Abstract
We have characterized a new strain, Bifidobacterium animalis subsp. lactis CECT 7953, obtained by random UV mutagenesis, which produces less acetic acid than the wild type (CECT 7954) in three different experimental settings: De Man-Rogosa-Sharpe broth without sodium acetate, resting cells, and skim milk. Genome sequencing revealed a single Phe-Ser substitution in the acetate kinase gene product that seems to be responsible for the strain's reduced acid production. Accordingly, acetate kinase specific activity was lower in the low acetate producer. Strain CECT 7953 produced less acetate, less ethanol, and more yoghourt-related volatile compounds in skim milk than the wild type did. Thus, CECT 7953 shows promising potential for the development of dairy products fermented exclusively by a bifidobacterial strain.
INTRODUCTION
Consumers are increasingly interested in supplemented foods that may provide benefits beyond their nutritional characteristics, known as functional foods. This has led to the development of many foods, mainly dairy based, with the inclusion of probiotic bacteria. Probiotics are live bacteria that can confer health benefits on the host (4). In the case of fermented foods, probiotics suffer from reduced viability because of processing (including manufacture and storage) and the harsh and competitive environment of the gastrointestinal tract.
Given their intrinsic resistance to both technological and physiological stresses, Bifidobacterium animalis subsp. lactis strains have been the probiotics of choice for inclusion in fermented foods (8, 13). Included in this group are sequenced strains DN 173010, BB-12, and HN019 (2, 3, 6, 9, 23). The suggested benefits of B. animalis subsp. lactis for human health, as reported in animal models, include decreased serum cholesterol levels, protection against colorectal cancer, regulation of gut transit time and constipation, and reduction of gut inflammation through the maintenance of a favorable balance of the microbiota (1, 10, 21, 24–26, 27).
Interestingly, we could not find any reference in the literature to dairy products fermented exclusively by bifidobacteria. Instead, bifidobacteria are added as adjunct cultures that do not participate metabolically in fermentation. Fermentation is conducted mainly by starter cultures of Streptococcus thermophilus and species of the genus Lactobacillus. There are two main difficulties arising from the production of foodstuffs from milk fermented by bifidobacteria. The first is the anaerobic metabolism of bifidobacteria, which grow slowly or not at all in the presence of oxygen, making it very expensive to include them in industrial processes. The second is that bifidobacteria produce large amounts of acetic acid via the glycolytic pathway. The first is overcome by using B. animalis subsp. lactis, which is more stress resistant than other strains. The second forms the basis of the present research. Here we describe the isolation and characterization of a bifidobacterial strain with a reduced ability to produce acetic acid.
MATERIALS AND METHODS
Growth conditions, mutant generation, and isolation.
To generate mutants, strain CECT 7954 was propagated in De Man-Rogosa-Sharpe (MRS) broth (Difco, Detroit, MI) supplemented with 0.05% (wt/vol) l-cysteine (Sigma, St. Louis, MO) (MRSC). Cultures were incubated at 37°C in an MG500 anaerobic chamber (Don Whitley Scientific, West Yorkshire, United Kingdom) with an atmosphere of 10% (vol/vol) H2, 10% CO2, and 80% N2. An overnight culture was used to inoculate 50 ml of fresh MRSC (1%, vol/vol). Exponential-phase cultures (A600 = 0.4) were washed twice with phosphate-buffered saline (PBS), resuspended in 5 ml PBS, poured onto a petri dish, and then exposed to UV light (UV radiation sterilization desk; JP Selecta, Barcelona, Spain) for 3 min. This exposure produced a viability loss of 15% (data not shown). Mutants with a decreased ability to incorporate sodium fluoroacetate were selected in MRSC supplemented with 1.8% (wt/vol) agar and 120 mM sodium fluoroacetate.
Selection of low acetate producers.
