Abstract
The complex symbiotic relationship between corals and their dinoflagellate partner Symbiodinium is believed to be sustained through close associations with mutualistic bacterial communities, though little is known about coral associations with bacterial groups able to fix nitrogen (diazotrophs). In this study, we investigated the diversity of diazotrophic bacterial communities associated with three common coral species (Acropora millepora, Acropora muricata, and Pocillopora damicormis) from three midshelf locations of the Great Barrier Reef (GBR) by profiling the conserved subunit of the nifH gene, which encodes the dinitrogenase iron protein. Comparisons of diazotrophic community diversity among coral tissue and mucus microenvironments and the surrounding seawater revealed that corals harbor diverse nifH phylotypes that differ between tissue and mucus microhabitats. Coral mucus nifH sequences displayed high heterogeneity, and many bacterial groups overlapped with those found in seawater. Moreover, coral mucus diazotrophs were specific neither to coral species nor to reef location, reflecting the ephemeral nature of coral mucus. In contrast, the dominant diazotrophic bacteria in tissue samples differed among coral species, with differences remaining consistent at all three reefs, indicating that coral-diazotroph associations are species specific. Notably, dominant diazotrophs for all coral species were closely related to the bacterial group rhizobia, which represented 71% of the total sequences retrieved from tissue samples. The species specificity of coral-diazotroph associations further supports the coral holobiont model that bacterial groups associated with corals are conserved. Our results suggest that, as in terrestrial plants, rhizobia have developed a mutualistic relationship with corals and may contribute fixed nitrogen to Symbiodinium.
INTRODUCTION
Corals are described as holobionts, where the coral animal hosts an array of mutualistic microorganisms, including the endosymbiotic dinoflagellate Symbiodinium, bacteria, archaea, and fungi (28, 44, 47, 48). These microorganisms inhabit the range of ecological niches provided by corals, such as the surface mucus layer, tissue layers, and the skeleton (6, 28, 30, 45, 48). Symbiotic relationships are based on the mutual exchange and control of nutritional resources. While interactions between Symbiodinium and coral hosts are well established, with Symbiodinium known to provide over 95% of fixed carbon to at least some coral hosts (42), the functional roles of diverse microbial communities that inhabit the coral holobiont are poorly understood. Documenting the bacterial communities associated with corals and elucidating the functional roles that they play in the corals' multipartner symbioses are the essential first steps in understanding processes involved in the establishment and structure of microbial communities and are critical to understanding the importance of bacterial communities for coral fitness.
Coral reefs have evolved in oligotrophic waters that are particularly poor in nitrogen (15). Although the success of coral reefs in such oligotrophic environments is largely dependent on symbioses between corals and Symbiodinium, the growth and abundance of these autotrophic dinoflagellates are also nitrogen limited (17). Gaseous nitrogen (N2) is present in relatively high concentrations in seawater, and therefore nitrogen fixation, the reduction of N2 to ammonia, is an important functional role that certain microorganisms, termed diazotrophs, might play in coral symbioses (36, 52, 58). Evidence is accumulating that diazotrophic organisms are more than just passive members of the coral microbial community and may potentially interact in a tight physiological relationship with both the coral animal and its associated microalgae and microbiota (26, 35, 36, 43, 52, 57). For example, acetylene reduction assays in early studies detected nitrogen fixation within coral samples (52, 58). More recently, cyanobacteria (32, 36) and other bacteria that possess genes for nitrogen fixation (26, 43, 57) have also been detected in coral tissues. Interestingly, Lesser et al. (35) and Olson et al. (43) both found that diazotrophs may have a close relationship with Symbiodinium. Lesser et al. (35) observed that the distribution of corals with symbiotic cyanobacteria was positively correlated with depth and suggested that cyanobacteria could sustain Symbiodinium nutrition in low-light environments. Olson et al. (43) found that the abundances of Symbiodinium and the dominant diazotrophic bacteria, which were closely related to the Vibrio genus, were positively correlated. Recent studies have also detected that both endosymbiotic algae and the coral host possess enzymes enabling ammonium assimilation (34, 55, 59); therefore, both could benefit from nitrogen fixation products. Additionally, coral microbiota, including archaea, are potentially involved in ammonium assimilation and other processes of nitrogen cycling, such as nitrification, ammonification, and denitrification (26, 53, 57).
