Abstract
To understand the etiology of exposure-related diseases and to establish standards for reducing the risks associated with working in contaminated environments, the exact nature of the bioaerosol components must be defined. Molecular biology tools were used to evaluate airborne bacterial and, for the first time, archaeal content of dairy barns. Three air samplers were tested in each of the 13 barns sampled. Up to 106 archaeal and 108 bacterial 16S rRNA genes per m3 of air were detected. Archaeal methanogens, mainly Methanobrevibacter species, were represented. Saccharopolyspora rectivirgula, the causative agent of farmer's lung, was quantified to up to 107 16S rRNA genes per m3 of air. In addition, a wide variety of bacterial agents were present in our air samples within the high airborne bioaerosol concentration range. Despite recommendations regarding hay preservation and baling conditions, farmers still develop an S. rectivirgula-specific humoral immune response, suggesting intense and continuous exposure. Our results demonstrate the complexity of bioaerosol components in dairy barns which could play a role in occupational respiratory diseases.
INTRODUCTION
Microbial flora from natural sources, such as water, soil, plants, and animals, are well known and studied (14, 19, 41, 43). Yet little information on bioaerosols is available. This lack of knowledge impedes the understanding of the bioaerosol-related respiratory diseases etiology (3, 18, 46). Indeed, the airway mucosa is a primary entry site for toxic and pathogenic factors, but what the workers are exposed to and how it impacts respiratory health are poorly addressed. From what is currently known, the presence of microbial components assessed with simple culture methods cannot explain the whole variety of respiratory diseases. Nonviable agents of bioaerosols, such as toxins and antigens, can induce sensitization or toxic diseases (3, 22). Moreover, due to a lack of standardization between air samplers used for exposure assessment purposes, the robustness of the sampling protocol is worrisome.
Complex environments, such as agricultural facilities, are used to study the causal effects of bioaerosol agents on respiratory health, since several sources of biological material potentially associated with lung diseases are present. As an example, our team discovered high concentrations of archaea in swine barn bioaerosols (33), revealing for the first time a human exposure to archaea through the airborne route. Those archaea (8) and their subcomponents (36) were revealed to have a significant immunogenic potential. Molecular biology is the most efficient approach allowing the evaluation of exposure to archaea and any other nonculturable microorganisms that can have an immunogenic effect on human health (6, 11, 26, 33). Indeed, molecular methods were used in several studies to characterize airborne bioaerosols from various agricultural environments (20, 35).
The main objective of this project was to characterize the bacterial and archaeal loads from the bioaerosols of dairy barns, with special emphasis on Saccharopolyspora rectivirgula (a farmer's lung agent) (13) and archaeal species known to have an immunogenic potential (8), using molecular approaches. Secondary objectives included a comparison between different types of air samplers, particle size-selective analysis of bioaerosols, and workers' IgG responses to S. rectivirgula as a marker of exposure.
MATERIALS AND METHODS
Sampling, collecting, and processing methods.
Airborne dust was sampled at one site each from 13 Holstein dairy barns (eastern Quebec, Canada). Three different air samplers were used, namely, the Institute of Occupational Medicine cassettes (SKC, Ancaster, ON, Canada) or IOM samplers, loaded with 25-mm-diameter gelatin membranes (SKC) and plugged into a Giliar-5 pump (Levitt-Sécurité Limitée, Dorval, QC, Canada), at 2 liters/min (50% cutoff size of 4.0 μm); the Coriolis (Bertin Technologies, Montigny-le-Bretonneux, France), which collects 100% of the particles of 4.4 μm, at 100 liters/min and loaded with 15 ml of 0.9% saline solution; and the NIOSH two-stage bioaerosol cyclone (BC 251) sampler (31, 42), plugged into an AirCon-2 pump (Gilian) and sampling at 10 liters/min for particulate size separation. The size distribution for each stage (50% cutoff size) of the NIOSH sampler was as follows: 2.1 μm for the first stage and 0.41 μm for the second stage. The third stage was composed of a 0.4-μm polycarbonate filter. Air samples from dairy barns were collected in dairy barns next to the cows during the winter season, 1 m above the floor and as far as possible from ventilation sources, for 4 to 5 h during morning or evening cow milking. One sampling per dairy barn was conducted.
Samples from IOM cassettes were treated as previously described (33). Liquid samples from the Coriolis samplers were divided into aliquots of 1.5 ml and were centrifuged (10 min, 21,000 × g, room temperature). Filters from the NIOSH samplers were transferred in sterile tubes in which 2 ml of NaCl solution (0.9%) containing 0.05% Tween 20 was added. This solution was also added to the first (2 ml) and second (1 ml) stages of this sampler. After homogenization by vortexing (15 min), the suspensions obtained from the three stages were divided into aliquots of 1 ml and centrifuged (10 min, 21,000 × g, room temperature). Pellets were kept at −20°C until their use for total DNA extraction.
In order to evaluate plasma IgG against S. rectivirgula as a marker of exposure, venous blood from 29 dairy barn workers and 35 control subjects (having no contact history with dairy barns) was sampled in Vacuette collection tubes (Greiner Bio-One North America, Monroe, NC). Plasma was separated from red blood cells by centrifugation and was stored at −80°C. All subjects signed an informed consent form approved by our institution's research ethics board (REB).
