Abstract
The use of thermostable cellulases is advantageous for the breakdown of lignocellulosic biomass toward the commercial production of biofuels. Previously, we have demonstrated the engineering of an enhanced thermostable family 8 cellulosomal endoglucanase (EC 3.2.1.4), Cel8A, from Clostridium thermocellum, using random error-prone PCR and a combination of three beneficial mutations, dominated by an intriguing serine-to-glycine substitution (M. Anbar, R. Lamed, E. A. Bayer, ChemCatChem 2:997–1003, 2010). In the present study, we used a bioinformatics-based approach involving sequence alignment of homologous family 8 glycoside hydrolases to create a library of consensus mutations in which residues of the catalytic module are replaced at specific positions with the most prevalent amino acids in the family. One of the mutants (G283P) displayed a higher thermal stability than the wild-type enzyme. Introducing this mutation into the previously engineered Cel8A triple mutant resulted in an optimized enzyme, increasing the half-life of activity by 14-fold at 85°C. Remarkably, no loss of catalytic activity was observed compared to that of the wild-type endoglucanase. The structural changes were simulated by molecular dynamics analysis, and specific regions were identified that contributed to the observed thermostability. Intriguingly, most of the proteins used for sequence alignment in determining the consensus residues were derived from mesophilic bacteria, with optimal temperatures well below that of C. thermocellum Cel8A.
INTRODUCTION
The last 2 decades have seen tremendous progress in research on conversion of cellulosic biomass to biofuels (29, 34). Nevertheless, many techno-economic challenges must be overcome before cellulosic fuel will be able to compete with corn ethanol and conventional sources of fossil fuel (17, 26). A major bottleneck in converting cellulose to fuels is the hydrolysis of plant cell wall biopolymers, especially the attack on the highly recalcitrant cellulose fibers (12). Enzymatic hydrolysis of cellulose involves the synergistic action of three classes of enzymes: endoglucanases which randomly cleave within the cellulose chain, exoglucanases which cleave the exposed chain ends, and β-glucosidases which cleave short cellodextrins, notably cellobiose, into glucose. The carbohydrate-active enzyme (CAZY) database groups these glycoside hydrolases (GH) into families according to sequence similarity and shared structural determinants (8).
Optimizing the biodegradation of lignocellulose substrates requires either the search for novel enzymes which are robust enough to withstand industrial processes or, alternatively, enzymes that can be engineered to enhance the desired qualities, such as high specific activity, low levels of end product inhibition, tolerance to broad ranges of temperature and pH, and inhibitors of degradation by-products (38). Using thermostable cellulases at high temperatures offers many advantages in the bioconversion process, which include an increase in specific activity, higher levels of stability, inhibition of contaminating microbial growth, an increase in mass transfer rate due to lower fluid viscosity, and greater flexibility in the bioprocess (37).
In the present work, we focused on endoglucanase Cel8A, one of the most prominent enzymes produced by the anaerobic thermophilic bacterium Clostridium thermocellum (23, 28, 40). This family 8 glycoside hydrolase is part of an extracellular multienzyme complex of cellulases, hemicellulases, and other carbohydrate-active enzymes, termed the cellulosome, which can degrade and solubilize crystalline cellulosic substrates in an efficient manner (5). The mature enzyme consists of a catalytic module, which folds into an (α/α)6 barrel formed by six inner and six outer α-helices (2), and a type I dockerin at its C terminus that serves as an anchor for attachment to the cellulosomal scaffoldin subunit via its resident type I cohesin modules.
Recently, we reported the construction of a Cel8A enzyme with enhanced thermostability using a directed evolution approach consisting of random PCR-based mutagenesis and recombination (4). The thermostability of Cel8A was also recently studied using specific substitutions of glycine and proline residues on the protein surface (35).
A different, potentially complementary approach takes advantage of the large number of available protein sequences. This semirational “consensus approach” is a well-established strategy to improve the thermostability and has been used successfully on both enzymatic and nonenzymatic proteins (3, 15, 16, 25, 32). The approach is based on the substitution of specific amino acids in a particular protein, with the most prevalent amino acid present at these positions among the homologous family members. A possible explanation for the stabilizing effect of consensus mutations based on analogy with statistical thermodynamics has been proposed by Steipe et al. (30). However, it was also shown that only some of the consensus mutations contribute to protein stability, while others destabilize the protein or are neutral (16). It is therefore suggested that a selection should be made in order to include only the beneficial mutations.
In the current study, we complemented the random mutagenesis strategy with a consensus approach to further investigate the protein sequence space for enzyme variants with enhanced thermostability and high specific activity. We used molecular dynamics (MD) analysis as a complementary tool to examine the effect of the beneficial mutations on the enzyme's dynamic stability and overall structure (7).
MATERIALS AND METHODS
Library construction.
Plasmid pET28aCel8A (4) containing the cel8A gene (NCBI accession no. AAA83521) from Clostridium thermocellum ATCC 27405 was used to construct the library. The cel8A gene was amplified and digested with DNase I (Sigma). The resulting 50- to 200-bp fragments were assembled by PCR in the presence of an equimolar mixture of 8 oligonucleotides (Table 1) encoding the consensus mutations (total of 10 pmol) as previously described (11). The resulting library was cloned into the pET28 vector using the NcoI and XhoI restriction sites.