Nitrogen fixation in diazotrophs is facilitated by the highly conserved nitrogenase enzyme complex (61). nifH is a segment of the gene that encodes the dinitrogenase iron protein, a subunit of the nitrogenase complex, and provides an ideal molecular target for gene-based phylogenetic characterization of diazotrophs because of agreement with 16S rRNA-based phylogeny (23, 56, 60, 61). In addition, there is little evidence of lateral gene transfer (61), which means that retrieval of nifH sequences has become a widely used approach for characterizing the diversity of diazotrophic communities in complex samples (12, 24, 38–40, 43, 56, 63).
In this study, we explored the diversity of N2-fixing bacteria associated with three coral species common on the Great Barrier Reef (GBR), using retrieval and analyses of nifH sequences. We tested for species specificity in coral-diazotroph associations through comparative analysis of the diversity of diazotrophic communities among three coral species located on the same reef, among three reef locations for each coral species, and finally within coral microhabitats, namely, mucus and tissue communities.
MATERIALS AND METHODS
Sample collection and processing.
Coral tissue and mucus samples were collected from three common coral species, Acropora millepora, Acropora muricata, and Pocillopora damicornis, at three midshelf reefs, Kelso reef (18°25′59″S,146°59′40″E), Knife reef (18°34′31″S, 147°34′5″E), and Davies reef (18°49′31″S, 147°38′50″E), in the central Great Barrier Reef (GBR) region in January 2009. Two coral colonies (biological replicates) of each species were sampled at each site. All coral colonies sampled were separated by at least 5 meters and were collected from depths of 5 to 10 meters.
To collect coral tissue, branches from the center of colonies were collected and placed in plastic bags underwater. At the surface, branches were first rinsed with autoclaved artificial seawater to remove exogenous microorganism contaminants from the ambient seawater column and mucus and then air brushed (80 lb/in2) to collect coral tissue as a slurry. The tissue slurry was homogenized, aliquoted into cryovials, and centrifuged at 13,000 × g to pellet the cellular material (supernatant was removed). Samples were then snap-frozen with liquid nitrogen and stored at −80°C until DNA extraction. Coral mucus was collected from only one of the two replicate colonies (for each species at each site). Mucus was aspirated in situ with a syringe and, at the surface, filtered through a Sterivex (0.2-μm) filter column (Millipore, MA), and frozen at −20°C shipboard before storage at −80°C. Seawater surrounding the sampled area (1 liter) was collected from Knife and Davies reefs and also filtered through Sterivex filters before storage at −80°C.
DNA extraction and purification.
Pellets of cellular material derived from coral tissue samples were resuspended in 0.5 ml sucrose extraction buffer (0.75 M sucrose, 40 mM EDTA, 50 mM Tris, pH 8.3), and total DNA was extracted following the protocol described by Bourne et al. (5). Extracted crude DNA was suspended in 30 μl sterile Milli-Q water and subsequently purified by passage through a 1.2% low-melting-point agarose gel, and high-quality DNA (2 kb) was cut from the gel and cleaned using the QIAquick gel extraction kit (Qiagen, Hilden, Germany), following the manufacturer's instructions. DNA was recovered in 30 μl sterile Milli-Q water, quantified using the NanoDrop ND1000 (NanoDrop Technologies), and stored at −20°C.
DNA from the coral mucus and seawater samples was extracted following a modified protocol described by Ceh et al. (8) and based on that described by Schauer et al. (50). DNA was purified as outlined for coral tissue samples.
PCR amplification of nifH gene and clone library preparation.
