Abstract
High hydrostatic pressure (HHP) processing is becoming a valuable nonthermal food pasteurization technique, although there is reasonable concern that bacterial HHP resistance could compromise the safety and stability of HHP-processed foods. While the degree of natural HHP resistance has already been shown to vary greatly among and within bacterial species, a still unresolved question remains as to what extent different food-borne pathogens can actually develop HHP resistance. In this study, we therefore examined and compared the intrinsic potentials for HHP resistance development among strains of Escherichia coli, Shigella flexneri, Salmonella enterica serovars Typhimurium and Enteritidis, Yersinia enterocolitica, Aeromonas hydrophila, Pseudomonas aeruginosa, and Listeria innocua using a selective enrichment approach. Interestingly, of all strains examined, the acquisition of extreme HHP resistance could be detected in only some of the E. coli strains, indicating that a specific genetic predisposition might be required for resistance development. Furthermore, once acquired, HHP resistance proved to be a very stable trait that was maintained for >80 generations in the absence of HHP exposure. Finally, at the mechanistic level, HHP resistance was not necessarily linked to derepression of the heat shock genes and was not related to the phenomenon of persistence.
INTRODUCTION
High hydrostatic pressure (HHP) processing is currently considered as one of the most promising nonthermal food preservation techniques and is being used for the commercial pasteurization of an increasing number of food products (7, 18). Typically, pressures in the range of 200 to 600 MPa are applied to inactivate food-borne pathogens and spoilage microorganisms, in order to enhance the safety and extend the shelf life of the product (5, 10, 17, 27). Since this process can be conducted at ambient temperature or in refrigerated conditions, products generally incur less deterioration of nutritional value, flavor, color, and texture than occurs with heating (19).
A sustained further exploitation of HHP technology in the food industry, however, requires a more profound understanding of the impact of pressure on microorganisms. In this context, for reasons still not understood, it appears that the susceptibility of vegetative bacteria to HHP varies significantly among different genera and species (2, 28), and even within species as well (4, 23, 31). Indeed, when exposure is to an identical HHP treatment (345 MPa, 5 min, 25°C), the difference in inactivation among different strains of Listeria monocytogenes or Escherichia coli O157:H7 can amount up to 3.5 and 5.6 log cycles, respectively (2). Moreover, studies with E. coli and L. monocytogenes have further revealed that HHP resistance seems to be a trait that can be readily acquired (9, 13, 34). As such, Karatzas et al. (13–15) obtained HHP-resistant mutants of L. monocytogenes Scott A from the small fraction of cells surviving a single HHP shock of 400 MPa (20 min). These spontaneous mutants were typically impaired in the CtsR regulator, which represses type 3 heat shock proteins, and de novo introduction of a ctsR-null allele correspondingly imposed HHP resistance on L. monocytogenes. Interestingly, because of a short but unstable tandem repeat tract in the ctsR gene, it was shown that ctsR mutants actually constituted a preexisting subpopulation in stationary-phase cultures of L. monocytogenes originating from a wild-type inoculum, which explained their isolation already after a single HHP shock (14, 15). In contrast, our group demonstrated the isolation of extremely HHP-resistant mutants of E. coli MG1655 (up to 2 GPa) during a selective enrichment approach based on consecutive cycles of increasingly severe HHP shocks with intermittent resuscitation and outgrowth of the surviving population (9, 34). While their level of HHP resistance by far exceeds that of the L. monocytogenes ctsR mutants, these E. coli MG1655 mutants similarly displayed derepression of a number of heat shock genes (1). Unfortunately, the genetic basis of this extreme HHP resistance in E. coli still remains elusive, although it can reasonably be anticipated that a number of different mutations are required and that the corresponding mutants concomitantly do not naturally preexist in a wild-type population of E. coli MG1655. Spurred by the apparent potential of E. coli MG1655 to develop extreme HHP resistance, and by the importance of this phenomenon for the efficacy and safety of HHP processing, we investigated how readily this trait could evolve in food-related pathogens and how stably it would be maintained in the absence of HHP exposure.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The different bacteria used in this work are listed in Table 1. E. coli 536 was kindly provided by Erick Denamur (Inserm U722, Paris, France), while the E. coli MG1655A7 mutant as well as its corresponding parental strain (i.e., E. coli MG21) was kindly provided by Nathalie Questembert-Balaban (Hebrew University, Jerusalem, Israel).
Table 1.