Table 1.
Primers used in this worka
| Primer name | Sequence (5′→3′) |
|---|---|
| Primers for library construction | |
| L101 M | GGTATGGGATACGGAATGCTTTTGGCGGTTTGC |
| D115G | CAGGCTTTGTTTGACGGTTTATACCGTTACGTA |
| L187I | ACATTGATAAACAATATTTACAACCATTGTGTA |
| Y224F | GCATGGTACAAAGTGTTTGCTCAATATACAGG |
| Y227Fb | CTTGTGTCTCCTGTAAATTGAGCATACACTTTG |
| G283P | GATGCTACACGTTACCCGTGGAGAACTGCCGTG |
| F293Y | GTGGACTATTCATGGTATGGTGACCAGAGAGC |
| I323L | GTTGACGGATACACACTGCAAGGTTCAAAAATTAG |
| Primers for mutagenesis | |
| G177P+1 | GGTGCAATAAACTACCCGCAGGAAGCAAGGACATTG |
| G177P+2 | CAATGTCCTTGCTTCCTGCGGGTAGTTTATTGCACC |
| G373P+1 | GAATATTACGGATATTACCCGAACAGCTTGAGACTG |
| G373P+2 | CAGTCTCAAGCTGTTCGGGTAATATCCGTAATATTC |
| L101 Mb | GCAAACCGCCAAAAGCATTCCGTATCCCATACC |
| Y224Fb | CCTGTATATTGAGCAAACACTTTGTACCATGC |
| G283Pb | CACGGCAGTTCTCCACGGGTAACGTGTAGCATC |
Codon changes are underlined.
Primer designed for complementary strand.
Screening for thermostable endoglucanase variants.
Transformed Escherichia coli cells derived from the library of Cel8A variants were spread onto LB plates containing 50 μg/ml kanamycin and incubated overnight at 37°C. Individual clones were selected and grown overnight at 37°C in 96-well plates containing 0.5 ml LB, 50 μg/ml kanamycin, and 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Proteins were extracted using PopCulture reagent (Novagen) according to the product manual. A sample of the extracted solution was diluted in 50 mM sodium acetate (pH 6.0) and incubated at various temperatures and time periods. Residual activity was determined using 1% carboxymethyl cellulose (CMC; medium viscosity; Sigma, St. Louis, MO) in 10 mM CaCl2 and 50 mM sodium acetate (pH 6.0) at 65°C. The amount of reducing sugars released by the enzyme was determined colorimetrically using the 3,5-dinitrosalicylic acid (DNS) reagent (19).
Site-directed and saturation mutagenesis.
Single-point mutations were generated in cel8A by using the QuikChange site-directed mutagenesis kit (Stratagene) and the primers listed in Table 1. To verify that only the designated mutations were inserted by the Pfu Turbo DNA polymerase, the full cel8A gene was sequenced. Saturation mutagenesis at amino acid position 323 of Cel8A was performed by constructing a library of all possible amino acids at that position using two degenerate primers, in which the mutation site was replaced with NNS (where N is A, G, C, or T and S is G or C). The pair of primers used was 5′-CGTTGACGGATACACANNSCAAGGTTCAAAAATTAG-3′ and 5′-CTAATTTTTGAACCTTGSNNTGTGTATCCGTCAACG-3′ (boldface letters indicate the degenerate nucleotides), which were designed to randomize position 323 in the nucleotide sequence.
Protein expression and purification.
For detailed analysis of Cel8A and variants, E. coli BL21(DE3) transformants were grown at 37°C in LB supplemented with 50 μg/ml kanamycin, until an optical density at 600 nm (OD600) of ∼0.8 was reached. Overexpression was induced by adding 0.5 mM IPTG; the cultures were grown for another 3 h and harvested (4,000 × g, 15 min, 4°C). The pellet was frozen overnight at −20°C. The cells were resuspended in Tris-buffered saline (TBS; 137 mM NaCl, 2.7 mM KCl, 25 mM Tris-HCl, pH 7.4) supplemented with 5 mM imidazole (Merck KGaA, Darmstadt, Germany) and protease inhibitor cocktail (1 mM phenylmethylsulfonyl fluoride [PMSF], 0.4 mM benzamidine, and 0.06 mM benzamide; Sigma-Aldrich, St. Louis, MO) and disrupted by sonication. The sonicate was heated for 30 min at 60°C and then centrifuged (20,000 × g, 30 min, 4°C). The soluble fraction was mixed with Ni-nitrilotriacetic acid (NTA) resin (Zephyr ProeomiX, Rehovot, Israel), supplemented with 5 to 10 mM imidazole, for 1 h in an Econo-pack column at 4°C (batch purification system). The column was then washed by gravity flow with 50 mM imidazole. Elution was performed using 100 mM imidazole followed by 250 mM imidazole. Fractions (2 ml) were collected and analyzed by SDS-PAGE. The fractions containing the purified proteins were pooled and dialyzed extensively against 50 mM sodium acetate buffer (pH 6.0). Protein concentrations were determined by spectrophotometeric absorbance (280 nm) using the calculated molar absorption coefficient of the protein. Samples were stored at 4°C and supplemented with 0.02% sodium azide or at −20°C with 50% glycerol.