Clone libraries were prepared for each of the 18 coral tissue, 8 coral mucus, and 2 seawater samples. For clone library construction, a 359-bp fragment of the nifH gene (nitrogenase Fe protein gene) was amplified using degenerate primers and the nested PCR approach described by Zehr and McReynolds (62). Briefly, to amplify the nifH fragment, 2 μl of template was added to a first PCR mixture (50 μl) containing 1× PCR buffer [Tris-Cl, KCl, (NH4)2SO4], 4.0 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 40 ng · μl−1 bovine serum albumin (New England BioLabs), 2.5 U of Taq polymerase (Scientifix), and 0.4 μM (each) primers NifH3 (5′-ATR TTR TTN GCN GCR TA-3′) and NifH4 (5′-TTY TAY GGN AAR GGN GG-3′). PCR amplification was performed in an Eppendorf Mastercycler (Eppendorf, Hamburg, Germany) thermocycler programmed with an initial heating step for 3 min at 95°C, followed by 25 cycles consisting of 95°C for 30 s, 57°C for 30 s, and 72°C for 45 s, followed by a final extension for 7 min at 72°C. After thermal cycling, the products (2 μl) were subjected to a second round of PCR, identical to the first but with primers nifH1 (5′-TGYGAYCCNAARGCNGA-3′) and nifH2 (5′-ANDGCCATCATYTCNCC-3′) and with 30 PCR cycles. For all PCR runs, negative controls were tested and no amplification was obtained.
PCR products from each sample were visualized by electrophoresis, excised, and purified using a QIAquick gel extraction kit (Qiagen) according to the manufacturer's instructions. Purified PCR products were ligated into the TOPO-TA cloning vector (Invitrogen, Carlsbad, CA), with subsequent transformation into competent Escherichia coli cells and selection by blue and white screening, according to the manufacturer's instructions. Individual white colonies were transferred to 96-well plates containing LB agar and ampicillin. Plates with the transformants were sealed and submitted to Macrogen Inc. (Seoul, South Korea) for plasmid extraction and sequencing using a model 3739 × I automatic sequencer (Applied Biosystems, Foster City, CA). Previous pilot sequencing and diversity indices demonstrated that the diversity of nifH genes in corals is low; therefore, only 48 clones were sequenced from each coral sample (mucus and tissue) from each species, and 96 clones were sequenced for each of the two seawater samples.
Sequencing and phylogenetic analysis.
DNA sequences were edited initially to exclude primer and vector sequences using the Sequencher program (Gene Codes Corp., MI). Sequences were imported into the ARB software package (version 5.2; Department of Microbiology, Technical University of Munich [http://www.arb-home.de/]), where they were translated into protein amino acid sequences and aligned to create a Phylip-formatted distance matrix. Analysis was performed at the protein level because of the low resolution for identity that exists at the DNA level for this gene. A distance matrix was analyzed using MOTHUR (version 1.20.1; Department of Microbiology and Immunology, The University of Michigan [http://www.mothur.org/wiki/]), where deduced amino acid sequences from each library were grouped into operational protein units (OPUs) with a distance threshold of 0.10 (90% similarity between sequences). A representative sequence of each OPU was selected and aligned to the large nifH database available (Marine Microbiology, University of California [http://www.es.ucsc.edu/∼wwwzehr/research/database]) and maintained in ARB. The OPUs were checked against the closest related sequence in GenBank using the online BLASTP function (http://www.ncbi.nlm.nih.gov/GenBank/index.html), and if not found in the ARB nifH database, they were imported and aligned. Phylogenetic trees from protein amino acid sequences were generated in ARB using a Phylip maximum-likelihood method (Phylip PROML), including bootstrap support of 1,000 replicates.
Statistical analysis.