Bacterial strains and plasmids used in this study
| Bacterium or plasmid | Characteristic | Source or reference |
|---|---|---|
| Bacteria | ||
| E. coli | ||
| MG1655 | Parental strain | 8 |
| DVL20 | HHP-resistant mutant of MG1655 | This work |
| MG21 | MG1655 zde-264::Tn10, parental strain of MG1655A7 | 29 |
| MG1655A7 | MG1655 zde-264::Tn10 hipA7, mutant of MG21 with increased persistence | 29 |
| ATCC 11775 | Parental strain | ATCCa |
| ATCC 43888 | Parental strain, O157:H7 serotype | ATCCa |
| DVL24 | HHP-resistant mutant of ATCC 43888 (O157:H7) | This work |
| 536 | Parental strain | 33 |
| DVL25 | HHP-resistant mutant of 536 | This work |
| S. Typhimurium LT2 | Parental strain | SGSCb |
| S. Enteritidis ATCC 13076 | Parental strain | ATCCa |
| S. flexneri LMG 10472 | Parental strain | BCCMc |
| Y. enterocolitica ATCC 9610 | Parental strain | ATCCa |
| A. hydrophila ATCC 7966 | Parental strain | ATCCa |
| P. aeruginosa PAO1 | Parental strain | 26 |
| L. innocua LMG 11387 | Parental strain | BCCMc |
| Plasmid | ||
| pAA212 | dnaK-gfp transcription fusion cloned in pFPV25 | 1 |
American Type Culture Collection, Manassas, Virginia.
Salmonella Genetic Stock Center, Calgary, Canada.
Belgian Coordinated Collection of Microorganisms, Ghent, Belgium.
Unless stated otherwise, stationary-phase cultures were obtained by aerobic growth with shaking (200 rpm) for 23 h in tryptone soy broth (TSB) (Oxoid, Basingstoke, United Kingdom) at 30°C (Pseudomonas aeruginosa, Aeromonas hydrophila, Yersinia enterocolitica, and Listeria innocua) or at 37°C (E. coli, Salmonella enterica serovars Typhimurium and Enteritidis, and Shigella flexneri). When necessary, a final concentration of 100 μg/ml ampicillin (Ap100; Applichem, Darmstadt, Germany) was used to select for the presence of pAA212 (encoding the promoter of the dnaK heat shock gene upstream of the green fluorescent protein gene [gfp]; i.e., PdnaK-gfp reporter) (Table 1).
During serial passaging (in the absence of HHP treatment), stationary-phase cultures were repeatedly diluted 1/10,000 in prewarmed TSB and grown overnight (24 h) to stationary phase again, until successive growth for ca. 80 generations was achieved (i.e., 6 growth cycles).
Treatment with HHP.
Cells from a stationary-phase culture were harvested by centrifugation (4,000 × g; 5 min) and resuspended in an equal volume of 0.85% KCl. Subsequently, 200 μl of the suspension was heat sealed in a sterile polyethylene bag after exclusion of the air bubbles and subjected to high hydrostatic pressure (HHP) (100 to 800 MPa) for 15 min in an 8-ml pressure vessel (Resato, Roden, The Netherlands), held at 20°C with an external water jacket connected to a cryostat. Please note that both the slow pressure increase (100 MPa/min) and the external water jacket attenuate adiabatic heating during pressure build-up, and conservative estimates indicate only a transient increase in sample temperature of ≤13°C at 800 MPa. Finally, decompression was almost instantaneous. After HHP treatment, the culture was used for determination of viability (see below). During the selection procedure for HHP-resistant mutants, an HHP-treated culture was also inoculated 1/100 into fresh, prewarmed TSB and regrown for 23 h at the appropriate temperature (30 or 37°C) prior to a next round of pressurization.
Determination of viability.
Appropriate dilutions of a sample were prepared in 0.85% KCl with 0.1% peptone (i.e., peptone physiological salt [PPS]) and subsequently spread plated (100 μl) or spot plated (5 μl) on tryptone soy agar (TSA). After 24 h of incubation at the optimal growth temperature for the respective bacterium (e.g., 30 or 37°C; see above), the plates were counted and the logarithmic reduction factor was calculated as log(N0/N), in which N0 and N represent the number of survivors in CFU per ml prior and after the HHP treatment, respectively. Please note that the detection limit was 10 or 200 CFU/ml for spread- or spot-plated samples, respectively.
Determination of growth curves.
In order to compare growth curves among different strains, the optical density at 600 nm (OD600) of growing cultures was measured over time using a Bioscreen C plate-reader system (Thermo Labsystems Oy, Helsinki, Finland). For each experiment conducted on the Bioscreen C system, stationary-phase precultures were grown overnight as described above, after which the culture was diluted to approximately 105 CFU/ml in prewarmed lysogeny broth (LB) (24). Next, 300 μl of the corresponding suspension was placed in a honeycomb well and incubated in the Bioscreen C system for a 24-h period at 37°C, with regular shaking and automatic OD600 measurements every 10 min. Growth curves were averaged across three replicate cultures, and standard variations were <8.5%.
Measurement of PdnaK-gfp expression.