Total DNA extraction.
A Qiagen QIAamp DNA extraction kit was used for total DNA extraction from airborne dust according to protocol 1, isolation of bacterial DNA from biological fluids, followed by protocol 2, DNA purification from tissues (37), with the required modification for bacteria (27, 33, 34). Cellular lysis was performed using proteinase K for 1.5 h.
Quantitative real-time PCR.
Quantitative real-time PCR was performed on a DNA Engine Opticon 2 (Bio-Rad, Mississauga, Canada). PCR for archaeal 16S rRNA genes was optimized for archaea-only amplification without bias for any archaeal phylum and was conducted using 0.5 μM A751F and A976R primers (5, 38) (Table 1). A total of 12.5 μl of iQ SYBR green Supermix (Bio-Rad Laboratories, Hercules, CA) and 2 μl of the DNA template were used in a 25-μl reaction mixture. The amplification program used for this PCR was as follows: one hold at 94°C for 5 min and then 35 cycles at 94°C for 10 s, 55.5°C for 20 s, fluorescence acquisition, and 72°C for 25 s, followed by one hold at 72°C for 10 min. A melting curve program was then performed to detect amplicon specificity by using the following program: 40°C to 94°C, read every 0.2 s, hold 1 s. Samples were considered positive for archaeal 16S rRNA genes when the melting temperature was around 88°C. Ten-fold serial dilutions of methanogenic archaea Methanosarcina mazei DNA (ATCC BAA-159D) were used for the standard curve. Data were acquired using the Opticon monitor software (Bio-Rad, version 2.02.24). The threshold was determined by the software with a standard deviation of 1.
Table 1.
Primers, probe, and GC clamps used in the study
| Primer, probe, or GC clamp | Nucleotide sequence (5′–3′) | Reference |
|---|---|---|
| A751F | CCG ACG GTG AGR GRY GAA | 5 |
| A976R | YCC GGC GTT GAM TCC AAT T | 38 |
| EUB F | GGT AGT CYA YGC MST AAA CG | 4 |
| EUB R | GAC ARC CAT GCA SCA CCT G | 4 |
| Probe EUB | FAM-TKC GCG TTG CDT CGA ATT AAW CCA C-TAMRA | 4 |
| Sac-86F | TGT GGT GGG GTG GAT GAG T | 40 |
| Sac-183R | ACC ATG CGG CAG AAT GTC CT | 40 |
| A333F | TCC AGG CCC TAC GGG | 38 |
| A751R(GC) | TTC RYC YCT CAC CGT CG | 5 |
| GC clamp archaea | CGC CCG CCG CGC CCC GCG CCC GGC CCG CCG CCC CCG CCC C | 32a |
| 341F | CCT ACG GGA GGC AGC AG | 32a |
| 907R | CCG TCA ATT CCT TTG AGT TT | 45 |
| GC clamp bacteria | CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG G | 32a |
PCR for bacterial 16S rRNA genes was conducted using the primers, probe (Table 1), and amplification program used by Bach et al. (4) with the iQ Supermix (Bio-Rad Laboratories, Hercules, CA). Ten-fold serial DNA dilutions of plasmid containing Escherichia coli (ATCC 25922) 16S rRNA gene sequences were used for the standard curve. Data analysis was performed as for archaeal detection. A quantitative PCR specific for S. rectivirgula was conducted using the primers used by Schäfer et al. (Table 1) (40). The amplification program was modified as follows: 3 min at 95°C, followed by 45 cycles of 30 s of denaturation at 98°C, 35 s of annealing at 59.6°C, and 45 s of elongation at 72°C. The standard curve was constructed using 10-fold serial dilutions of S. rectivirgula full 16S rRNA gene amplicons.
Denaturing gradient gel electrophoresis (DGGE).
PCR for archaeal 16S rRNA genes was conducted using 0.5 μM A333F and A751R(GC) primers (5, 38) (Table 1), 3.5 mM MgCl2, 100 μM dNTP, 5% (vol/vol) dimethyl sulfoxide (DMSO), 2.5 U Taq (Promega) polymerase, and 2 μl DNA template in a 50-μl reaction mixture. The amplification program used for this PCR was as follows: one hold at 94°C for 45 s and then 35 cycles at 94°C for 30 s, 54.8°C for 105 s, and 72°C for 1 min, followed by one hold at 72°C for 10 min. PCR for bacterial 16S rRNA genes was conducted using 341F(GC) and 907R primers (Table 1), and the amplification program used for this PCR was performed as previously described (25). PCR products were revealed by 1% agarose electrophoresis gel, and the amount of amplified DNA was quantified using a Bio-Rad molecular mass ladder and GenTools software (SynGen, Cambridge, United Kingdom) as previously described (25).