Diversity parameters of the nifH gene for each sample (tissue, mucus, and seawater) were generated in MOTHUR (version 1.20.1; Department of Microbiology and Immunology, The University of Michigan [http://www.mothur.org/wiki/]) using a 90% protein sequence similarity and included the Chao1 nonparametric richness estimates (9) and the Shannon-Weaver (51) and Simpson (54) indices of diversity. To determine patterns emerging from diazotrophic bacterial profiles from coral tissue, mucus, and seawater samples, principal-component analysis (PCA) was plotted using PAST statistical software (version 2.12) (22; http://palaeo-electronica.org/2001_1/past/issue1_01.htm). For tissue samples, the two biological replicates for each coral species at each site were pooled together. For all data analyses, only OPUs representing greater than 5 sequences were included.
Accession numbers.
The nifH sequences from the representative OPUs in this study have been deposited in GenBank under nucleotide and protein sequence accession numbers JN601397 to JN601421 and AEU12166 to AEU12190, respectively.
RESULTS
Diversity of nifH sequences.
A total of 1,344 high-quality nifH sequences were retrieved from coral mucus, coral tissue, and surrounding seawater samples. Retrieved nifH sequences had low relative diversity, and therefore sequences from replicate sample types were grouped, resulting in a total of 26 distinct OPUs across all samples. For mucus samples, between 6 and 10 OPUs were retrieved, with many being shared by 2 or more of the 3 coral species. Richness estimators (ACE and Chao1) revealed that the sampling effort was sufficient and that the diversity of the nifH sequences was well covered in these mucus samples (Table 1). Shannon-Weaver and Simpson indices provided corroborative evidence that the diversity in coral mucus samples was low, with values ranging from only 1.59 to 1.70 and 0.22 to 0.23, respectively, among coral species. Diversity parameters for coral tissue samples displayed more variability than those for mucus samples, with between 6 and 17 OPUs identified, many of which were shared across coral species. Interestingly, tissue samples of A. millepora revealed the lowest number of OPUs (6 OPUs) and the lowest Shannon-Weaver index (1.5), despite having the highest sequencing effort (280 retrieved nifH sequences). In contrast, Pocillopora damicornis (15 retrieved OPUs) and Acropora muricata (17 OPUs) showed a greater diversity and had the highest ACE and Chao estimator values, suggesting that the full diversity of the nifH gene was not encompassed by the sampling effort for these two species. Not surprisingly, nifH gene diversity in seawater samples was the highest overall, with 23 OPUs recovered. The high diversity of nifH sequences recovered from seawater samples was also observed in rarefaction analysis, which did not reach saturation (see Fig. S1 in the supplemental material). In contrast, mucus samples appeared to be highly saturated, and tissue samples approached an asymptote with sampling effort (see Fig. S1 in the supplemental material).
Table 1.
Number of OPUs and diversity estimates for nifH-deduced protein sequences from mucus and tissue samples of three coral species
| Library | No. of clones analyzed | No. of OPUs observed | ACE estimator | Chao1 estimator | Simpson index | Shannon-Weaver index |
|---|---|---|---|---|---|---|
| Acropora millepora | ||||||
| Tissue | 280 | 6 | 6 | 6 | 0.24 | 1.51 |
| Mucus | 145 | 10 | 10.19 | 11 | 0.23 | 1.70 |
| Pocillopora damicornis | ||||||
| Tissue | 257 | 15 | 70.51 | 22.50 | 0.15 | 2.07 |
| Mucus | 133 | 6 | 6 | 6 | 0.22 | 1.60 |
| Acropora muricata | ||||||
| Tissue | 230 | 17 | 38.43 | 24 | 0.17 | 1.99 |
| Mucus | 107 | 8 | 11.21 | 11 | 0.23 | 1.59 |
| Seawater | 197 | 23 | 33.22 | 34.25 | 0.26 | 1.96 |
Phylogeny of nifH sequences.