Strains of E. coli were first transformed with pAA212 (encoding PdnaK-gfp) by electroporation and subsequently purified. For measurement of basal PdnaK-gfp expression, cultures of the corresponding strains were grown to stationary phase in LB Ap100 at 37°C prior to analysis of GFP fluorescence. For measurement of induction by heat shock, on the other hand, stationary cultures were diluted 1/100 in fresh prewarmed LB Ap100 and further incubated at 37°C until late exponential phase (OD600 = 0.6). Portions (1 ml) of these cultures were subsequently pelleted by centrifugation (4,000 × g; 5 min) and resuspended in the same volume of fresh prewarmed LB, after which an aliquot (500 μl) was placed in a water bath at 50°C for 15 min. Heat-shocked and untreated control cultures were maintained at 37°C for 5 h prior to measurement of GFP fluorescence.
For measurement of GFP fluorescence, 200-μl samples were transferred to microplate wells and placed in a Fluoroscan Ascent FL (Thermo Labsystems, Brussels, Belgium), after which fluorescence at 520 nm was measured using an excitation wavelength of 480 nm. The obtained fluorescence values were subsequently divided by the OD600 of the same sample and expressed as relative fluorescence units (i.e., RFU = fluorescence/OD600).
Determination of persistent fraction with ampicillin.
To determine the fraction of persister cells, the method as described by De Groote et al. (6) was adapted. Briefly, stationary-phase cultures were diluted 1/100 in 4 ml of fresh, prewarmed LB, containing 100 μg/ml of ampicillin. Cultures were subsequently further incubated at 37°C with shaking (200 rpm) for 5 h, after which samples were serially diluted in PPS and plated on LB agar for the determination of survivors (corresponding to the persisters).
RESULTS
Development of HHP resistance in different food-borne pathogens.
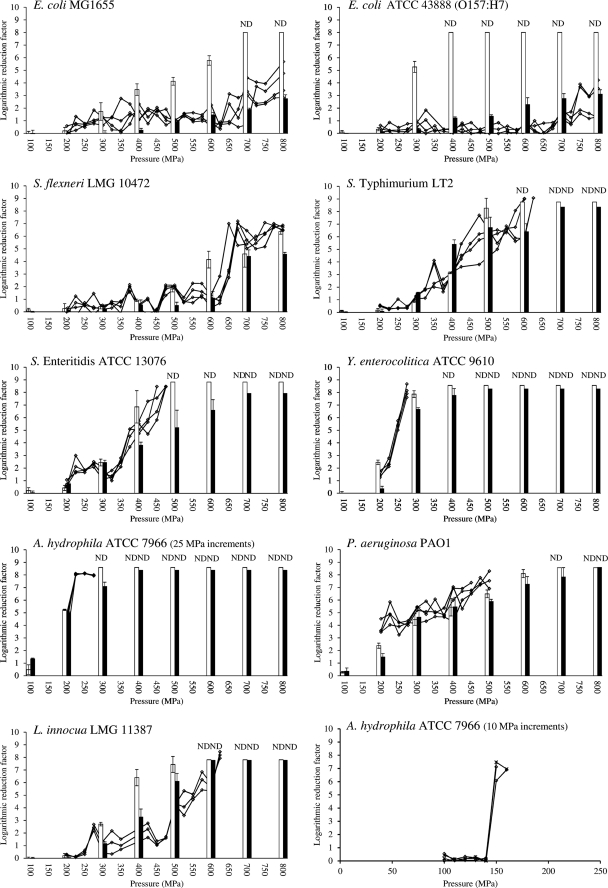
To examine their intrinsic abilities to evolve extreme HHP resistance, different food-borne pathogens (including E. coli, S. flexneri, S. Typhimurium, S. Enteritidis, Y. enterocolitica, A. hydrophila, P. aeruginosa, and L. innocua) were individually subjected to iterative cycles of increasingly severe HHP shocks with intermittent outgrowth of the survivors. Starting from an initial exposure of 200 MPa, the pressure was incrementally increased by 25 MPa each cycle, and this selection regimen was maintained until bacterial growth was no longer observed after HHP treatment or until the maximum pressure of the equipment (i.e., 800 MPa) was reached. For each of the examined strains, four independent cultures underwent the above selection procedure, after which 10 purified clones out of each evolved culture were individually examined for HHP resistance. Of these mutants, HHP inactivation of the most resistant derivative was compared to that of its corresponding parental strain (Fig. 1).
Fig 1.
Selective enrichment of different food-borne pathogens toward HHP resistance (at 20°C). For each strain, four independent axenic cultures were iteratively exposed to progressively intensifying pressure shocks (15 min; 20°C), with intermittent resuscitation and growth of the survivors. Line graphs (-♢-) present the inactivation during the stepwise selection regimen with 10- or 25-MPa increments and are expressed as the logarithmic reduction factor [i.e., log(N0/N)]. At the end of the selection regimen, the most HHP-resistant clone that had enriched in the corresponding cultures was isolated, and its acquired HHP resistance (black bars) was determined and compared to that of the original parent strain (white bars). Results shown in the bars are expressed as mean values ± standard deviations of three independent replicates. ND as well as the abrupt end of a line graph at pressures below 800 MPa indicate that the corresponding N fell below the detection limit, which was 10 or 200 CFU/ml for results shown in bars or line graphs, respectively.