CD measurements.
Circular dichroism (CD) spectra were recorded on a Chirascan circular dichroism spectrometer (Applied Photophysics, Surrey, United Kingdom) in a 1-mm-path-length cuvette. The proteins were used at a ∼5 μM concentration in 50 mM sodium acetate buffer (pH 6.0). To construct the melting curves, the samples were heated at a rate of 1°C per min from 55 to 95°C and the CD ellipticity signal at 222 nm, which showed the maximal change with temperature, was monitored.
Enzymatic assays.
Endoglucanase activity was measured by incubating the purified enzymes with 0.5% (wt/vol) CMC or phosphoric acid-swollen cellulose (PASC) at 65°C for 1 h with occasional shaking. Activities were determined by assaying the release of reducing sugars by the DNS method. Enzyme activity was expressed as units (U). One unit of endoglucanase activity corresponds to the release of 1 μmol of glucose equivalent per hour (28).
Thermal stability of enzymes.
Enzyme thermostability was monitored in 50 mM sodium acetate buffer (pH 6.0). Enzymes were incubated at 85°C for various periods, and residual endoglucanase activity was determined. The rate of inactivation was determined by removing samples for assay of enzymatic activity. The inactivation rate constant was calculated using the equation log(% remaining activity) = 2.303 × Kin × t, where Kin is the inactivation constant and t is time (14), and the enzyme half-lives were deduced from the plots.
Molecular dynamics studies.
Mutant structures were generated with the Mutator module of the Visual Molecular Dynamics (VMD) program, using Protein Data Bank (PDB) entry 1CEM as a template (13). The NAMD/VMD programs were used for molecular dynamics simulation and trajectory analysis (24) with the CHARMM27 force field (18). The psfgen module in VMD was employed to generate a protein structure file. Subsequently, the structures were solvated in a box of water (TIP3P water model) with a 10-Å buffering distance and ionized using sodium (Na+) and chloride (Cl−) counterions at a concentration of 0.1 mol/liter to achieve charge neutrality and to mimic the biological environment. All systems were subjected to minimization for 10,000 steps using the conjugate gradient method.
Simulations were performed prior to MD runs; the systems were equilibrated for 500 ps with a time step of 2 fs while maintaining the temperature by velocity rescaling. The data collection stages were 4 ns in length. At this stage, the temperature control was established via Langevin dynamics. At least 2 runs for each mutant and native structure were performed, and the trajectory files during the 4-ns period were collected and analyzed.
RESULTS AND DISCUSSION
Construction and screening of libraries containing consensus mutations.
The amino acid sequence of Cel8A from C. thermocellum was used to identify 18 homologous sequences in GenBank (Table 2). The sequences were selected based on amino acid identity values of 30 to 60%. The sequences were aligned using the ClustalW algorithm, and either consensus positions or most abundant positions were determined. Overall, Cel8A differed in 8 positions from the consensus sequence (Fig. 1). It should be noted that many of the proteins used for the alignment were from mesophilic bacteria, e.g., Clostridium cellulolyticum and Flavobacterium johnsoniae, with an optimal temperature well below that of Cel8A from C. thermocellum.
Table 2.
cel8A sequences used for consensus alignment
| Species name | Optimum growth temp (°C) | GenBank accession no. |
|---|---|---|
| Bacillus cereus BDRD-Cer4 | <40a | ZP_04257004 |
| Bacillus pseudomycoides DSM 12442 | 28 | ZP_04154406 |
| Clostridium cellulolyticum H10 | 34 | YP_002505089 |
| Clostridium josui | 45 | sp_P37701 |
| Clostridium papyrosolvens DSM 2782 | 30a | ZP_05497268 |
| Cytophaga hutchinsonii ATCC 33406 | 27 | YP_679442 and YP_680303 |
| Ethanoligenens harbinense YUAN-3 | 35 | ZP_06472621 |
| Flavobacterium johnsoniae UW101 | 30 | YP_001193374 |
| Hahella chejuensis KCTC 2396 | 37 | YP_437840 |
| Herpetosiphon aurantiacus ATCC 23779 | 30 | YP_001543073 and YP_001543074 |
| Marinomonas sp. MWYL1 | 30 | YP_001342550 |
| Opitutus terrae PB90-1 | 37 | YP_001819146 |
| Paenibacillus fukuinensis | 30 | BAB64835 |
| Paenibacillus sp. Y412MC10 | 55–65a | YP_003240630 |
| Planctomyces maris DSM 8797 | 30 | ZP_01854832 |
| Plesiocystis pacifica SIR-1 | 28 | ZP_01905268 |
Data inferred from closely related strains.
Fig 1.
Protein sequence of Cel8A from C. thermocellum. Consensus mutations are indicated (in bold).