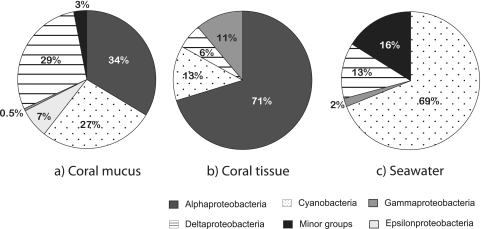
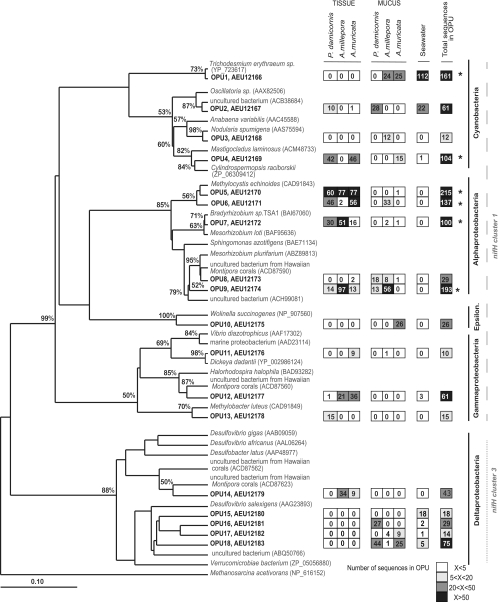
Comparisons of the class composition of diazotrophic bacteria among sample types (Fig. 1) and phylogenetic analysis (Fig. 2) both revealed that diazotrophic bacterial communities differed among coral tissue, coral mucus, and seawater libraries. Mucus libraries comprised very heterogeneous diazotrophic types and shared several OPUs (OPUs 1, 2, 16, 17, and 18) with seawater libraries (Fig. 2). In contrast, the Alphaproteobacteria class dominated coral tissue libraries combined for the three coral species (Fig. 1), and these did not share any OPUs with seawater libraries (Fig. 2), which were dominated by Cyanobacteria (Fig. 1).
Fig 1.
Class-level compositions of coral nifH clone libraries. Percent abundances per clone library from coral mucus (414 clones) (a), coral tissue sequences (767 clones) (b), and seawater (195 clones) (c) are shown. Minor groups represent OPUs that had fewer than 5 sequences and were not further analyzed.
Fig 2.
Phylogeny and composition of the nifH operational protein units (OPU) representing more than 10 sequences (cutoff, 0.1) (n = 1,347 sequences). OPUs from this study are represented in bold with associated protein accession numbers. The maximum-likelihood phylogenetic tree was generated in ARB and includes bootstrap support of 1,000 iterations. The tree was rooted with the nifH protein sequence of an archaeon (Methanosarcina acetivorans). The heat map to the right of the tree represents the total number of sequences retrieved for each OPU for each coral species (mucus and tissue separately) and seawater, with highest values (>50) in black and lowest (<5) in white. OPUs representing over 100 sequences are indicated with asterisks. Sequences from known phylogenies are indicated by species names and protein GenBank accession numbers in parentheses. Classes of bacteria are indicated to the far right of tree (Epsilon. stands for Epsilonproteobacteria), and nifH clusters are indicated with different dashed lines.
Coral mucus-derived sequences were taxonomically diverse, with the majority of sequences distributed among the Cyanobacteria, Deltaproteobacteria, and Alphaproteobacteria classes (Fig. 1). In these three classes of bacteria, the most abundant OPUs in terms of sequence numbers were OPU 18 (71 sequences), which demonstrated 100% amino acid sequence identity with an uncultured member of Deltaproteobacteria (ABQ50766), OPU 9 (69 sequences), which was closely related to an uncultured member of Alphaproteobacteria (ACH99081) (98% similarity), and OPU 1 (50 sequences), which was closest (99% similarity) to the Trichodesmium erythraeum nifH sequence (Fig. 2; Table 2). Of the remaining mucus-derived sequences, only one was found in the Gammaproteobacteria class, and 26 sequences were found in OPU 10, the only OPU belonging to the Epsilonproteobacteria class (Fig. 1 and 2). These last sequences had 89% similarity to a Wolinella succinogenes nifH sequence (Table 2).