Profiles for amplified DNA biodiversity were produced by denaturing gradient gel electrophoresis (DGGE) on a DCode system (Bio-Rad) as previously described (33). A total of 100 ng of amplified DNA was loaded in 8% (archaea) and 6% (bacteria) polyacrylamide gels with 25 to 65% and 30 to 55% denaturing gradients for archaea and bacteria gels, respectively (from top to bottom). The electrophoresis and DNA fragment staining were carried out as previously described (33). DNA from polyacrylamide gel bands was excised using a micropipette tip and put into PCR mix for reamplification. Amplicons were sequenced (CHUL Research Centre) and proofread with FinchTV 1.4.0 software (Geospiza; Perkin Elmer, Seattle, WA). Amplicons that did not result in high-quality sequences were cloned into Qiagen pDrive cloning vector using the Qiagen PCR cloning kit. The DNA constructions were transformed into homemade E. coli DH5α competent cells with the electroporation method using the Bio-Rad Gene Pulser apparatus (Bio-Rad Laboratories) equipped with a Bio-Rad pulse controller as described by the manufacturer (Bio-Rad Laboratories, La Jolla, CA).
Phylogenetic analysis.
GelCompar II software version 6.5 (Applied Maths, Austin, TX) was used to normalize and compare DGGE profiles from each barn with hierarchical clustering to join similar profiles into groups (24). A 3% tolerance in the band position was applied. Similarity among profiles was calculated with the Jaccard coefficient to assess the frequency of DGGE bands in dairy barns sampled. Clustering was performed with the unweighted-pair group method using average linkages (UPGMA).
Each DNA sequence obtained was compared to sequences available in databases by using BLASTN (2) from the National Center of Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). Sequences' affiliations to known genera or species were assessed regarding their similarity.
Antigen-specific immunoglobulin G detection.
As previously described, total IgGs specific for Saccharopolyspora rectivirgula from plasma of dairy barn workers and control subjects were measured by indirect enzyme-linked immunosorbent assay (ELISA) (10). Plasma samples from workers and control subjects were diluted 1/500 before they were added to the antigen (crude extract)-coated plate, prepared as previously described (15). Blank, negative, and positive controls were also added to each plate. The enzyme reaction was stopped when the positive control for each plate reached the same optical density (OD). Plasma samples were considered positive (1+) for the antigen when the optical density was higher than the OD 95% confidence interval of the control samples. The reaction intensity from control subjects and workers was scored from − (no reactivity) to 4+ (strong reactivity) depending on the OD obtained. A score of 2+ corresponded to an OD from samples that doubled the control sample average, whereas 3+ and 4+ ODs tripled (3+) or quadrupled (4+) it. Samples were classified as negative (−), total positive (1+ to 4+), or strongly positive (3+ and 4+). Although the plasma IgG should be specific to the S. rectivirgula antigen, cross-reactions between epitopes from different antigens could not be prevented in this technique.
Statistical analysis.
Quantitative data were expressed using the average and the standard error of the mean (SEM). Total archaeal and bacterial data from the three different samplers were analyzed using a mixed analysis of variance (ANOVA) with two experimental factors, one associated with the comparison between samplers (factor fixed) and one associated with dairy barns and analyzed as a random effect (blocking variable). The same statistical approach was used for data comparison among the three NIOSH stages. A compound symmetry structure was used to take into account the dependency among bacteria and archaea in dairy barns. Data from workers and controls were analyzed using Student's t test. For some variables, values were log10 transformed to stabilize variances. Reported P values are based on these transformations. The normality assumption was verified using Shapiro-Wilk tests after a Cholesky factorization. Brown and Forsythe's variation of Levene's test was used to verify the homogeneity of variances. Relationships between variables were expressed using Pearson's correlation coefficient. The results were considered significant with P values of ≤0.05. All analyses were conducted using the statistical package SAS, version 9.2 (SAS Institute Inc., Cary, NC).
RESULTS
High concentrations of airborne archaea and bacteria in dairy barns.
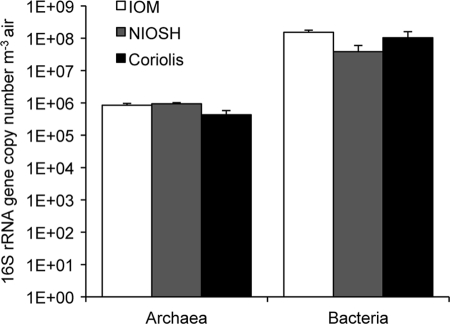
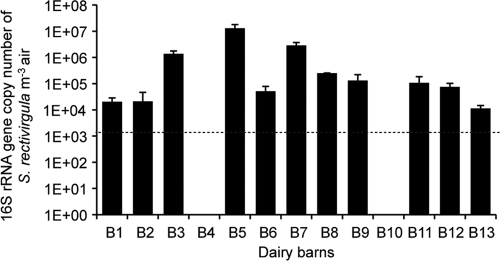
Airborne archaea and bacteria from dairy barns were quantified by total DNA extracts (Fig. 1). The primer set of A751F and A976R and the optimized PCR protocol were validated from the study by Baker et al. (5) with five different archaeal species. DNA from three species was provided by the American Type and Culture Collection (ATCC), namely, Aeropyrum pernix (ATCC 700893D), Thermoplasma volcanium (ATCC 51530D), and Methanosarcina mazei (ATCC BAA-159D). The last two species, Methanobrevibacter smithii and Methanosphaera stadtmanae, were graciously provided by R. Forster, and DNA was extracted as explained above. These primers and protocol were successful in amplifying variable regions V4 to V5 of archaeal 16S rRNA genes without bias for any archaeal phylum and without bacterial amplification. The standard curve was linear from 2 × 100 to 2 × 105 gene copies per reaction (efficiency = 86.21%, r2 = 0.997). High concentrations of archaeal and bacterial 16S rRNA genes were detected in the dairy barns' air. Similar data were obtained with the three samplers tested (Fig. 1). Averages of 8.5 × 105 archaeal and 1.5 × 108 bacterial 16S rRNA genes were detected per m3 of air in the 13 dairy barns sampled (IOM sampler) (Fig. 1). Archaeal detection ranged from 8.6 × 104 to 3.5 × 106 16S rRNA genes per m3 of air, while the amount of bacteria ranged from 1.81 × 106 to 1.05 × 109 16S rRNA genes per m3 of air (IOM sampler). In addition to broad-spectrum quantification of bacterial DNA, we also performed agent-specific detection of S. rectivirgula since bioaerosols from dairy barns have historically been shown to contain this agent, a cause of the farmer's lung disease. Detection of S. rectivirgula by quantitative PCR showed high differences between the amounts of this bacterium in each dairy barn sampled. Indeed, an average of 1.4 × 106 16S rRNA genes of S. rectivirgula per m3 of air was found, but this level ranged from below the limit of detection (LOD) to 1.3 × 107 gene copy numbers per m3 of air (Fig. 2).