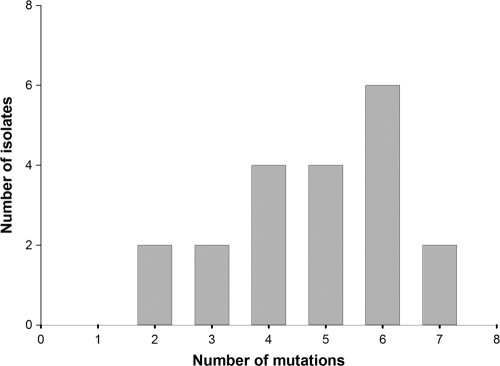
We designed 8 oligonucleotide primers, each containing a single codon replacement of the Cel8A gene with the matching consensus residue, and used in vitro recombination to produce an assembly of the different combinations of mutations. The resultant library was cloned into the pET28a expression vector and expressed in E. coli. The diversity of the genes in the resulting unselected library is presented in Fig. 2.
Fig 2.
Distribution of consensus mutations in the shuffled library. Several isolates contained between 1 and 2 additional missense mutations that were not included in the analysis.
All of the planned consensus mutations were observed in the library, but in half of the genes one to two unplanned point mutations appeared that were introduced during the DNA fragment assembly. Preliminary experiments for detection of active enzymes on a two-layer CMC-containing plate revealed that over 90% were active enzymes. Therefore, an initial screening step for activity prior to the thermostability screening was not necessary.
The retention of endoglucanase activity was measured after heating the samples for 15 min at 82°C. After heating the wild-type (WT) Cel8A endoglucanase sample at this temperature, the enzyme retained approximately 40% of its activity (4). Enzymes which showed enhanced thermostability after the heat treatment were considered candidates for further analysis. In various methods which use random mutagenesis in order to generate thermostable mutants, large numbers of clones have to be screened before identifying the desired mutants (4). Using the consensus approach, it is possible to screen significantly fewer clones and still acquire thermostable variants. Here, we screened fewer than 600 clones before identifying 11 thermostable mutants that showed considerably higher residual activity after heat treatment than the wild-type Cel8A. The cel8A gene from each of the positive clones was sequenced in order to determine the mutation(s) responsible for the increased thermostability.
Identification of mutations responsible for thermostability.
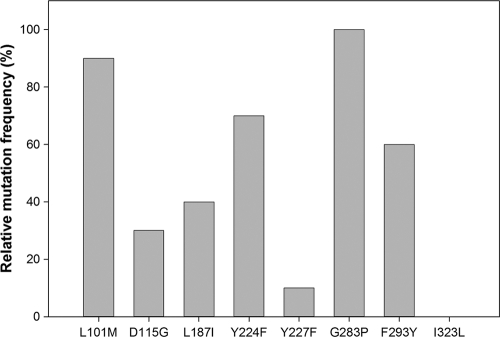
The consensus approach enabled the identification of thermostable variants by combining multiple consensus mutations in different combinations. In order to determine which amino acid substitutions were responsible for the increased thermostability, we determined the frequency of the individual consensus mutations in these variants. The results, presented in Fig. 3, indicated that 7/8 of the consensus mutations appear in all of the stable variants, but three of the mutations (L101M, Y224F, and G283P) are the most prevalent and were pursued further.
Fig 3.
Frequency of mutations in thermostable mutants. Ten of the most stable mutants were sequenced and mapped.
We performed site-directed mutagenesis in order to determine the individual contribution of each of the three mutations. The results showed that a single mutation (G283P) was sufficient to produce the thermostable variant. The other two mutations did not contribute to its thermostability, and these variants showed wild-type levels of residual activity (data not shown). Interestingly, mutation I323L did not appear in any of the stable mutants but appeared in all the library variants that were completely inactivated upon a heat challenge (1.2% ± 1.8% of the initial activity), although high initial activity was preserved (101.2% ± 4.7% compared to initial activity of wild-type Cel8A). It is intriguing that such a conservative mutation could lead to such a dramatic effect. The I323 position is located on a surface loop, and it was speculated that it might be significant for thermostability, as seen in previous work on Cel8A (36). Nevertheless, analysis of the thermostability of variants generated by site saturation mutagenesis at the 323 site did not indicate any other amino acid that could contribute to thermostability.
It has been previously shown that introduction of a proline at key sites could contribute to protein stabilization (1, 10, 31, 39). The G283P substitution occurred in the first turn of the α-helix 8. It was therefore interesting to determine whether the substitution of proline for glycine in other similar locations could improve the enzyme thermostability. Two glycines (G177 and G373) near the N cap of helices 5 and 12 that were not conserved in the Cel8A family were chosen for replacement by proline via site-directed mutagenesis. Each glycine residue was individually mutated to proline, and both enzymes were tested for activity and thermostability. The results showed that both enzymes maintained their initial activity levels (108.2% ± 2.6% and 96.4% ± 2.8% for the G177P and G373P mutants, respectively) but had significant reductions in residual activity compared to that of the wild-type enzyme (23% ± 1.5% for the G177P mutant and undetectable levels for the G373P mutant). This indicates that the G283P mutation may be a unique case of thermostabilization and cannot be generalized to glycines in the N cap of other helices of Cel8A.
Additive effect of the G283P mutation to the previously engineered thermostable Cel8A.