Table 2.
Affiliations of representative nifH-deduced protein sequences grouped into OPUs (all samples pooled) and which contain 10 or more sequences
| Total no. of sequences in OPU | Source OPU, protein accession no. | Closest relative (accession no.) | Alignment (bp) | Similarity (%)a | Taxonomic description |
|---|---|---|---|---|---|
| 161 | OPU 1, AEU12166 | Trichodesmium erythraeum (YP_723617) | 117/118 | 99 | Cyanobacteria |
| 61 | OPU 2, AEU12167 | Uncultured bacterium (ACB38684) | 116/118 | 98 | Cyanobacteria |
| 12 | OPU 3, AEU12168 | Nodularia spumigena (AAS75594) | 106/108 | 98 | Cyanobacteria |
| 104 | OPU 4, AEU12169 | Cylindrospermopsis raciborskii (ZP_06309412) | 114/119 | 96 | Cyanobacteria |
| 215 | OPU 5, AEU12170 | Methylocystis echinoides (CAD91843) | 115/118 | 97 | Alphaproteobacteria |
| 137 | OPU 6, AEU12171 | Methylocystis echinoides (CAD91843) | 115/118 | 97 | Alphaproteobacteria |
| 100 | OPU 7, AEU12172 | Bradyrhizobium sp. (BAI67060) | 117/118 | 99 | Alphaproteobacteria |
| 29 | OPU 8, AEU12173 | Uncultured bacterium from Hawaiian Montipora corals (ACD87590) | 115/118 | 97 | Alphaproteobacteria |
| 193 | OPU 9, AEU12174 | Uncultured bacterium (ACH99081) | 116/118 | 98 | Alphaproteobacteria |
| 26 | OPU 10, AEU12175 | Wolinella succinogenes (NP_907560) | 118/132 | 89 | Epsilonproteobacteria |
| 10 | OPU 11, AEU12176 | Dickeya dadantii (YP_002986124) | 118/119 | 99 | Gammaproteobacteria |
| 61 | OPU 12, AEU12177 | Uncultured bacterium from Hawaiian Montipora corals (ACD87560) | 114/119 | 96 | Gammaproteobacteria |
| 15 | OPU 13, AEU12178 | Methylobacter luteus (CAD91849) | 114/119 | 96 | Gammaproteobacteria |
| 43 | OPU 14, AEU12179 | Uncultured bacterium from Hawaiian Montipora corals (ACD87623) | 112/119 | 94 | Deltaproteobacteria |
| 18 | OPU 15, AEU12180 | Desulfovibrio salexigens (AAG23893) | 96/109 | 88 | Deltaproteobacteria |
| 29 | OPU 16, AEU12181 | Uncultured bacterium (ABQ50766) | 103/119 | 87 | Deltaproteobacteria |
| 14 | OPU 17, AEU12182 | Uncultured bacterium (ABQ50766) | 103/119 | 87 | Deltaproteobacteria |
| 75 | OPU 18, AEU12183 | Uncultured bacterium (ABQ50766) | 119/119 | 100 | Deltaproteobacteria |
Protein sequences were aligned to their closest relative using protein BLAST (Blastp) results for affiliations.
The seawater libraries were dominated by Cyanobacteria (Fig. 1), with more than half of the total sequences (112 of 192 sequences) belonging to OPU 1, which was affiliated with Trichodesmium erythraeum (99% amino acid sequence identity), and 22 other sequences being affiliated with an uncultured member of Cyanobacteria (ACB38684) (Fig. 2; Table 2). The remaining seawater sequences were found in the Deltaproteobacteria class (nifH cluster 3), with OPU 15, which had the highest numbers of seawater sequences in this class, being only distantly affiliated with Desulfovibrio salexigens nifH (88% protein sequence similarity) (Fig. 2 and Table 2). Sixteen percent of seawater libraries were represented by single retrieved clones, further indicating high sequence diversity in seawater samples (Fig. 1), though this diversity was not explored further.