Fig 1.
Airborne archaeal and bacterial 16S rRNA gene concentrations from dairy barn samples (average ± SEM), as determined by quantitative PCR. Comparison between IOM, NIOSH, and Coriolis air samplers (n = 13 each). LOD, 4 × 102 16S rRNA genes for archaea and 2 × 103 16S rRNA genes for bacteria. No statistical differences between results from the three air samplers tested (archaea, P = 0.3135; bacteria, P = 0.4981).
Fig 2.
Airborne S. rectivirgula 16S rRNA gene concentrations from dairy barn samples (average ± SEM), as determined by quantitative PCR from the IOM air sampler (n = 13). The dotted line at 2 × 103 indicates the LOD of the PCR.
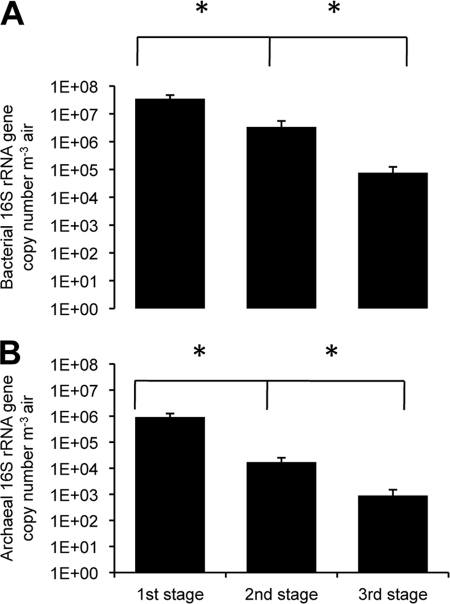
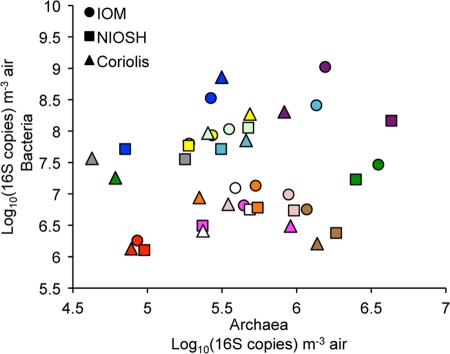
Bioaerosol size distribution studied with the NIOSH sampler showed that the majority of microorganisms sampled were found in the first stage, i.e., particles with an aerodynamic diameter of over 2.1 μm (Fig. 3). Indeed, averages of 9.2 × 105, 1.7 × 104, and 9.0 × 102 archaeal 16S rRNA genes per m3 of air were found in stages 1 to 3, respectively. The same trend was observed for bacteria, with averages of 3.5 × 107, 3.4 × 106, and 7.7 × 104 16S rRNA genes per m3 of air found in stages 1 to 3, respectively. There was no correlation between the quantity of airborne archaea and that of airborne bacteria in dairy barns, with a correlation coefficient (r) of 0.2629 for the IOM sampler, 0.0839 for the NIOSH sampler, and 0.0464 for the Coriolis sampler (Fig. 4).
Fig 3.
Size distribution of particles in the dairy barns: airborne archaeal and bacterial concentrations 16S rRNA gene from dairy barn samples (average ± SEM), as determined by quantitative PCR. Comparison between the three NIOSH sampler stages (n = 13 each). LOD, 1 × 102 16S rRNA genes for archaea and 6 × 102 16S rRNA genes for bacteria. *, P < 0.005.
Fig 4.
Lack of correlation between the concentrations of 16S rRNA genes of archaea and bacteria in dairy barns. Comparison between IOM (r = 0.2629, P = 0.3853), NIOSH (r = 0.0839, P = 0.7852), and Coriolis (r = 0.0464, P = 0.8803) air samplers (n = 13 each). Data from the same barn are represented by the same color. LOD, 4 × 102 16S rRNA genes for archaea and 2 × 103 16S rRNA genes for bacteria.