One hundred colonies able to grow in the presence of 120 mM sodium fluoroacetate were isolated following 48 h of incubation in MRSC plates. Mutant colonies were grown in MRS without sodium acetate (Difco). Acetic acid production was measured by high-performance liquid chromatography with an Alliance 2690 injector module, a Photodiode Array PDA 996 detector (210 nm), a 410 Differential Refractometer detector, and the Empower software (Waters, Milford, MA). Filtered (0.22-μm pore size) supernatant aliquots of 50 μl were injected into an ICSep ION-300 ion-exchange column (Transgenomic, San Jose, CA). H2SO4 (8.5 mN) was used as the mobile phase at 65°C, with a flow rate of 0.4 ml/min.
Genome sequencing and strain comparison.
Total DNA was extracted using a DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany), with a protocol modified for Gram-positive bacteria. Sequencing was performed at Macrogen Inc., Seoul, South Korea, using an Illumina GAIIx 36-bp paired-end approach according to standard Illumina protocols. Of the >12 million 36-bp-long reads generated for each strain, 99.6% (CECT 7954) and 99.8% (CECT 7953) were successfully mapped to the B. animalis subsp. lactis DSM10140 complete genome sequence (RefSeq accession no. NC_012815.1) using Maq software (11) (default parameters). This resulted in 237× (CECT 7954) or 219× (CECT 7953) genome coverage. Single nucleotide polymorphisms (SNPs) were then identified and filtered using the Maq option SNPfilter -q 40 -w 5 -N 2 -d 3 -D 256 -n 20 -Q 40, as recommended for bacterial genomes (11). Bioinformatic computation was performed at the Clúster de Modelización Científica, Oviedo University (http://cms.uniovi.es).
Acetate kinase assay.
Acetate kinase activity was determined in the direction of acetyl-phosphate formation. First, 10-ml cultures of each of the bifidobacterial strains grown to the mid-exponential phase (about 5 × 108 CFU/ml). Cell extracts were obtained by sonication on ice using a CV17 sonicator (VibraCell; Sonics and Materials, Newtown, CT) and centrifuged (5 min, 16,000 × g, 4°C) to remove cell debris. Protein concentrations in the different extracts were measured with the BCA Protein Assay kit (Pierce, Rockford, IL). Mixtures of 1 ml containing 50 mM Tris-HCl adjusted to pH 7.5, 10 mM MgSO2, 750 mM acetate, 1.16 M NH2OH·HCl adjusted to pH 7.5 with KOH were preincubated for 3 min at 60°C with 500 μg of total bacterial protein. The reaction was started by the addition of 30 μl ATP (333 mM in 1 M Tris) and stopped after 60 min of incubation at 37°C by transferring 300 μl of the assay mixture into 750 μl of 77 mM FeCl3, in 1 M HCl. Acetohydroxamate was quantified at 540 nm using a series of different acetyl-phosphate concentrations as standards. One unit of activity was defined as the amount of protein producing 1 nmol of hydroxamate per min. Results were expressed as the mean of three independent measurements.
Glucose consumption and end product formation.
To precisely determine glucose consumption and organic acid production, buffered cell suspensions where obtained as described previously (19). Briefly, 10-ml cultures of strains CECT 7954 and CECT 7953 at an optical density at 600 nm of 0.6, grown in MRSC without sodium acetate, were collected by centrifugation, washed, and resuspended in 10 ml of 33 mM potassium phosphate buffer, pH 5.6, containing 25 mM glucose. Cells were incubated for 4 h at 37°C in anaerobic jars, and the supernatants were analyzed by high-performance liquid chromatography as described before. Glucose and organic acid levels are presented as the means of at least three independent experiments. In addition, the acetic/lactic acid ratios were calculated, as well as the total carbon balance of the bifid shunt expressed as moles of carbon formed/moles of glucose consumed using the following formula: (2 × [acetic acid] + 3 × [lactic acid] + 2 × [ethanol] + 1 × [formic acid])/(6 × [glucose]).
Manufacture of a bifidobacterium-fermented milk.