Recently, we reported the construction of a Cel8A enzyme derivative with enhanced thermostability by using random error-prone PCR and a combination of beneficial mutations (4). The engineered triple mutant (TM; K276R/S329G/S375T) exhibited increased thermostability while maintaining wild-type levels of activity. It was therefore interesting to introduce the G283P mutation into the triple mutant to determine whether it could contribute in a positive manner to its stability. Site-directed mutagenesis was thus performed on the TM variant to create the quadruple-mutant (QM) variant (K276R/G283P/S329G/S375T). The enzymes were purified to homogeneity, and their properties were determined and compared with those of the wild-type enzyme. A 3.3°C increase in the melting temperature (Tm) of the QM variant relative to that of the TM variant (Table 3) was observed, which demonstrates that the thermostabilizing effect of G283P is additive.
Table 3.
Thermostability of the wild type and thermostable mutants
| Enzyme | Mutation | Tmapp (°C)a | ΔTm (°C) | Rate constant Kin (min−1)b |
|---|---|---|---|---|
| Cel8A | None | 80.7 | 1.558 | |
| G283P | G283P | 84.2 | 3.5 | 1.318 |
| TM | K276R, S329G, S375T | 86.9 | 6.2 | 0.239 |
| QM | K276R, G283P, S329G, S375T | 90.2 | 9.5 | 0.115 |
Tmapp is the approximate midpoint temperature of melting determined by CD spectroscopy at 222 nm.
The inactivation rate constants were deduced from plots of ln(% residual activity) versus time at 85°C.
Enzymatic characterization of the thermostable mutants.
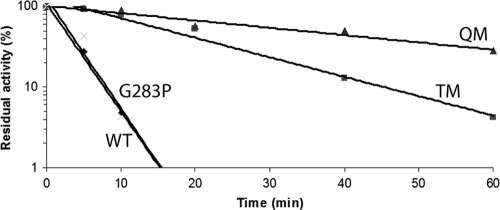
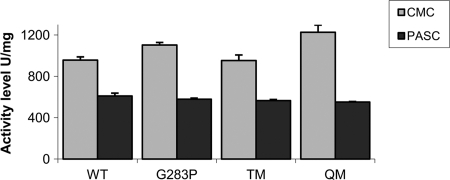
Kinetic analysis of the thermal inactivation of the wild-type Cel8A and the mutants at 85°C followed first-order kinetics (Fig. 4). Thermal melting points are listed in Table 3. The introduction of a single proline at the second residue of helix 8 increased the Tm by 3.5°C. The introduction of the mutation into the TM showed an additive effect, further increasing the Tm from 86.9°C to 90.2°C. The Kin of the QM variant was found to be 0.115 min−1, which is 14-fold lower than that of wild-type Cel8A. The half-life of the QM variant at 85°C was determined to be 34 min, compared to 2.5 min for the wild-type Cel8A and 16 min for the TM variant (4). At this elevated temperature, the G283P variant shows a half-life similar to that of wild-type Cel8A. The specific activities of the mutants were determined on both CMC and phosphoric acid-swollen cellulose (PASC). As shown in Fig. 5, both the G283P and the QM variants demonstrate similar specific activities on PASC compared to those of the TM and wild-type enzymes (4). The G283P mutation demonstrated an increase in activity on CMC either as a single mutation or in combination with the TM variant (QM). The stability profile of the mutants was also determined at pH values of 3.0 to 9.0 and showed residual activities similar to that of the wild-type Cel8A enzyme (data not shown).
Fig 4.
Kinetics of thermal inactivation of Cel8A variants at 85°C. The residual activities were measured at different time points. Shown are Cel8A mutants G283P, TM, and QM (see Table 3). Wild-type Cel8A (WT) served as a control. The activity of unheated enzymes was taken as 100%. Each point represents the mean of duplicate determinations.
Fig 5.
Specific activities of wild-type Cel8A (WT) and thermostable mutants on CMC and PASC. Enzymes were incubated with 0.5% (wt/vol) solutions at 65°C for 1 h.
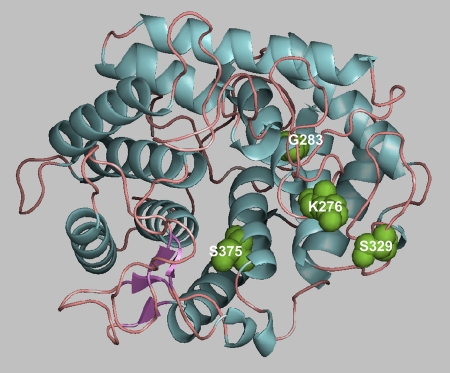
Structural analysis of the mutations.
The CD spectra of the thermostable variants were analyzed to determine whether the mutation affected the secondary structure. The results showed that there were no significant changes between the wild-type enzyme and the mutants with increased thermostability. Figure 6 shows the three-dimensional structure of Cel8A as determined by Alzari et al. (2) and the positions where the mutations were introduced in the present work. The explanation for the increased thermostability appears to lie in the reduction of the conformational freedom of the protein backbone in its unfolded state. It has been reported that introduction of prolines at key positions, namely, the first turn of the α-helix, the second site of the β-turn, and in flexible loops, can stabilize proteins. Indeed, in many thermostable proteins there is an increase in the number of prolines at the N terminus of α-helices (33). Because the proline residue has a pyrrolidine ring, the backbone conformation of proline is constrained. The φ and ψ values of the proline residue are restricted, and in addition, the φ and ψ values of the preceding residue are also limited (27). Several reports demonstrate the contribution of prolines in the first turn of α-helices for thermostabilization. For example Tk-RNase HII from the hyperthermophile Thermococcus kodakaraensis was thermostabilized by the introduction of prolines at the N terminus of α-helices. Barley α-glucosidase was thermostabilized by replacing the Thr340 residue with proline in the first turn of an α-helix (20).