Methanogens and various species of bacteria aerosolized in dairy barns.
Airborne archaeal and bacterial species detected in dairy barns were identified using PCR-DGGE on 16S rRNA genes from both domains. Since similar archaeal and bacterial biodiversity profiles were observed on DGGE for every sampler (data not shown), we used only total DNA from IOM cassettes for DGGE analysis. For archaeal analysis (Table 2), 17 different band classes were observed among DGGE profiles from the 13 dairy barns sampled. Clustering within archaeal DGGE profiles from each barn showed high homology (80% similarity and higher) for 7 barns out of 13, and the percentage similarity was lower than 40% for 4 barns out of 13. As suspected, the five extracted and sequenced bands had high identity homology (95 to 98%) with the Methanobacteriaceae group. The genus Methanobrevibacter represented 100% of all DGGE bands sequenced. M. smithii, detected in 2 barns out of 13, and Methanobrevibacter ruminantium, detected in every barn sampled, were the archaeal species identified by sequencing (Table 2).
Table 2.
Sequence matches for bands from DGGE gels containing dairy barn archaeal and bacterial DNA
| Type of DNA | Band no. | Frequency (no. of positive samples)a | Most similar sequence | bp | % similarity |
|---|---|---|---|---|---|
| Archaeal | 1 | 5 | Methanobrevibacter sp. JQ267743 | 302 | 97 |
| 2 | 2 | Methanobrevibacter smithii JQ267744 | 313 | 97 | |
| 3 | 9 | Methanobrevibacter sp. JQ267745 | 300 | 95 | |
| 4 | 12 | Methanobrevibacter ruminantium JQ267746 | 305 | 95 | |
| 5 | 13 | Methanobrevibacter ruminantium JQ267747 | 316 | 98 | |
| Bacterial | 1 | 13 | Staphylococcus gallinarum JQ267748 | 570 | 99 |
| 1 | 13 | Crocebacterium ilecola JQ267749 | 549 | 99 | |
| 2 | 12 | Oxalobacter sp. JQ267750 | 507 | 97 | |
| 3 | 13 | Agrobacterium tumefaciens JQ267751 | 543 | 99 | |
| 4 | 12 | Clostridium quinii JQ267752 | 542 | 99 | |
| 4 | 12 | Staphylococcus sp. JQ267753 | 564 | 99 | |
| 5 | 2 | Agrobacterium sp. JQ267754 | 435 | 94 | |
| 6 | 12 | Corynebacterium variabile JQ267755 | 550 | 98 | |
| 7 | 13 | Corynebacterium xerosis JQ267756 | 484 | 98 | |
| 8 | 13 | Corynebacterium xerosis JQ267757 | 482 | 95 |
Out of 13 total barns.
The characterization of airborne bacterial species from dairy barns (Table 2) revealed a DGGE profile of 22 different band classes, from which 8 were extracted and sequenced. All dairy barns' bacterial DGGE profiles clustered together with 65 to 92% similarity. Two of the sequenced bands (1 and 4) included DNA from two different bacterial species. DGGE bands had identity homology (94 to 99%) with various bacterial species, namely, Staphylococcus gallinarum, Crocebacterium ilecola, Oxalobacter sp., Agrobacterium tumefaciens, Clostridium quinii, Staphylococcus sp., Agrobacterium sp., Corynebacterium variabile, and Corynebacterium xerosis. All of the bacterial DGGE-sequenced bands except for band 5 were found in 12 or 13 barns out of 13 (Table 2).
Dairy barn workers were sufficiently exposed to airborne S. rectivirgula to induce a humoral response.
S. rectivirgula is a major airborne microorganism found in dairy barns, and, consequently, the workers' airways can be exposed to this microorganism. We actually found up to 107 16S rRNA genes of S. rectivirgula per m3 of air in this work environment. Thus, we determined if plasma of workers from the 13 dairy barns visited contained IgG specific to this antigen, a method often used by our team to confirm exposure (10, 12, 32). A total of 85.7% of plasma samples from control subjects had a negative IgG response to S. rectivirgula, compared to 62.1% for samples from dairy barn workers. Total positive samples reached 14.3% for control subjects and 37.9% for workers. From these positive samples, only 2.9% were considered strongly positive for control subjects, compared to 10.3% for the workers. Overall, a higher number of workers than control subjects had positive plasma S. rectivirgula-specific IgGs (Table 3).
Table 3.
IgG specific for S. rectivirgula in plasma of dairy barn workers compared to that in plasma of controlsa
| Population (n) | No. of subjects (%) with each immune response intensity |
||
|---|---|---|---|
| Negative | Total positive | Strongly positive | |
| Control subjects (35) | 30 (85.7) | 5 (14.3) | 1 (2.9) |
| Workers (29) | 18 (62.1) | 11 (37.9) | 3 (10.3) |
Frequency distribution between workers and control subjects is significantly different (P = 0.0427).