Only Bifidobacterium animalis subsp. lactis CECT 7954 or CECT 7953 was used for the manufacture of fermented milk. Commercial semiskim milk supplemented with yeast extract (Bacto yeast extract; Difco); containing 4.05% (wt/vol) protein, 4.5% (wt/vol) carbohydrates, 1.6% (wt/vol) fats, and 0.11% (wt/vol) calcium; and supplemented with 1% skim milk (Difco) was used after Tyndallization. Both strains were separately propagated (1% wt/vol) in the milk overnight at 37°C and in aerobiosis and used to inoculate freshly prepared milk. Milk fermentations were carried out in aerobiosis at 37°C until they reached a pH of 5.0 ± 0.1, and afterwards, fermented milk samples were stored at 4° for 24 h. Bacterial counts and pHs were then determined, as well as other parameters, such as residual lactose and organic acid production (as described before). Volatile compound production was determined through a dynamic headspace extraction and gas chromatography-mass spectrometry analysis, according to the method of Ruas-Madiedo and coworkers (18). Results were expressed as millimolar concentrations of sugar and organic acids and as micrograms of volatile compounds per gram.
Statistical analysis.
Throughout this study, data were subjected to one-way analysis of variance with the SPSS 18.0 software (SPSS Inc., Chicago, IL) using the factor “strain” with two categories, CECT 7954 and CECT 7953.
RESULTS AND DISCUSSION
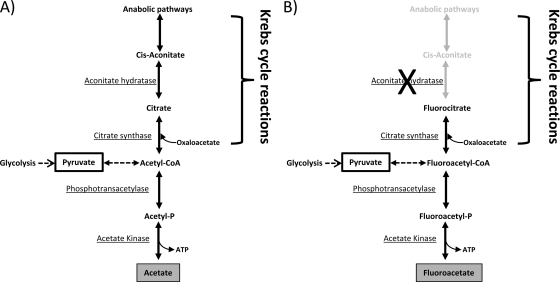
In the present work, we discuss a new strategy for selecting bifidobacteria with reduced acetic acid-producing capacity, which may be of industrial interest for the elaboration of new (and potentially functional) dairy products. We have chosen strain CECT 7954, a spontaneous extracellular polysaccharide-producing strain that was isolated in our lab from a dairy isolate. This strain is able to grow in the presence of oxygen in both static and agitated cultures and is thus of interest to industry and a good candidate for the study of potential human health benefits. Figure 1 depicts the strategy for mutant selection. Bifidobacteria obtain some ATP equivalents at a substrate level through the glycolytic pathway by coupling acetic acid production from acetyl phosphate (Fig. 1A). Sodium fluoroacetate is incorporated by acetate kinase to the central metabolism of bifidobacteria and irreversibly inhibits aconitate hydratase, an enzyme of the Krebs cycle, as shown in Fig. 1B (17). According to the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/kegg2.html), B. animalis subsp. lactis contains the information for some of the Krebs cycle reactions, which is the starting point of some anabolic pathways, including the biosynthesis of branched-chain amino acids. Thus, inhibition of aconitate hydratase inhibits these anabolic reactions and is lethal to bifidobacteria. We aimed to select mutants harboring mutations in one of the genes involved in acetic acid production and therefore a decreased ability to incorporate sodium fluoroacetate. A similar strategy has previously been used to select low acetate producers of Thermoanaerobacter thermohydrosulfuricus (14).
Fig 1.
Schematic representation of the strategy used for low acetate producers. (A) Metabolic pathway of acetic acid production in B. animalis subsp. lactis DSM10140. Pathways obtained from KEGG. (B) Uptake of fluoroacetate through the central metabolism of bifidobacteria irreversibly inhibits aconitate hydratase, blocking anabolic pathways and leading to bacterial death.