Selection of E. coli MG1655 toward extreme HHP resistance was already demonstrated earlier (9, 34), and this strain was included as a positive control in this study. In agreement with those earlier findings, high levels of piezotolerance were reproducibly (i.e., in all four of the independently evolved lineages) achieved after 25 consecutive cycles of HHP treatment (Fig. 1). While survival of the E. coli MG1655 parental strain already fell below the detection limit at 700 MPa (i.e., >8.0 log inactivation), its most HHP-resistant derivative (i.e., DVL20) obtained in this study only decreased 2.8 log cycles at 800 MPa (Fig. 1).
Subsequently, we examined HHP resistance development in E. coli O157:H7 and some other food-borne pathogens from related Enterobacteriaceae, such as S. flexneri, S. Typhimurium, S. Enteritidis, and Y. enterocolitica (Fig. 1). Interestingly, of these strains, E. coli O157:H7 readily developed HHP resistance to a similar extent as E. coli MG1655, even though the original parental strain was markedly more HHP sensitive. Furthermore, the wild-type strain of S. flexneri, a species that is in fact indistinguishable from E. coli (11), already exhibited an exceptional HHP resistance prior to the imposed selection regimen, although this resistance did not further develop to the same extent as in E. coli MG1655 and O157:H7 and resulted in 6.4-log cycle inactivation at 800 MPa (Fig. 1, white bars). In contrast, however, the evolving lineages of S. Typhimurium, S. Enteritidis, and Y. enterocolitica failed to survive the imposed selection regimen well before 800 MPa could be reached. As such, only limited HHP resistance could be raised in the most resistant derivatives of S. Typhimurium (6.4-log cycle inactivation at 600 MPa) and S. Enteritidis (6.6-log cycle inactivation at 600 MPa), while Y. enterocolitica failed to develop any notable HHP resistance at pressures of >400 MPa (Fig. 1).
Interestingly, HHP resistance likewise failed to develop in A. hydrophila, P. aeruginosa PAO1, and the Gram-positive bacterium L. innocua subjected to the same selection procedure (Fig. 1). Since A. hydrophila appeared to be the most HHP-sensitive parental strain, being completely inactivated already after a 300-MPa shock, we wondered whether the failure to develop HHP resistance could have been the result of a too-severe selection procedure, rather than this strain's intrinsic inability to evolve HHP resistance. For this reason, we subjected A. hydrophila to a less stringent selection procedure by (i) starting the selection regimen at 100 MPa instead of 200 MPa and (ii) lowering the pressure increment after each cycle from 25 to only 10 MPa. However, despite this attenuation, still no increased HHP resistance could be observed, and after 7 cycles of HHP treatments, no surviving cells could be detected (Fig. 1).
Development of HHP resistance in different E. coli strains.
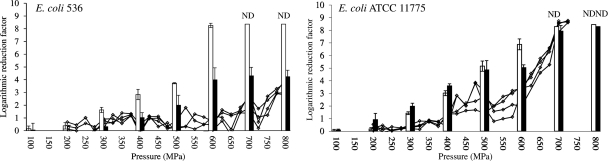
Since the potential to acquire extreme HHP resistance so far seemed restricted to E. coli MG1655 and O157:H7 (Fig. 1), we decided to examine additional E. coli strains to check whether this ability is universal among isolates of this species. As such, HHP resistance development was examined in E. coli 536 and ATCC 11775 (Fig. 2). Interestingly, while the former readily developed HHP resistance (4.2-log cycle inactivation at 800 MPa), strain ATCC 11775 consistently failed to do so, indicating that not all E. coli isolates share the predisposition to become extremely HHP resistant.
Fig 2.
Selective enrichment of different E. coli strains toward HHP resistance (at 20°C). For each strain, four independent axenic cultures were iteratively exposed to progressively intensifying pressure shocks (15 min; 20°C), with intermittent resuscitation and growth of the survivors. Line graphs (-♢-) present the inactivation during the stepwise selection regimen with 25-MPa increments and are expressed as the logarithmic reduction factor [i.e., log(N0/N)]. At the end of the selection regimen, the most HHP-resistant clone that had enriched in the corresponding cultures was isolated and its acquired HHP resistance (black bars) was determined and compared to that of the original parent strain (white bars). Results shown in the bars are expressed as mean values ± standard deviations of three independent replicates. ND as well as the abrupt end of a line graph at pressures below 800 MPa indicate that the corresponding N fell below the detection limit, which was 10 or 200 CFU/ml for results shown in bars or line graphs, respectively.
