometer. A naphthalene oxidation assay was performed to assay for monooxygenase activity (3, 4). PDN3 produced 2.5- to 3-times-higher levels of naphthol compared to those of E. coli DH5α and Agrobacterium tumefaciens strain C58 (data not shown). Another interesting result was the short-term production of nitrite, which may be an oxidation product of ammonia, by PDN3 in response to TCE addition, thereby indicating the presence of some unknown oxygenase that can be induced on addition of TCE.
Fig 2.
Time course of TCE degradation and chloride production by PDN3 in chloride-free medium. Trichloroethylene was added at a concentration of 76.3 μM to 20 ml chloride-free medium in 125-ml Erlenmeyer flasks with screw caps and Teflon Mininert valves to prevent evaporation of TCE. The cultures were incubated at 150 rpm at 30°C. The filled diamonds indicate TCE removal by strain PDN3, the filled circles indicate TCE in the uninoculated control, the filled squares indicate chloride production from TCE by PDN3, the open circles indicate chloride in the uninoculated control with TCE, and the open squares indicate chloride in the uninoculated control without TCE.
Genetic analysis.
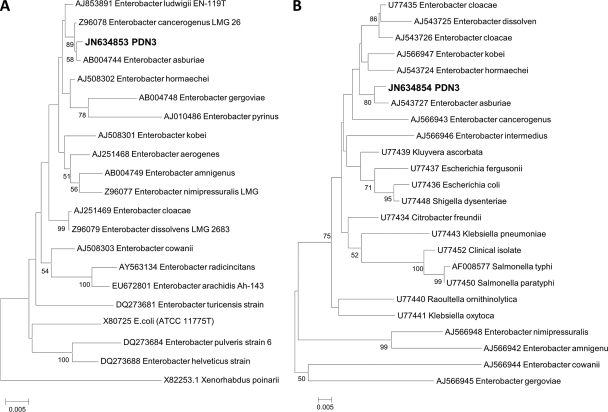
Genomic DNA was prepared from PDN3, and PCR was performed using the 16S rRNA primers 8F and 1492R (7, 8). The RNA polymerase β subunit-encoding gene (rpoB) primers were CM7 and CM31b (22). DNA sequence comparisons with public sequence databases were performed. Phylogenetic analysis was done using CLUSTAL W software. Evolutionary distance matrixes were constructed using the algorithm of Jukes and Cantor (34), and the evolutionary trees for the data sets were inferred from the neighbor-joining method (28) by using MEGA version 4.0.1 (33). 16S rRNA (1,425/1,430 nucleotides) and rpoB (1,000/1,008 nucleotides) gene sequences were 99% identical with the sequence of Enterobacter asburiae (HQ242719) (Fig. 3). In previous studies, E. asburiae strains have been reported as endophytes and rhizospheric microbes associated with sweet potato, cotton, bean, and cucumber (1, 24). The facultative anaerobes Enterobacter sp. strain MS-1 and Pantoea agglomerans have so far been reported to reductively dechlorinate perchloroethylene (PCE), thereby converting it to dichloroethene (DCE). This occurs only under strictly anoxic conditions (14, 29). Additionally, the attempts to amplify monooxygenase genes using conserved PCR primer sets were unsuccessful, suggesting that PDN3 uses a not-yet-discovered pathway for TCE degradation.
Fig 3.
Phylogenetic relationships between the endophyte PDN3 and the related bacterial species. The neighbor-joining dendrogram was derived from 16S rRNA (A) and β subunit of RNA polymerase-encoding gene (rpoB) (B) sequence distance matrixes (Jukes-Cantor). Bootstrap confidence levels (expressed as percentages of 1,000 replications) greater than 50% are indicated at the internodes.
To summarize, we have shown that Enterobacter sp. PDN3, a novel endophyte isolated from poplar, is able to effectively dechlorinate TCE without the addition of inducers, such as toluene or phenol. Bioremediation of TCE-contaminated sites using this strain seems to be a promising, cost-effective remediation strategy. Another approach is to use this strain to inoculate trees, such as poplars and willows, and test for increased TCE removal and degradation. Experiments are under way in our lab to evaluate endophyte-assisted phytoremediation of TCE by PDN3.