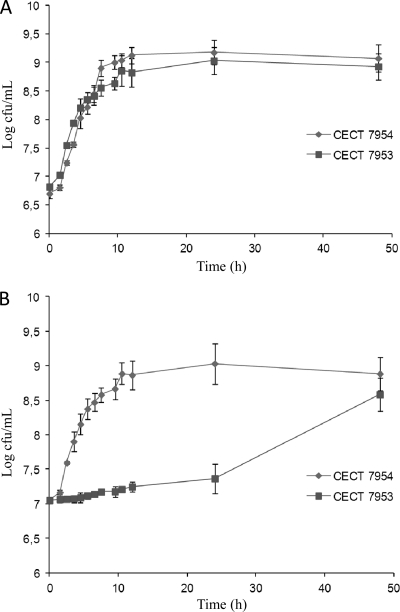
Among the colonies obtained, only one strain, designated CECT 7953, showed reduced acetic acid production compared with that of the wild type. The resistance of the remaining strains may have been induced by mutations in multidrug systems, as proposed for T. thermohydrosulfuricus (14). Stationary-phase cultures of strain CECT 7953 in MRSC elaborated without sodium acetate produced 4.3-fold less acetic acid than wild-type strain CECT 7954 (3.73 ± 0.17 versus 16.10 ± 1.01 mM, respectively) after 24 h of growth. Consequently, the lactic/acetic acid ratio increased from 0.4 (CECT 7954) to 1.8 (CECT 7953), resembling the metabolic profile of a homofermentative lactic acid bacterium. Interestingly, the growth of CECT 7953 was delayed by 24 h if pyruvate was used as the sole carbon source (Fig. 2). In this sense, acetate kinase mutants of T. thermohydrosulfuricus were unable to use pyruvate as a carbon and energy source due to an inability to produce ATP at the substrate level in the reaction catalyzed by acetate kinase (14). This supports the concept of a partial block in the acetic acid production pathway.
Fig 2.
Growth curves of CECT 7954 and CECT 7953 in MRSC without sodium acetate and with glucose (A) or sodium pyruvate (B) as a carbon and energy source.
To determine the genetic changes induced by UV mutagenesis in strain CECT 7953, we sequenced the genomes of both strains. Genome comparison identified 45 SNPs in CECT 7954 and 49 in CECT 7953 with respect to strain DSM10140 (data not shown). We focused on the four differential SNPs contained in the CECT 7953 genome, which are listed in Table 1. One SNP was located in the intergenic region between the d-tyrosyl-tRNA deacylase and phage-related integrase genes. Two others were found within open reading frames but produced silent mutations. Finally, the fourth SNP was located in the acetate kinase gene and resulted in a serine-to-phenylalanine substitution at position 154 of the protein. This SNP was confirmed by conventional PCR using four different forward primers (AckA-Dir [5′-TCGCGAATTCATGACGTG-3′], AckA-Dir2 [5′-TTGACGGCCACTACAAGTAC-3′], AckA-Dir3 [5′-GCACCTATGCGTTGAACAAG-3′], and AckA-Dir4 [5′-TTCCAGAGTTCCTCGGCATTG-3′]) in combination with the reverse primer AckA-Rev (5′-CCGGATTTCGTGGGTATG-3′).
Table 1.
Differential SNPs in the CECT 7953 genome with respect to strain CECT 7954
| Position in DSM genome | SNP |
Triplet change |
Amino acid change |
Function | Accession no.b | |||
|---|---|---|---|---|---|---|---|---|
| DSM, CECT 7954 | CECT 7953 | DSM, CECT 7954 | CECT 7953 | DSM, CECT 7954 | CECT 7953 | |||
| 579761 | G | A | TTCa | TTTa | Phe | Phe | MalF-type ABC sugar transport systems permeasea | ACS47432 |
| 1111792 | C | T | TCC | TTC | Ser | Phe | Acetate kinase | ACS47901 |
| 1424380 | C | A | GTGa | GTTa | Val | Val | Cell division protein FtsZa | ACS48156 |
| 1362250 | T | C | Intergenic DNAc | |||||
Serine is a polar amino acid with a hydroxyl group in its side chain, whereas phenylalanine is nonpolar because of its hydrophobic benzyl side chain. This single amino acid change is therefore likely to be responsible for the phenotype of CECT 7953. This mutation was responsible for a decrease in acetate kinase activity, as deduced from specific enzymatic assays performed with cells grown to the mid-exponential phase (CECT 7954, 8.34 ± 0.45 U/mg; CECT 7953, 5.88 ± 0.45 U/mg; P < 0.01). This was also reflected in the metabolic footprint of the strains. As shown in Table 2, resting cells of CECT 7953 produced less acetic acid after 4 h of incubation, supporting the reduced acetate kinase activity observed by enzymatic assays.