As acquisition of extreme HHP resistance was observed to coincide with increased expression of heat shock proteins in three previously evolved mutants of E. coli MG1655 (1, 9), we wondered whether or not this link was maintained in the HHP-resistant mutants from different E. coli strains obtained in this study. We therefore used promoter activity stemming from a PdnaK-gfp reporter plasmid as a reliable proxy for derepression of the heat shock regulon (Table 2). Please note that all examined parental and mutant strains exhibited increased PdnaK-gfp activity after heat shock, which (i) validated reporter fusion functionality in these different backgrounds and (ii) demonstrated that the heat shock regulon was not completely derepressed in any of the derived mutants. Interestingly, in contrast to current and earlier results with E. coli MG1655 (1) (Table 2), the obtained HHP-resistant mutant of E. coli O157:H7 (i.e., DVL24) did not exhibit constitutively increased PdnaK-gfp levels compared to its parent strain. This suggests that different mechanisms to evolve extreme HHP resistance in the cell might exist and that these mechanisms do not necessarily depend on the constitutive derepression of heat shock genes.
Table 2.
Basal and heat shock-induced expression of PdnaK-gfp in indicated backgrounds of E. coli
| Straina | RFUb,c noninduced | RFUb,c induced by heat shock | Ratio of parental strain vs HHP-resistant mutant | Ratio of induced vs noninduced strain |
|---|---|---|---|---|
| E. coli MG1655/pAA212 (parental strain) | 4.9 ± 0.2 | 28.1 ± 1.2 | 1.0 | 5.7 |
| E. coli DVL20/pAA212 (HHP-resistant mutant) | 7.5 ± 0.7 | 13.6 ± 0.5 | 1.5 | 1.8 |
| E. coli O157:H7/pAA212 (parental strain) | 3.6 ± 0.1 | 11.8 ± 0.5 | 1.0 | 3.3 |
| E. coli DVL24/pAA212 (HHP-resistant mutant) | 3.2 ± 0.2 | 10.3 ± 0.4 | 0.9 | 3.2 |
Please note that E. coli 536 and its HHP-resistant mutant (DVL25) were not included, as we were unable to transform them with plasmid pAA212.
RFU, relative fluorescence units.
All data shown are mean values ± standard deviations from an experiment with three independent replicates.
Burden and stability of the HHP resistance trait.
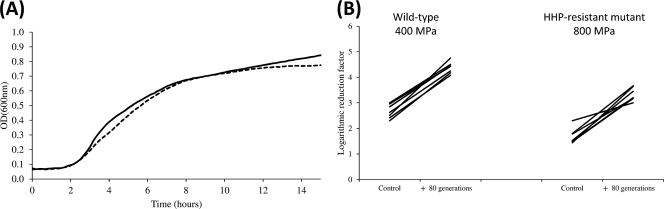
Subsequently, we examined the stability of extreme HHP resistance in the evolved mutants, as this important issue has never been addressed before. First of all, the general fitness levels of wild-type E. coli MG1655 and its HHP-resistant mutant derived in this study (i.e., DVL20) were compared based on their growth curves in standard conditions. Since no significant differences in growth characteristics were observed, the selected HHP-resistant E. coli mutant appears not to carry important physiological handicaps under these conditions (Fig. 3A). Moreover, similar results were obtained for the isolated HHP-resistant mutants of E. coli O157:H7 (i.e., DVL24) and 536 (i.e., DVL25) (data not shown).
Fig 3.
(A) Growth curves of E. coli MG1655 (straight line) and its corresponding HHP-resistant mutant (dotted line) (i.e., DVL20), monitored as OD600 in LB growth medium at 37°C. Growth curves were averaged across three replicate cultures. Standard variations were below <8.5% and were not included. (B) Change in logarithmic reduction factor of seven independent axenic cultures of both E. coli MG1655 (treated at 400 MPa, 15 min, 20°C) and its HHP-resistant mutant (DVL20; treated at 800 MPa, 15 min, 20°C) after growth to stationary phase for 24 h (i.e., control) or for an additional ca. 80 generations (i.e., + 80 generations) in the absence of HHP exposure. Please note that lines are used to connect data points from the same lineage.
Subsequently, we examined how stable the trait of HHP resistance can be maintained in the absence of HHP as a selective force. Therefore, we determined the residual HHP resistance of wild-type MG1655 and its resistant mutant (i.e., DVL20) after subculturing seven independent lineages of each strain for ca. 80 generations in the absence of any HHP stress. Interestingly, although subculturing seemed to cause a general decrease in HHP resistance in both the wild type (on average 1.7-fold-increased inactivation at 400 MPa) and the mutant strain (on average 2.0-fold-increased inactivation at 800 MPa), the HHP-resistant mutant did not at all revert to parental HHP sensitivity (Fig. 3B). Together, these findings indicate that acquisition of extreme HHP resistance does not constitute a burden for the cell and that this trait is not readily lost in the absence of the selective pressure exerted by HHP.
Investigating a possible link between HHP resistance and persistence in E. coli.