Fig 6.
The positions of the residues that were replaced in the QM in the Cel8A structure (PDB code 1CEM) are shown.
MD analysis of mutations.
With the selected subset of proteins, MD simulations were performed to determine the effect of the selected mutations on overall protein stability. Several authors have previously utilized detailed MD studies to understand protein stability. Parthasarathy and his coworkers have carried out a preliminary analysis suggesting that the crystallographically determined atomic displacement parameters might actually convey information regarding thermal stability (21). They have highlighted the variation of the crystallographic B factors (Debye-Waller factors) from the core to the surface of proteins and found the amino acid compositions in high B-factor value regions of the polypeptide chains. Similarly, the mean square displacements of nonhydrogen atoms can be monitored by using Debye-Waller factors from the output of MD simulations (6). In another study, the mean square displacements of each residue were investigated via MD to determine the thermal stability change in thioredoxins (22). In that study, the authors confirmed, via MD, the experimental observation that replacement of residues reduces backbone flexibility, thereby stabilizing the protein. They were also able to correlate the increase in protein stabilization caused by single-amino-acid replacements. Dynamic studies of different proteins performed by Daggett and Levitt (9) using high-temperature MD, in which they checked the mean square displacements of the residues, suggest that loop regions in proteins undergo the largest deviations and that at higher temperatures these may be the regions of the protein that unfold first during thermal denaturation.
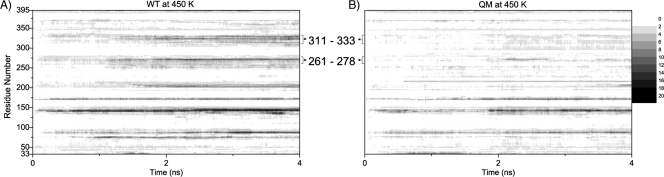
In the current study, MD simulations were performed at elevated temperatures to investigate the impact of given mutations on the thermodynamic stability of Cel8A. As a measure of thermostability, root mean square deviation (RMSD) was used. RMSD during simulation represents the relative distance of a given structure to the minimized initial structure. Root mean square fluctuation (RMSF) is the measurement of fluctuations per amino acid during simulation. RMSF represents the flexibility of a given amino acid over trajectory at defined temperatures. A more comprehensive description of the enzyme thermostability at elevated temperatures can be obtained by plotting the RMSD values as a function of time and residue number. This is depicted in Fig. 7, where the RMSD plots at 450 K for the wild-type enzyme are compared to the QM variant. The x axis represents the time, and the y axis is the residue number. The gray scale shows the RMSD values on the z axis. Black-colored regions have higher RMSD values and are more flexible. Both the WT and QM show an increase in structural changes, but two regions, between amino acids 261 and 278 and between 311 and 333, show a notable difference. These regions may represent the first loops which undergo unfolding during heat treatment. Analysis of trajectories at high temperatures for the WT structure revealed that six different regions of the WT protein became more unstable during simulations. These regions are amino acids 75 to 100, 135 to 150, 172 to 175, 200 to 225, 261 to 278, and 311 to 333. Compared to the QM structure simulation at high temperature (Fig. 7), we have observed that the first 4 of these regions showed a delayed increase (approximately 2 ns) in RMSD. On the other hand, the last two regions (amino acids 261 to 278 and 311 to 333) showed no sign of unfolding in terms of RMSD during the 4-ns simulation, suggesting that these regions are responsible for increased thermostability of variant QM.
Fig 7.
Contour plots of RMSD (Angstrom) values for the Cα atom of residues (33 to 395) during simulation time (ns) for the wild type (WT) (A) and the quadruple mutant (QM) (B). Graphs are grayscaled from white (0) to black (20) with increments of 2. The regions between amino acids 261 and 278 and between 311 to 333 are labeled. Significant destabilizations of these loops are observed after a 1-ns simulation time for the WT structure at 450 K (dark-shaded regions). On the other hand, the cumulative effect of the given mutations for the QM structure significantly enhances the stabilization of these loops (lighter shades of gray) at 450 K.
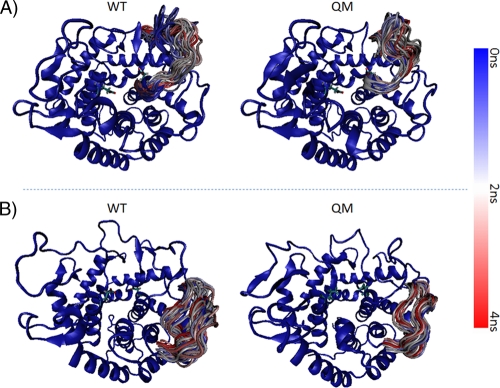
Moreover, detailed analysis of the RMSF was carried out using trajectory data. Although overall comparison of RMSF showed similar patterns for the mutant and native proteins (data not shown), the regions between amino acids 261 to 278 (Fig. 8A) and 311 to 333 (Fig. 8B) are clearly more rigid for the QM structure compared to that of the native protein at 450 K. These regions have no impact on structure in terms of RMSF at 300 K (data not shown), but the mutations rendered these regions more rigid and stable at higher temperatures. Structural analysis indicated that the two regions are close to the active-site cleft. The glucopyranoside ring of the substrate is within hydrogen bonding distance of Asp278 (2), which we have found is in the more rigid region at elevated temperature as a result of the described mutations. Rigidity of the region between amino acids 261 and 278 directly affects the hydrogen bonding distance for Asp278 at elevated temperatures and increases the thermostability of the given mutants.