Table 2.
Glucose consumed, production of metabolic end products, acetic/lactic acid ratios, and carbon balance of buffered resting cells of B. animalis subsp. lactis CECT 7954 and low acetic acid producer CECT 7953a
| Strain | Glucose consumed (mM) | Organic acid formation (mM) |
Ethanol formation (mM) | Acetic/lactic acid ratio | Carbon balance | ||
|---|---|---|---|---|---|---|---|
| Lactic acid | Formic acid | Acetic acid | |||||
| CECT 7954 | 2.83 ± 0.65 | 1.73 ± 0.09 | 0.04 ± 0.01 | 3.97 ± 0.18b | 0.02 ± 0.01 | 2.29 ± 0.12b | 0.77 ± 0.05 |
| CECT 7953 | 2.36 ± 0.43 | 1.75 ± 0.08 | 0.03 ± 0.01 | 3.21 ± 0.09 | 0.01 ± 0.00 | 1.83 ± 0.09 | 0.82 ± 0.04 |
Results are expressed as means ± standard deviations of at least three independent experiments.
P < 0.01.
One of the common uses of bifidobacteria in human nutrition is inclusion in dairy products. However, this is very challenging from a practical point of view. First, bifidobacteria produce large amounts of acetic acid through sugar fermentation, conferring undesirable organoleptic properties on the final products. Second, the genus Bifidobacterium is composed mainly of strictly anaerobic members, with some exceptions, which makes their industrial use difficult (12). In fact, B. animalis subsp. lactis is widely present in commercial probiotic fermented milk samples (7) due to its good technological tolerance (22). For these reasons, bifidobacteria have been traditionally used in fermented milk samples as adjunct cultures, not participating directly in milk fermentation (13). To our knowledge, there is no fermented product on the market containing exclusively bifidobacteria due to these metabolic peculiarities, so we studied the performance of strains CECT 7954 and CECT 7953 grown in milk.
In general, strain CECT 7954 grew better than strain CECT 7953 in milk, although both were able to coagulate milk after 12 or 18 h of incubation, respectively. In both cases, viable counts after fermentation/storage of the fermented milk samples were over 108 CFU/ml (Table 3), higher than the bifidobacterium levels reported in commercial fermented milk samples (104 to 107 CFU/ml). This might supply the daily intake of viable bacteria, which is 109 in the case of probiotic strains (15), but it requires further research on the optimal supplementation of milk to favor bifidobacterial growth. Although no beneficial effects have been proved for any of the strains, these viable numbers make very promising the accomplishment of future work directed to the study of potential benefits of milk fermented by strain CECT 7953. However, care should be taken in the sense that, in our experimental approach, skim milk was supplemented with yeast extract. Thus, a direct comparison with commercial fermented milk samples cannot be performed.
Table 3.
Different parameters of milk fermented exclusively by strain CECT 7954 or CECT 7953a
| Feature | CECT 7954 | CECT 7953 |
|---|---|---|
| Residual lactose concn (mM) | 116.70 ± 2.52 | 118.00 ± 2.19 |
| Lactic acid concn (mM) | 61.00 ± 10.00b | 38.00 ± 14.00 |
| Acetic acid concn (mM) | 75.48 ± 16.13b | 41.60 ± 5.49 |
| Acetaldehyde concn (μg/g) | 3.52 ± 1.15b | 1.11 ± 0.71 |
| Ethanol concn (μg/g) | 421.34 ± 12.25b | 128.42 ± 28.49 |
| Diacetyl concn (μg/g) | 0.39 ± 0.17b | 1.39 ± 0.32 |
| Final pH | 4.18 ± 0.06b | 4.78 ± 0.09 |
| Log no. of CFU/ml | 9.2 ± 0.1b | 8.6 ± 0.2 |
After fermentation (pH ∼5.0), fermented milk samples were kept at 4°C for 24 h. Results are expressed as means ± standard deviations of at least three independent experiments.