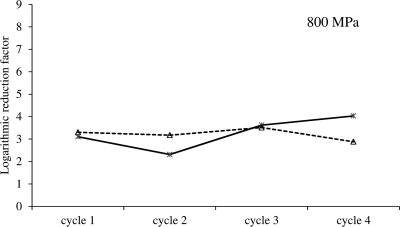
Interestingly, despite the ability of a resistant E. coli population to survive a severe HHP shock, the vast majority (>90% or 1 log cycle) of its cells still fail to survive at relatively mild pressures (Fig. 1 and 2, black bars). In this context, it should be noted that it is unlikely that the surviving cells would be genetically different from the inactivated ones, as they individually grow up to yield a population with an HHP sensitivity similar to that of their parental one. Furthermore, selective enrichment experiments aimed to select for increased survival at a given dose of HHP (i.e., through repeated cycling at 800 MPa) were unable to affect this phenotype (Fig. 4). As all cells inevitably experience exactly the same HHP shock, these observations would imply that the mechanism of HHP resistance is absent or not at work in the majority of the mutant population. As such, this feature is reminiscent of the phenomenon of bacterial persistence, in which a small fraction of transiently metabolically dormant cells in a clonal population display increased resistance to inimical stresses such as antibiotics (16, 22).
Fig 4.
Selective enrichment of two independent cultures of E. coli DVL20 toward increased HHP resistance at 800 MPa (20°C). The two axenic cultures were iteratively exposed to HHP shocks of 800 MPa (15 min, 20°C), with intermittent resuscitation and growth. Line graphs (----△---- and  ) present the inactivation after each HHP shock and are expressed as log(N0/N).
) present the inactivation after each HHP shock and are expressed as log(N0/N).
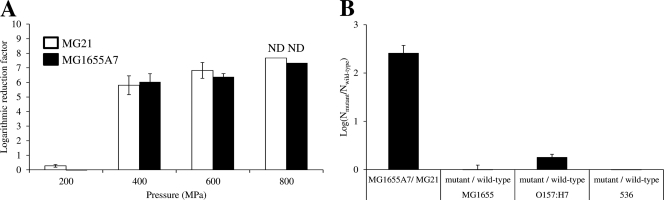
In order to examine the possible involvement of persistence also in HHP resistance, HHP inactivation of an E. coli MG1655 hipA7 mutant (i.e., MG1655A7), exhibiting a 1,000-fold-increased fraction of persister cells (21), was compared to that of its parental strain (i.e., MG21) (29) (Fig. 5A). However, increasing the persister fraction did not affect the number of survivors after any of the HHP shocks imposed. Conversely, we also determined the number of persisters present in populations of E. coli MG1655, O157:H7, 536, and their corresponding HHP-resistant mutants (DVL20, DVL24, and DVL25), based on the capacity of such cells to resist ampicillin-induced cell lysis (16) (Fig. 5B). The hipA7 mutant was included as a positive control to validate the assay and displayed a ca. 255-fold increase in persisters in our assay. However, of the HHP-resistant E. coli mutants, only DVL24 (derived from E. coli O157:H7) showed a significant 1.8-fold increase in persister cells (Fig. 5B), which is nevertheless too modest to explain its extreme HHP resistance.
Fig 5.
(A) Logarithmic reduction factor [i.e., log(N0/N)] of E. coli MG21 (□) and its hipA7 derivative (i.e., MG1655A7; with increased persister fraction) (■) after HHP treatment at the indicated pressures (15 min, 20°C). ND indicates that the corresponding N fell below the detection limit of 10 CFU/ml. (B) Difference in amount of persister cells among indicated wild-type and derived mutant strains of E. coli, expressed as log(Nmutant/Nwild-type) in which Nmutant and Nwild-type represent the persister fraction (in CFU/ml) of the corresponding populations. Please note that the MG1655 hipA7 mutant (i.e., MG1655A7) was included as a positive control. All data shown are averages ± standard deviations from an experiment with three independent cultures of each strain.
DISCUSSION
The increasing use of HHP pasteurization in the food industry necessitates a better understanding of bacterial resistance development against this stress. While our previous studies have addressed the extent of extreme HHP resistance in E. coli (9, 34), this study reports on the emergence and stability of extreme HHP resistance in different food-borne pathogens.
During examination of the intrinsic capacity of these bacteria to develop extreme HHP resistance when selected for, we surprisingly found that acquisition of this trait seemed limited to some strains of E. coli, as it was virtually absent from Y. enterocolitica, A. hydrophila, P. aeruginosa, and L. innocua. Moreover, while most of the E. coli strains reproducibly evolved to readily survive 800 MPa, close relatives and important pathogens such as S. Typhimurium and S. Enteritidis achieved only limited resistance to pressures up to 600 MPa. A notable exception in this context was posed by S. flexneri, a phylogenetically indistinguishable relative of E. coli, of which the wild-type strain already exhibited extreme HHP resistance prior to the imposed selection regimen, further underscoring the finding that especially strains of E. coli are poised to become extremely HHP resistant. It should be noted in this regard that E. coli and Shigella spp. harbor a number of important pathogens whose increased HHP survival might compromise the safety of HHP processing in food industry (3, 12, 32). In fact, a distinct representative of one of these pathogens, i.e., the O157:H7 isolate designated ATCC 43888, was shown to readily evolve explicit HHP resistance.