Nucleotide sequence accession numbers.
The sequences for the 16S rRNA gene and the rpoB gene were deposited in GenBank under accession numbers JN634853 and JN634854, respectively.
ACKNOWLEDGMENTS
We thank Jud Isebrands for providing the dormant cuttings, C. Andrew James for reviewing this article, Stuart Strand for providing the IC, Keith Stewart for help with the IC, and the Mass Spectrometry Center at the University of Washington for analyzing the samples on the GC-TOF mass spectrometer.
This work was funded through the NSF Environmental Engineering grant CBET-0829027.
Footnotes
Published ahead of print 24 February 2012
REFERENCES
- 1. Asis CA, Jr, Adachi K. 2004. Isolation of endophytic diazotroph Pantoea agglomerans and nondiazotroph Enterobacter asburiae from sweet potato stem in Japan. Lett. Appl. Microbiol. 38:19–23 [DOI] [PubMed] [Google Scholar]
- 2. Barac T, et al. 2004. Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile, organic pollutants. Nat. Biotechnol. 22:583–588 [DOI] [PubMed] [Google Scholar]
- 3. Brusseau GA, Tsien HC, Hanson RS, Wackett LP. 1990. Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation 1:19–29 [DOI] [PubMed] [Google Scholar]
- 4. Chu KH, Alvarez-Cohen L. 1998. Effect of nitrogen source on growth and trichloroethylene degradation by methane-oxidizing bacteria. Appl. Environ. Microbiol. 64:3451–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dey K, Roy P. 2009. Degradation of trichloroethylene by Bacillus sp.: isolation strategy, strain characteristics, and cell immobilization. Curr. Microbiol. 59:256–260 [DOI] [PubMed] [Google Scholar]
- 6. Doty SL. 2008. Enhancing phytoremediation through the use of transgenics and endophytes. New Phytol. 179:318–333 [DOI] [PubMed] [Google Scholar]
- 7. Doty SL, et al. 2009. Diazotrophic endophytes of native black cottonwood and willow. Symbiosis 47:23–33 [Google Scholar]
- 8. Doty SL, et al. 2005. Identification of an endophytic Rhizobium in stems of Populus. Symbiosis 39:27–36 [Google Scholar]
- 9. Doty SL, et al. 2007. Enhanced phytoremediation of volatile environmental pollutants with transgenic trees. Proc. Natl. Acad. Sci. U. S. A. 104:16816–16821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dower WJ, Miller JF, Ragsdale CW. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Glick BR. 2010. Using soil bacteria to facilitate phytoremediation. Biotechnol. Adv. 28:367–374 [DOI] [PubMed] [Google Scholar]
- 12. Goodner B, et al. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323–2328 [DOI] [PubMed] [Google Scholar]
- 13. Gordon M, et al. 1998. Phytoremediation of trichloroethylene with hybrid poplars. Environ. Health Perspect. 106:1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holliger C, Wohlfarth G, Diekert G. 1998. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22:383 [Google Scholar]
- 15. James CA, et al. 2009. A mass balance study of the phytoremediation of perchloroethylene-contaminated groundwater. Environ. Pollut. 157:2564–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kang JW, et al. 2010. Mammalian cytochrome CYP2E1 triggered differential gene regulation in response to trichloroethylene (TCE) in a transgenic poplar. Funct. Integr. Genomics 10:417–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuritz T, Wolk CP. 1995. Use of filamentous cyanobacteria for biodegradation of organic pollutants. Appl. Environ. Microbiol. 61:234–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Landa AS, et al. 1994. Cometabolic degradation of trichloroethylene by Pseudomonas cepacia G4 in a chemostat with toluene as the primary substrate. Appl. Environ. Microbiol. 60:3368–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee W, Wood TK, Chen W. 2006. Engineering TCE-degrading rhizobacteria for heavy metal accumulation and enhanced TCE degradation. Biotechnol. Bioeng. 95:399–403 [DOI] [PubMed] [Google Scholar]