Statistically significantly different (P < 0.01).
Milk fermented by strain CECT 7953 was characterized by lower concentrations of final metabolites, in direct agreement with its poorer growth. Smaller amounts of volatile compounds were also detected, among them acetate and ethanol. In addition, milk fermented by strain CECT 7953 contained higher levels of acetaldehyde, a metabolite associated with yoghurt-like flavors (16). Compared to those in classical fermented milk containing bifidobacteria, acetate levels still remained over 30 times greater, reflecting the metabolic activity of the bifidobacteria (20). Acetate is one of the main mechanisms by which bifidobacteria protect against enteropathogens (5). Thus, the natural presence of acetic acid in milk fermented by strain CECT 7953 might mediate beneficial effects, although this speculative point is reserved for further research.
In conclusion, strain CECT 7953 and strains derived from it in the future might be applied in the manufacture of new types of fermented milk products based exclusively on bifidobacterial fermentation. The probiotic vector is of paramount importance, and it is recognized that fermented milk can protect probiotics from the harsh conditions of the human gut (15). With the use of low acetate producer bifidobacteria, these beneficial microorganisms can be delivered while they are still metabolically active in less acidic end products.
ACKNOWLEDGMENTS
Borja Sánchez was the recipient of a Juan de la Cierva postdoctoral contract from the Spanish Ministerio de Ciencia e Innovación. This research was supported by grants AGL2007-61805 and RM2010-00012-00-00 from the Spanish Ministerio de Ciencia e Innovación.
The methods and strains described in this work are protected under Spanish patent P201131138.
Footnotes
Published ahead of print 2 March 2012
REFERENCES
- 1. Alhaj OA, Peters AC, Tatham AS, Kanekanian AD. 2010. Hypocholesterolaemic effect of Bifidobacterium animalis subsp. lactis (Bb12) and trypsin casein hydrolysate. Food Chem. 123:430–435 [Google Scholar]
- 2. Barrangou R, et al. 2009. Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04. J. Bacteriol. 191:4144–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chervaux C, et al. 2011. Genome sequence of the probiotic strain Bifidobacterium animalis subsp. lactis CNCM I-2494. J. Bacteriol. 193:5560–5561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. FAO/WHO 2001. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. World Health Organization, Geneva, Switzerland: http://www.who.int/foodsafety/publications/fs_management/en/probiotics.pdf Accessed 14 February 2012 [Google Scholar]
- 5. Fukuda S, et al. 2011. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469:543–547 [DOI] [PubMed] [Google Scholar]
- 6. Garrigues C, Pedersen MB, Johansen E. 2010. Complete genome sequence of Bifidobacterium animalis subsp. lactis BB-12, a widely consumed probiotic strain. J. Bacteriol. 192:2467–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gueimonde M, et al. 2004. Viability and diversity of probiotic Lactobacillus and Bifidobacterium populations included in commercial fermented milks. Food Res. Int. 37:839–850 [Google Scholar]
- 8. Jayamanne VS, Adams MR. 2006. Determination of survival, identity and stress resistance of probiotic bifidobacteria in bio-yoghurts. Lett. Appl. Microbiol. 42:189–194 [DOI] [PubMed] [Google Scholar]
- 9. Kim JF, et al. 2009. Genome sequence of the probiotic bacterium Bifidobacterium animalis subsp. lactis AD011. J. Bacteriol. 191:678–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Le Leu RK, Hu Y, Brown IL, Young GP. 2009. A combination of Bifidobacterium lactis and resistant starch can protect against colorectal cancer development in rats. Gastroenterology 136:A762–A763 [DOI] [PubMed] [Google Scholar]