Fig 8.
Sampling of loop conformations from MD trajectory. Cartoon representations of wild-type (WT) and quadruple-mutant (QM) structures from MD trajectory are superimposed on the starting conformation (colored blue) at 450 K. To demonstrate the increased rigidity of the loop residues 261 to 278 (A) and 311 to 333 (B) for QM relative to WT, these residues are colored according to time scale (blue for 0 ns, white for 2 ns, and red for 4 ns), and representative snapshots from the trajectory (every 80 ps) are superimposed.
The results presented here demonstrate the utility of the consensus approach as a viable tool for enhancement of the thermostability characteristics of an innately thermostable endoglucanase. Introducing the G283P mutation to increase the thermostability of the TM variant generated by a random library resulted in further enhancement of its thermostability properties. This study underscores the value of combining several complementary mutagenesis approaches in order to better explore the sequence space available for enzyme improvement.
ACKNOWLEDGMENTS
This research was supported by grants from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel (grant no. 966/09 and 159/07 to E.A.B. and R.L., respectively), by the Israel Science Foundation, by a grant from the Israel Ministry of Science (IMOS), by the Weizmann Institute of Science Alternative Energy Research Initiative, and by the establishment of an Israeli Center of Research Excellence (I-CORE) managed by the Israel Science Foundation. E.A.B. is the incumbent of The Maynard I. and Elaine Wishner Chair of Bio-Organic Chemistry.
Footnotes
Published ahead of print 2 March 2012
REFERENCES
- 1. Allen MJ, Coutinho PM, Ford CF. 1998. Stabilization of Aspergillus awamori glucoamylase by proline substitution and combining stabilizing mutations. Protein Eng. 11:783–788 [DOI] [PubMed] [Google Scholar]
- 2. Alzari PM, Souchon H, Dominguez R. 1996. The crystal structure of endoglucanase CelA, a family 8 glycosyl hydrolase from Clostridium thermocellum. Structure 4:265–275 [DOI] [PubMed] [Google Scholar]
- 3. Amin N, et al. 2004. Construction of stabilized proteins by combinatorial consensus mutagenesis. Protein Eng. Des. Sel. 17:787–793 [DOI] [PubMed] [Google Scholar]
- 4. Anbar M, Lamed R, Bayer EA. 2010. Thermostability enhancement of Clostridium thermocellum cellulosomal endoglucanase Cel8A by a single glycine substitution. ChemCatChem 2:997–1003 [Google Scholar]
- 5. Bayer EA, Lamed R, White BA, Flint HJ. 2008. From cellulosomes to cellulosomics. Chem. Rec. 8:364–377 [DOI] [PubMed] [Google Scholar]
- 6. Baysal C, Atilgan AR. 2005. Relaxation kinetics and the glassiness of native proteins: coupling of timescales. Biophys. J. 88:1570–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beckham GT, Bomble YJ, Bayer EA, Himmel ME, Crowley MF. 2011. Applications of computational science for understanding enzymatic deconstruction of cellulose. Curr. Op. Biotechnol. 22:231–238 [DOI] [PubMed] [Google Scholar]
- 8. Cantarel BL, et al. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daggett V, Levitt M. 1993. Protein unfolding pathways explored through molecular dynamics simulations. J. Mol. Biol. 232:600–619 [DOI] [PubMed] [Google Scholar]
- 10. Goihberg E, et al. 2007. A single proline substitution is critical for the thermostabilization of Clostridium beijerinckii alcohol dehydrogenase. Proteins 66:196–204 [DOI] [PubMed] [Google Scholar]
- 11. Herman A, Tawfik DS. 2007. Incorporating synthetic oligonucleotides via gene reassembly (ISOR): a versatile tool for generating targeted libraries. Protein Eng. Des. Sel. 20:219–226 [DOI] [PubMed] [Google Scholar]
- 12. Himmel ME, et al. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804–807 [DOI] [PubMed] [Google Scholar]
- 13. Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–8, 27-8 [DOI] [PubMed] [Google Scholar]
- 14. Kotzia GA, Labrou NE. 2009. Engineering thermal stability of l-asparaginase by in vitro directed evolution. FEBS J. 276:1750–1761 [DOI] [PubMed] [Google Scholar]
- 15. Lehmann M, et al. 2000. From DNA sequence to improved functionality: using protein sequence comparisons to rapidly design a thermostable consensus phytase. Protein Eng. 13:49–57 [DOI] [PubMed] [Google Scholar]
- 16. Lehmann M, et al. 2002. The consensus concept for thermostability engineering of proteins: further proof of concept. Protein Eng. 15:403–411 [DOI] [PubMed] [Google Scholar]