Interestingly, one of the examined E. coli isolates (i.e., ATCC 11775) reproducibly failed to acquire HHP resistance, and the observation that the ability to develop HHP resistance is not at all common among different bacterial species, nor obligatorily shared among closely related strains of E. coli, seems to suggest the existence of a particular set of genes from which this trait is able to evolve. As such, the possibility cannot be excluded that specific isolates of other relevant species could harbor the correct genetic predisposition to become HHP resistant. Clearly, identification of this genetic signature will shed light on the evolution and mechanism of HHP resistance and might in the future even serve to more rapidly identify strains able to develop HHP resistance when exposed to this stress.
Once acquired, HHP resistance in E. coli did not seem to impair cellular fitness nor revert during at least 80 generations in the absence of any HHP exposure, underscoring both the limited burden and genetic stability of this trait. Surprisingly, however, serial culturing for 80 generations in the absence of HHP did increase HHP sensitivity in both the wild type and HHP-resistant mutants alike, although the latter strain remained abundantly more resistant than its original parent. While the underlying mechanism behind this observation remains unclear, prolonged growth in liquid broth during serial passaging has previously been shown to select for an altered genetic constitution (20, 30). As serial passaging is also an integral part of the employed regimen selecting for HHP resistance, it is tempting to think that the development of this trait would even be more rapid and pronounced in the absence of the apparent counterselection imposed by subculturing.
Interestingly, it was also observed that not all HHP-resistant E. coli mutants displayed the anticipated constitutive derepression of the heat shock regulon (as indicated by the PdnaK-gfp reporter) that had previously been proposed as a possible mechanism for HHP resistance (1). Furthermore, complementary to this observation, a recently evolved heat-resistant mutant of E. coli MG1655 (34) exhibited constitutive derepression of the heat shock regulon (ca. 5-fold induction of PdnaK-gfp compared to its parental strain; data not shown) without displaying a marked increase in HHP resistance. As such, while it has been shown that the heat shock response can transiently increase HHP resistance in E. coli (1), the causal relationship between constitutive derepression of heat shock proteins and mutationally acquired extreme HHP resistance remains unclear.
Since the majority of cells within an extremely HHP-resistant population still fail to survive relatively mild pressures (∼400 MPa) and thus seem to remain pressure sensitive, we decided to specifically examine the putative involvement of persistence in HHP resistance. Persistence is a well-established phenomenon in antimicrobial therapy and refers to a small number of cells within a clonal population that are refractory to the effects of inimical treatments (16, 21, 22). Interestingly, this specific form of resistance stems from the transient metabolic dormancy of persister cells, which is assumed to attenuate stress-induced deregulation of cellular physiology. Although the exact molecular mechanism behind persistence remains unsolved, a particular mutation in the hipA gene (yielding the hipA7 allele) drastically increases the fraction of persisters in a population (25). Nevertheless, E. coli cells carrying this mutation and displaying a ca. 255-fold-increased persistence to ampicillin exposure did not show any increased HHP resistance. Conversely, none of the HHP-resistant mutants of E. coli isolated in this study (i.e., DVL20, -24, and -25) showed a markedly increased persister subpopulation that could otherwise explain their resilience to HHP stress. As such, it is unlikely that the metabolic dormancy that is characteristic for persisters, and enables them to survive antibiotic treatment, plays any role in HHP survival.
Among the bacteria tested in this study, the capacity to develop extreme HHP resistance was limited to the species of E. coli, although not all of its isolates share this characteristic. Once acquired, HHP resistance appears to be a stable trait with no obvious impact on cellular fitness. Furthermore, HHP resistance is not necessarily linked to derepression of the heat shock response as previously proposed, and further research is necessary to mechanistically understand and perhaps predict the emergence of HHP resistance in bacteria.
ACKNOWLEDGMENTS
We thank Nathalie Questembert-Balaban and Erick Denamur for kindly sharing strains.
This work was supported by a doctoral fellowship of the Flemish Agency for Innovation by Science and Technology (IWT-Vlaanderen; to D.V.) and a grant from the Research Fund of the Catholic University of Leuven (KU Leuven STRT1/10/036; to A.A.).