DISCUSSION
Unlike most natural microbial habitats, such as water and soil, bioaerosols have not been well studied and characterized. Since hay and straw are important sources of microorganisms in dairy barns, most studies have focused on fungus, endotoxin, and actinomycete airborne contamination (16, 28, 30). However, as reported earlier in swine barns (33, 34), using molecular biology tools, we demonstrated that bioaerosol biodiversity is much more complex than expected. In the current study, we detected airborne archaeal species, which can be immunogenic agents (8), for the first time in dairy barns. Moreover, we characterized the airborne bacterial diversity of dairy barns, focusing on the occupational respiratory diseases. Results show strong similarities between data from different air samplers. It was also revealed that, despite modern hay baling and management practices, dairy barn workers are exposed to high concentrations of S. rectivirgula, the causative agent for farmer's lung, and a significant proportion of workers develop humoral responses against that bioaerosol agent. These results provide the first culture-independent data on dairy barn bioaerosol composition. They highlight the lack of knowledge on bioaerosol agents in working environments and the importance of a better biodiversity assessment.
Results from bioaerosol studies that were obtained with different air samplers are comparable. Indeed, quantitative data from the IOM, the NIOSH, and the Coriolis samplers were similar (Fig. 1). Qualitative data from DGGE profiles of different samplers also showed similar data (data not shown). These results are not surprising considering the fact that the majority of the particles captured had an aerodynamic diameter of 2.1 μm (Fig. 3), which is in the capture range of all three samplers (1, 7, 31). Therefore, even while using different particulate trapping systems, data were similar, so results from different studies can be compared. No matter which air sampler is used on the field, analysis should lead to similar results.
Bioaerosols of dairy barns contain high concentrations of various species of bacteria and archaea. According to King et al., Methanobrevibacter (RO and SGMT clades) and Methanosphaera are the two main archaeal genera found in the rumens of Holstein cows. Two different species of Methanobrevibacter were detected by PCR-DGGE, including Methanobrevibacter smithii, an immunogenic species of archaea (8). Even if Methanosphaera stadtmanae is one of the main species found in cows' rumens, it has not been detected in the air of the dairy barns sampled in this study. However, according to Yu et al., PCR-DGGE archaeal biodiversity results depend on the primer set used (44). The main bacterial genera in the rumens of Holstein cows are the Cytoflaga-Flavobacter-Bacteroides phylum, Prevotella, Ruminobacter, and Clostridium (43). The majority of our bacterial airborne results did not include these species, which are probably due to bacterial sources other than cow manure. Indeed, a recent study on bioaerosols' emission from an open-freestall dairy barn operation (17) showed a wide bacterial diversity. As suspected, airborne archaeal and bacterial biodiversity showed high similarity within dairy barns sampled, although clustering percentages were higher for the bacterial analysis. This can be explained by the higher number of archaeal bands that appeared sporadically on the gel compared to that of bacteria. Interestingly, we found no correlation between bacterial and archaeal bioaerosols in dairy barns (Fig. 4), while this correlation was found to be positive in swine barn facilities (33). This can be explained by the consistency of airborne archaeal concentrations between the different dairy barns sampled (Fig. 4). According to Jeyanathan et al., the archaeal community in cows' rumens is constant, less variable, and less diverse than the bacterial community (26). Indeed, the archaeal source, the cows' rumens, is the most constant source of bioaerosols in a dairy barn. Bacterial sources in dairy barns are probably more diversified (hay, straw, water, manure, grain) than those in swine barns, justifying the lack of correlation between archaeal and bacterial quantities in dairy barns. These results increase the knowledge on dairy barns' biological burden. What are the potential impacts of these newly found airborne microorganisms on the respiratory health of workers? Can they play a synergic role with other bioaerosol components in lung diseases?
Dairy barn workers are still exposed to S. rectivirgula, the causal agent of farmer's lung. Even though recommendations are frequently made to farmers about hay preservation to minimize the risk of contamination by the actinomycete (9, 21, 23, 39), S. rectivirgula is still present in high concentrations in the air in dairy barns (Fig. 2). Indeed, up to 107 airborne S. rectivirgula CFU per m3 were found by culture in Quebec dairy barns in 1999 (16) and up to 1 million 16S S. rectivirgula rRNA genes per m3 were found in this study using molecular biology. Bioaerosol contamination by a high concentration of S. rectivirgula cells has led to significantly higher production of antigen-specific IgG in the plasma of workers compared to that of the control subjects (Table 3). Because the amount of plasma S. rectivirgula-specific IgG is used as one of the diagnostic tools for farmer's lung (29), dairy barn workers are still at risk for developing this airway disease.
Conclusion.
We demonstrated that dairy barn bioaerosols contain high concentrations of various species of methanogenic archaea and bacteria, especially M. smithii, an immunogenic archaeal species, and S. rectivirgula, which is the causal agent of farmer's lung. These results improve our knowledge of aerobiological burden in this work environment. This study reveals the consistency in the results obtained from three different air samplers. Dairy barn workers are exposed to S. rectivirgula despite the recommendations for hay management. A strong similarity between data from various air samplers is observed. This is the first study on dairy barn bioaerosols using molecular biology, which allowed the discovery of airborne archaea that may be involved in the etiology of agriculture-related respiratory diseases.