- 17. Lynd LR, et al. 2008. How biotech can transform biofuels. Nat. Biotechnol. 26:169–172 [DOI] [PubMed] [Google Scholar]
- 18. Mackerell AD, Jr, Feig M, Brooks CL., III 2004. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25:1400–1415 [DOI] [PubMed] [Google Scholar]
- 19. Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426–428 [Google Scholar]
- 20. Muslin EH, Clark SE, Henson CA. 2002. The effect of proline insertions on the thermostability of a barley alpha-glucosidase. Protein Eng. 15:29–33 [DOI] [PubMed] [Google Scholar]
- 21. Parthasarathy S, Murthy MR. 2000. Protein thermal stability: insights from atomic displacement parameters (B values). Protein Eng. 13:9–13 [DOI] [PubMed] [Google Scholar]
- 22. Pedone EM, Bartolucci S, Rossi M, Saviano M. 1998. Computational analysis of the thermal stability in thioredoxins: a molecular dynamics approach. J. Biomol. Struct. Dyn. 16:437–446 [DOI] [PubMed] [Google Scholar]
- 23. Petre J, Longin R, Millet J. 1981. Purification and properties of an endo-β-1,4-glucanase from Clostridium thermocellum. Biochimie 63:629–639 [DOI] [PubMed] [Google Scholar]
- 24. Phillips JC, et al. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Polizzi KM, Chaparro-Riggers JF, Vazquez-Figueroa E, Bommarius AS. 2006. Structure-guided consensus approach to create a more thermostable penicillin G acylase. Biotechnol. J. 1:531–536 [DOI] [PubMed] [Google Scholar]
- 26. Ragauskas AJ, et al. 2006. The path forward for biofuels and biomaterials. Science 311:484–489 [DOI] [PubMed] [Google Scholar]
- 27. Schimmel PR, Flory PJ. 1968. Conformational energies and configurational statistics of copolypeptides containing l-proline. J. Mol. Biol. 34:105–120 [DOI] [PubMed] [Google Scholar]
- 28. Schwarz WH, Grabnitz F, Staudenbauer WL. 1986. Properties of a Clostridium thermocellum endoglucanase produced in Escherichia coli. Appl. Environ. Microbiol. 51:1293–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sims RE, Mabee W, Saddler JN, Taylor M. 2010. An overview of second generation biofuel technologies. Bioresour. Technol. 101:1570–1580 [DOI] [PubMed] [Google Scholar]
- 30. Steipe B, Schiller B, Pluckthun A, Steinbacher S. 1994. Sequence statistics reliably predict stabilizing mutations in a protein domain. J. Mol. Biol. 240:188–192 [DOI] [PubMed] [Google Scholar]
- 31. Tian J, et al. 2010. Enhanced thermostability of methyl parathion hydrolase from Ochrobactrum sp. M231 by rational engineering of a glycine to proline mutation. FEBS J. 277:4901–4908 [DOI] [PubMed] [Google Scholar]
- 32. Viader-Salvado JM, et al. 2010. Design of thermostable beta-propeller phytases with activity over a broad range of pHs and their overproduction by Pichia pastoris. Appl. Environ. Microbiol. 76:6423–6430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Watanabe K, Hata Y, Kizaki H, Katsube Y, Suzuki Y. 1997. The refined crystal structure of Bacillus cereus oligo-1,6-glucosidase at 2.0 Å resolution: structural characterization of proline-substitution sites for protein thermostabilization. J. Mol. Biol. 269:142–153 [DOI] [PubMed] [Google Scholar]
- 34. Wilson DB. 2009. Cellulases and biofuels. Curr. Opin. Biotechnol. 20:295–299 [DOI] [PubMed] [Google Scholar]
- 35. Yi ZL, Pei XQ, Wu ZL. 2010. Introduction of glycine and proline residues onto protein surface increases the thermostability of endoglucanase CelA from Clostridium thermocellum. Bioresour. Technol. 102:3636–3638 [DOI] [PubMed] [Google Scholar]
- 36. Yi ZL, Wu ZL. 2010. Mutations from a family-shuffling-library reveal amino acid residues responsible for the thermostability of endoglucanase CelA from Clostridium thermocellum. Biotechnol. Lett. 32:1869–1875 [DOI] [PubMed] [Google Scholar]
- 37. Zamost BL, Nielsen HK, Starnes RL. 1991. Thermostable enzymes for industrial applications. J. Ind. Microbiol. Biotechnol. 8:71–81 [Google Scholar]
- 38. Zhang YH, Himmel ME, Mielenz JR. 2006. Outlook for cellulase improvement: screening and selection strategies. Biotechnol. Adv. 24:452–481 [DOI] [PubMed] [Google Scholar]
- 39. Zhou C, Xue Y, Ma Y. 2010. Enhancing the thermostability of α-glucosidase from Thermoanaerobacter tengcongensis MB4 by single proline substitution. J. Biosci. Bioeng. 110:12–17 [DOI] [PubMed] [Google Scholar]
- 40. Zverlov VV, Kellermann J, Schwarz WH. 2005. Functional subgenomics of Clostridium thermocellum cellulosomal genes: identification of the major catalytic components in the extracellular complex and detection of three new enzymes. Proteomics 5:3646–3653 [DOI] [PubMed] [Google Scholar]