Footnotes
Published ahead of print 17 February 2012
REFERENCES
- 1. Aertsen A, et al. 2004. Heat shock protein-mediated resistance to high hydrostatic pressure in Escherichia coli. Appl. Environ. Microbiol. 70:2660–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alpas H, et al. 1999. Variation in resistance to hydrostatic pressure among strains of foodborne pathogens. Appl. Environ. Microbiol. 65:4248–4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ashida H, et al. 2011. Shigella are versatile mucosal pathogens that circumvent the host innate immune system. Curr. Opin. Immunol. 23:448–455 [DOI] [PubMed] [Google Scholar]
- 4. Benito A, Ventoura G, Casadei M, Robinson T, Mackey B. 1999. Variation in resistance of natural isolates of Escherichia coli O157 to high hydrostatic pressure, mild heat, and other stresses. Appl. Environ. Microbiol. 65:1564–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Considine K, Kelly A, Fitzgerald G, Hill C, Sleator R. 2008. High-pressure processing-effects on microbial food safety and food quality. FEMS Microbiol. Lett. 281:1–9 [DOI] [PubMed] [Google Scholar]
- 6. De Groote VN, et al. 2009. Novel persistence genes in Pseudomonas aeruginosa identified by high-throughput screening. FEMS Microbiol. Lett. 297:73–79 [DOI] [PubMed] [Google Scholar]
- 7. Gould G. 2000. Preservation: past, present and future. Br. Med. Bull. 56:84–96 [DOI] [PubMed] [Google Scholar]
- 8. Guyer M, Reed R, Steitz J, Low K. 1981. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb. Symp. Quant. Biol. 45:135–140 [DOI] [PubMed] [Google Scholar]
- 9. Hauben KJA, et al. 1997. Escherichia coli mutants resistant to inactivation by high hydrostatic pressure. Appl. Environ. Microbiol. 63:945–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoover D, Metrick C, Papineau A, Farkas D, Knorr D. 1989. Biological effects of high hydrostatic-pressure on food microorganisms. Food Technol. 47:99–107 [Google Scholar]
- 11. Jin Q, et al. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30:4432–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 13. Karatzas KA, Bennik MH. 2002. Characterization of a Listeria monocytogenes Scott A isolate with high tolerance towards high hydrostatic pressure. Appl. Environ. Microbiol. 68:3183–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karatzas KA, Valdramidis VP, Bennik MH. 2005. Contingency locus in ctsR of Listeria monocytogenes Scott A: a strategy for occurrence of abundant piezotolerant isolates within clonal populations. Appl. Environ. Microbiol. 71:8390–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karatzas KA, et al. 2003. The CtsR regulator of Listeria monocytogenes contains a variant glycine repeat region that affects piezotolerance, stress resistance, motility and virulence. Mol. Microbiol. 49:1227–1238 [DOI] [PubMed] [Google Scholar]
- 16. Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13–18 [DOI] [PubMed] [Google Scholar]
- 17. Knorr D. 1993. Effects of high-hydrostatic-pressure processes on food safety and quality. Food Technol. 47:156–161 [Google Scholar]
- 18. Knorr D. 1999. Novel approaches in food-processing technology: new technologies for preserving foods and modifying function. Curr. Opin. Biotechnol. 10:485–491 [DOI] [PubMed] [Google Scholar]
- 19. Knorr D, Mertens B. 1992. Development of nonthermal processes for food preservation. Food Technol. 46:123–134 [Google Scholar]
- 20. Langridge GC, et al. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 19:2308–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48–56 [DOI] [PubMed] [Google Scholar]
- 22. Lewis K. 2010. Persister cells. Annu. Rev. Microbiol. 64:357–372 [DOI] [PubMed] [Google Scholar]
- 23. Malone A, Chung Y, Yousef A. 2006. Genes of Escherichia coli O157:H7 that are involved in high-pressure resistance. Appl. Environ. Microbiol. 72:2661–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25. Moyed H, Bertrand K. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155:768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nicas T, Hancock R. 1980. Outer-membrane protein H-1 of Pseudomonas aeruginosa involvement in adaptive and mutational resistance to ethylenediaminetetraacetate, polymyxin-B, and gentamicin. J. Bacteriol. 143:872–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Patterson MF. 2005. Microbiology of pressure-treated foods. J. Appl. Microbiol. 98:1400–1409 [DOI] [PubMed] [Google Scholar]
- 28. Patterson MF, Quinn M, Simpson R, Gilmour A. 1995. Sensitivity of vegetative pathogens to high hydrostatic-pressure treatment in phosphate-buffered saline and foods. J. Food Prot. 58:524–529 [DOI] [PubMed] [Google Scholar]
- 29. Pearl S, Gabay C, Kishony R, Oppenheim A, Balaban NQ. 2008. Nongenetic individuality in the host-phage interaction. PLoS Biol. 6:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Philippe N, Crozat E, Lenski RE, Schneider D. 2007. Evolution of global regulatory networks during a long-term experiment with Escherichia coli. Bioessays 29:846–860 [DOI] [PubMed] [Google Scholar]
- 31. Robey M, et al. 2001. Variation in resistance to high hydrostatic pressure and rpoS heterogeneity in natural isolates of Escherichia coli O157:H7. Appl. Environ. Microbiol. 67:4901–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith JL, Fratamico PM, Gunther NW. 2007. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 4:134–163 [DOI] [PubMed] [Google Scholar]
- 33. Touchon M, et al. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vanlint D, et al. 2011. Rapid acquisition of Gigapascal-high-pressure resistance by Escherichia coli. mBio 2:e00130–e00110 [DOI] [PMC free article] [PubMed] [Google Scholar]