ACKNOWLEDGMENTS
This work was supported by Institut de Recherche Robert-Sauvé en Santé et Sécurité au Travail (IRSST) grant 099-864 (C.D.) and Canadian Center for Health and Safety in Agriculture (CCHSA) grant CDA-66151 (C.D.). P.B.L. is a CIHR Strategic Training Fellow in PHARE and received a CIHR-Quebec Respiratory Health Training Program short-term training scholarship. C.D. and D.M. are members of the FRSQ Respiratory Health Network, and C.D. is an FRSQ Senior Scholar.
We thank William G. Lindsley (CDC/NIOSH) for providing NIOSH air samplers, Gaëtane Racine for performing blood sampling on volunteers, Serge Simard for the statistical analyses, Robert Forster for providing archaeal species, and every person (dairy barn workers and control subjects) who volunteered to participate in this study.
Footnotes
Published ahead of print 24 February 2012
REFERENCES
- 1. ACGIH 1999. Particle size-selective sampling for health-related aerosols. American Conference of Governmental Industrial Hygienists, Cincinnati, OH [Google Scholar]
- 2. Altschul SF, et al. 1997. Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. ATS 1998. Respiratory health hazards in agriculture. American Thoracic Society, Medical Section of the American Lung Association, New York, NY [Google Scholar]
- 4. Bach HJ, Hartmann A, Schloter M, Munch JC. 2001. PCR primers and functional probes for amplification and detection of bacterial genes for extracellular peptidases in single strains and in soil. J. Microbiol. Methods 44:173–182 [DOI] [PubMed] [Google Scholar]
- 5. Baker GC, Smith JJ, Cowan DA. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541–555 [DOI] [PubMed] [Google Scholar]
- 6. Bates ST, et al. 2011. Examining the global distribution of dominant archaeal populations in soil. ISME J. 5:908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bertin Technologies Coriolis brochure. Bertin Technologies, Montigny-le-Bretonneux, France [Google Scholar]
- 8. Blais Lecours P, et al. 2011. Immunogenic properties of archaeal species found in bioaerosols. PLoS One 6:e23326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Canadian Centre for Occupational Health and Safety 15 October 2008, posting date Farmer's lung. http://www.ccohs.ca/oshanswers/diseases/farmers_lung.html
- 10. Cayer MP, et al. 2007. Identification of mycobacteria in peat moss processing plants: application of molecular biology approaches. Can. J. Microbiol. 53:92–99 [DOI] [PubMed] [Google Scholar]
- 11. Chapelle FH, et al. 2002. A hydrogen-based subsurface microbial community dominated by methanogens. Nature 415:312–315 [DOI] [PubMed] [Google Scholar]
- 12. Cormier Y, Israel-Assayag E, Bedard G, Duchaine C. 1998. Hypersensitivity pneumonitis in peat moss processing plant workers. Am. J. Respir. Crit. Care Med. 158:412–417 [DOI] [PubMed] [Google Scholar]
- 13. Dakhama A, Hegele RG, Laflamme G, Israel-Assayag E, Cormier Y. 1999. Common respiratory viruses in lower airways of patients with acute hypersensitivity pneumonitis. Am. J. Respir. Crit. Care Med. 159:1316–1322 [DOI] [PubMed] [Google Scholar]
- 14. Delmont TO, et al. 2011. Accessing the soil metagenome for studies of microbial diversity. Appl. Environ. Microbiol. 77:1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duchaine C, Meriaux A, Brochu G, Bernard K, Cormier Y. 1999. Saccharopolyspora rectivirgula from Quebec dairy barns: application of simplified criteria for the identification of an agent responsible for farmer's lung disease. J. Med. Microbiol. 48:173–180 [DOI] [PubMed] [Google Scholar]
- 16. Duchaine C, Meriaux A, Brochu G, Cormier Y. 1999. Airborne microflora in Quebec dairy farms: lack of effect of bacterial hay preservatives. Am. Ind. Hyg. Assoc. J. 60:89–95 [DOI] [PubMed] [Google Scholar]
- 17. Dungan RS. 2012. Use of a culture-independent approach to characterize aerosolized bacteria near an open-freestall dairy operation. Environ. Int. 41:8–14 [DOI] [PubMed] [Google Scholar]
- 18. Eduard W. 1997. Exposure to non-infectious microorganisms and endotoxins in agriculture. Ann. Agric. Environ. Med. 4:179–186 [Google Scholar]
- 19. Ennahar S, Cai Y, Fujita Y. 2003. Phylogenetic diversity of lactic acid bacteria associated with paddy rice silage as determined by 16S ribosomal DNA analysis. Appl. Environ. Microbiol. 69:444–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fallschissel K, Kampfer P, Jackel U. 2009. Direct detection of salmonella cells in the air of livestock stables by real-time PCR. Ann. Occup. Hyg. 53:859–868 [DOI] [PubMed] [Google Scholar]
- 21.Farm Safety Association. Farmer's lung: it takes your breath away! 1990. http://nasdonline.org/static_content/documents/1663/d001538.pdf.