- 20. Leibman KC, Hindman D. 1964. Modification of the Fujiwara reaction for determination of polyhalogenated organic compounds. Anal. Chem. 36:348–350 [Google Scholar]
- 21. Matin A, Little CD, Fraley CD, Keyhan M. 1995. Use of starvation promoters to limit growth and selectively enrich expression of trichloroethylene- and phenol-transforming activity in recombinant Escherichia coli. Appl. Environ. Microbiol. 61:3323–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mollet C, Drancourt M, Raoult D. 1997. rpoB sequence analysis as a novel basis for bacterial identification. Mol. Microbiol. 26:1005–1011 [DOI] [PubMed] [Google Scholar]
- 23. Pflugmacher U, Averhoff B, Gottschalk G. 1996. Cloning, sequencing, and expression of isopropylbenzene degradation genes from Pseudomonas sp. strain JR1: identification of isopropylbenzene dioxygenase that mediates trichloroethene oxidation. Appl. Environ. Microbiol. 62:3967–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quadt-Hallamnn A, Benhamou N, Kloepper JW. 1997. Bacterial endophytes in cotton: mechanisms of entering the plant. Can. J. Microbiol. 43:577 [Google Scholar]
- 25. Reith JF, van Ditmarsch WC, Ruiter TD. 1974. An improved procedure for application of the Fujiwara reaction in the determination of organic halides. Analyst 99:652–656 [Google Scholar]
- 26. Rosenblueth M, Martínez-Romero E. 2006. Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 19:827–837 [DOI] [PubMed] [Google Scholar]
- 27. Ryan RP, Germaine K, Franks A, Ryan DJ, Dowling DN. 2008. Bacterial endophytes: recent developments and applications. FEMS Microbiol. Lett. 278:1–9 [DOI] [PubMed] [Google Scholar]
- 28. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 29. Sharma PK, McCarty PL. 1996. Isolation and characterization of a facultatively aerobic bacterium that reductively dehalogenates tetrachloroethene to cis-1,2-dichloroethene. Appl. Environ. Microbiol. 62:761–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shields MS, Reagin MJ. 1992. Selection of a Pseudomonas cepacia strain constitutive for the degradation of trichloroethylene. Appl. Environ. Microbiol. 58:3977–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Suttinun O, Muller R, Luepromchai E. 2010. Cometabolic degradation of trichloroethene by Rhodococcus sp. strain L4 immobilized on plant materials rich in essential oils. Appl. Environ. Microbiol. 76:4684–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Suttinun O, Müller R, Luepromchai E. 2009. Trichloroethylene cometabolic degradation by Rhodococcus sp. L4 induced with plant essential oils. Biodegradation 20:281–291 [DOI] [PubMed] [Google Scholar]
- 33. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 34. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weyens N, et al. 2010. Endophytic bacteria improve phytoremediation of Ni and TCE co-contamination. Environ. Pollut. 158:2422–2427 [DOI] [PubMed] [Google Scholar]
- 36. Weyens N, et al. 2010. Potential of the TCE-degrading endophyte Pseudomonas putida W619-TCE to improve plant growth and reduce TCE phytotoxicity and evapotranspiration in poplar cuttings. Environ. Pollut. 158:2915–2919 [DOI] [PubMed] [Google Scholar]
- 37. Weyens N, et al. 2011. Endophytes and their potential to deal with co-contamination of organic contaminants (toluene) and toxic metals (nickel) during phytoremediation. Int. J. Phytoremediation 13:244–255 [DOI] [PubMed] [Google Scholar]
- 38. Weyens N, et al. 2009. Bioaugmentation with engineered endophytic bacteria improves contaminant fate in phytoremediation. Environ. Sci. Technol. 43:9413–9418 [DOI] [PubMed] [Google Scholar]