- 11. Li H, Ruan J, Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Masco L, Crockaert C, Van Hoorde K, Swings J, Huys G. 2007. In vitro assessment of the gastrointestinal transit tolerance of taxonomic reference strains from human origin and probiotic product isolates of Bifidobacterium. J. Dairy Sci. 90:3572–3578 [DOI] [PubMed] [Google Scholar]
- 13. Masco L, Huys G, De Brandt E, Temmerman R, Swings J. 2005. Culture-dependent and culture-independent qualitative analysis of probiotic products claimed to contain bifidobacteria. Int. J. Food Microbiol. 102:221–230 [DOI] [PubMed] [Google Scholar]
- 14. Mayer MAG, Bronnenmeier K, Schwarz WH, Schertler C, Staudenbauer WL. 1995. Isolation and properties of acetate kinase-negative and phosphotransacetylase-negative mutants of Thermoanaerobacter thermohydrosulfuricus. Microbiology 141:2891–2896 [Google Scholar]
- 15. Ouwehand AC, Salminen S, Isolauri E. 2002. Probiotics: an overview of beneficial effects. Antonie Van Leeuwenhoek 82:279–289 [PubMed] [Google Scholar]
- 16. Pinto SM, Clemente MDG, De Abreu LR. 2009. Behaviour of volatile compounds during the shelf life of yoghurt. Int. J. Dairy Technol. 62:215–223 [Google Scholar]
- 17. Proudfoot AT, Bradberry SM, Vale JA. 2006. Sodium fluoroacetate poisoning. Toxicol. Rev. 25:213–219 [DOI] [PubMed] [Google Scholar]
- 18. Ruas-Madiedo P, Hernández-Barranco A, Margolles A, CGde los Reyes-Gavilán 2005. A bile salt-resistant derivative of Bifidobacterium animalis has an altered fermentation pattern when grown on glucose and maltose. Appl. Environ. Microbiol. 71:6564–6570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sánchez B, et al. 2005. Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 187:5799–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sánchez B, Fernández-Garcia M, Margolles A, CGde los Reyes-Gavilán Ruas-Madiedo P. 2010. Technological and probiotic selection criteria of a bile-adapted Bifidobacterium animalis subsp. lactis strain. Int. Dairy J. 20:800–805 [Google Scholar]
- 21. Savard P, et al. 2011. Impact of Bifidobacterium animalis subsp. lactis BB-12 and, Lactobacillus acidophilus LA-5-containing yoghurt, on fecal bacterial counts of healthy adults. Int. J. Food Microbiol. 149:50–57 [DOI] [PubMed] [Google Scholar]
- 22. Simpson PJ, Stanton C, Fitzgerald GF, Ross RP. 2005. Intrinsic tolerance of Bifidobacterium species to heat and oxygen and survival following spray drying and storage. J. Appl. Microbiol. 99:493–501 [DOI] [PubMed] [Google Scholar]
- 23. Sun ZH, et al. 2010. Complete genome sequence of probiotic Bifidobacterium animalis subsp. lactis strain V9. J. Bacteriol. 192:4080–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tabbers MM, et al. 2011. Fermented milk containing Bifidobacterium lactis DN-173 010 in childhood constipation: a randomized, double-blind, controlled trial. Pediatrics 127:e1392–e1399 [DOI] [PubMed] [Google Scholar]
- 25. Taipale T, et al. 2011. Bifidobacterium animalis subsp. lactis BB-12 in reducing the risk of infections in infancy. Br. J. Nutr. 105:409–416 [DOI] [PubMed] [Google Scholar]
- 26. Veiga P, et al. 2010. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc. Natl. Acad. Sci. U. S. A. 107:18132–18137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Waller PA, et al. 2011. Dose-response effect of Bifidobacterium lactis HN019 on whole gut transit time and functional gastrointestinal symptoms in adults. Scand. J. Gastroenterol. 46:1057–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]