- 22. Fischer G, Schwalbe R, Ostrowski R, Dott W. 1998. Airborne fungi and their secondary metabolites in working places in a compost facility. Mycoses 41:383–388 [DOI] [PubMed] [Google Scholar]
- 23. Fontanier F, et al. 2008. Le séchage en grange du foin conditionné en grosses bottes. Pôle Fromager AOC Massif Central; http://www.vienne.chambagri.fr/fileadmin/publication/Intranet/WikiConseil/Machinisme/Documents/PUBLI_Sechage_en_grange_du_foin_en_bottes.pdf [Google Scholar]
- 24. Fromin N, et al. 2002. Statistical analysis of denaturing gel electrophoresis (dge) fingerprinting patterns. Environ. Microbiol. 4:634–643 [DOI] [PubMed] [Google Scholar]
- 25. Gilbert Y, Veillette M, Duchaine C. 2010. Metalworking fluids biodiversity characterization. J. Appl. Microbiol. 108:437–449 [DOI] [PubMed] [Google Scholar]
- 26. Jeyanathan J, Kirs M, Ronimus RS, Hoskin SO, Janssen PH. 2011. Methanogen community structure in the rumens of farmed sheep, cattle and red deer fed different diets. FEMS Microbiol. Ecol. 76:311–326 [DOI] [PubMed] [Google Scholar]
- 27. Just N, et al. 2011. Bacterial diversity characterization of bioaerosols from cage-housed and floor-housed poultry operations. Environ. Res. 111:492–498 [DOI] [PubMed] [Google Scholar]
- 28. Kullman GJ, et al. 1998. Organic dust exposures from work in dairy barns. Am. Ind. Hyg. Assoc. J. 59:403–413 [DOI] [PubMed] [Google Scholar]
- 29. Lacasse Y, et al. 2003. Clinical diagnosis of hypersensitivity pneumonitis. Am. J. Respir. Crit. Care Med. 168:952–958 [DOI] [PubMed] [Google Scholar]
- 30. Lee SA, et al. 2006. Personal exposure to airborne dust and microorganisms in agricultural environments. J. Occup. Environ. Hyg. 3:118–130 [DOI] [PubMed] [Google Scholar]
- 31. Lindsley WG, Schmechel D, Chen BT. 2006. A two-stage cyclone using microcentrifuge tubes for personal bioaerosol sampling. J. Environ. Monit. 8:1136–1142 [DOI] [PubMed] [Google Scholar]
- 32. Meriaux A, Cormier Y, Pageau P, Israel-Assayag E, Duchaine C. 2006. Sensitization to airborne molds and its health effects in peat moss processing plant workers. J. Occup. Environ. Hyg. 3:442–447 [DOI] [PubMed] [Google Scholar]
- 32a. Muyzer G, de waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nehme B, et al. 2009. Culture-independent characterization of archaeal biodiversity in swine confinement building bioaerosols. Appl. Environ. Microbiol. 75:5445–5450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nehme B, Letourneau V, Forster RJ, Veillette M, Duchaine C. 2008. Culture-independent approach of the bacterial bioaerosol diversity in the standard swine confinement buildings, and assessment of the seasonal effect. Environ. Microbiol. 10:665–675 [DOI] [PubMed] [Google Scholar]
- 35. Oppliger A, Charriere N, Droz PO, Rinsoz T. 2008. Exposure to bioaerosols in poultry houses at different stages of fattening; use of real-time PCR for airborne bacterial quantification. Ann. Occup. Hyg. 52:405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patel GB, Zhou H, Ponce A, Chen W. 2007. Mucosal and systemic immune responses by intranasal immunization using archaeal lipid-adjuvanted vaccines. Vaccine 25:8622–8636 [DOI] [PubMed] [Google Scholar]
- 37. Qiagen 2010. QIAamp DNA mini and blood mini handbook, 3rd ed Qiagen, Toronto, ON, Canada [Google Scholar]
- 38. Reysenbach A-L, Pace NR. 1995. Archaea: a laboratory manual—thermophiles. Cold Spring Harbor Laboratory Press, New York, NY [Google Scholar]
- 39. Samson M, Choinière Y. 2007. Recommandations pour les entrepôts à foin, p 27–29 Équi-Libre. http://www.yveschoiniere.com/pdfs/articles/recommandations-entrepots-foin.pdf
- 40. Schafer J, Kampfer P, Jackel U. 2011. Detection of Saccharopolyspora rectivirgula by quantitative real-time PCR. Ann. Occup. Hyg. 55:612–619 [DOI] [PubMed] [Google Scholar]
- 41. Venter JC, et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74 [DOI] [PubMed] [Google Scholar]
- 42. Verreault D, et al. 2011. Detection of airborne lactococcal bacteriophages in cheese manufacturing plants. Appl. Environ. Microbiol. 77:491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Whitford MF, Forster RJ, Beard CE, Gong J, Teather RM. 1998. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe 4:153–163 [DOI] [PubMed] [Google Scholar]
- 44. Yu Z, Garcia-Gonzalez R, Schanbacher FL, Morrison M. 2008. Evaluations of different hypervariable regions of archaeal 16S rRNA genes in profiling of methanogens by archaea-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 74:889–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu Z, Morrison M. 2004. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:4800–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhiping W, et al. 1996. Exposure to bacteria in swine-house dust and acute inflammatory reactions in humans. Am. J. Respir. Crit. Care Med. 154:1261–1266 [DOI] [PubMed] [Google Scholar]