Seventy-one percent of sequences retrieved from coral tissues (539 sequences) belonged to the Alphaproteobacteria class (Fig. 1). Interestingly, all Alphaproteobacteria sequences were found in only 4 OPUs (OPU 5, OPU 6, OPU 7, and OPU 9), and these OPUs were all affiliated with Rhizobiales, a group of symbiotic diazotrophs that are found in the root nodules of legume plants (Fabaceae) (Fig. 2). OPU 7 (97 tissue sequences) was most closely affiliated (99%) to the nifH amino acid sequence of Bradyrhizobium sp. strain TSA1, and OPU 9 (124 tissue sequences) was closest taxonomically to an uncultured Alphaproteobacteria (ACH99081) nifH sequence (Fig. 2; Table 2). Interestingly, OPU 5 (214 tissue sequences), which was the most abundant OPU in all libraries, and OPU 6 (104 tissue sequences), which was the third most abundant group, were both affiliated (97% amino acid identity) with a known methanotroph (type I), Methylocystis echinoides. OPU 13, a member of Gammaproteobacteria, was also closely affiliated with the methanotroph (type II) Methylobacter luteus (CAD91849) nifH sequence, although this OPU comprised only 15 retrieved sequences. The remaining 29% of coral tissue-derived nifH sequences were distributed among 3 classes, the Cyanobacteria, the Gammaproteobacteria, and the Deltaproteobacteria (Fig. 1). Eighty-eight sequences derived from coral tissues were closely related (96% sequence identity) to a nifH sequence from the freshwater cyanobacterium, Cylindrospermopsis raciborskii. Interestingly, OPU 14 and OPU 12 were both most closely related to sequences that were found in the only other study to investigate nifH sequences in corals (43) (Fig. 2; Table 2). Another OPU that was closely related to a sequence found by Olson et al. (43) was OPU 8, which constituted 36 mucus-derived sequences and 2 tissue-derived sequences.
Specificity of coral diazotrophic assemblages.
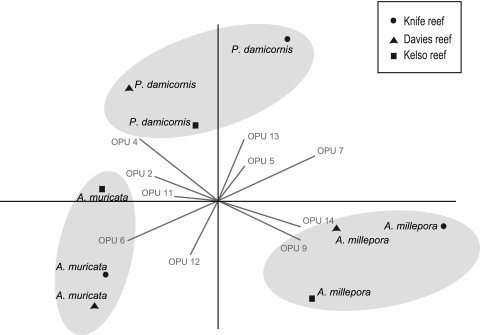
PCA comparison of diazotroph diversity associated with replicate coral tissue from the three coral species (Fig. 3) and with mucus from the three coral species and seawater samples from reefs samples (see Fig. S2 in the supplemental material) revealed clear patterns in bacterial communities among coral species. Tissue samples derived from colonies of the same coral species grouped together regardless of reef sampling location, with the first two components explaining 65% of the variation (Fig. 3). Although tissue samples from the three species were dominated by rhizobium-like sequences and all possessed similar abundances of sequences grouped in OPU 5, the relative abundances of other coral species-specific OPUs separated diazotrophic communities into three distinct groups corresponding to the three species (Fig. 3). For example, OPU 13 (15 sequences) was retrieved only from P. damicornis libraries, whereas OPU 11 (9 sequences) was retrieved only from A. muricata tissue libraries (Fig. 2 and 3). Both P. damicornis and A. muricata libraries contained sequences belonging to OPU 4, which was affiliated with a cyanobacterium closely related to Cylindrospermopsis raciborskii, but no cyanobacterial sequences were retrieved from A. millepora tissue samples. Sequences closely related to a methanotroph-derived nifH sequence grouped in OPU 6 and were abundant in both P. damicornis and A. muricata tissue libraries. A. millepora samples were clearly separated from those of the two other coral species in the PCA by their higher abundance of sequences grouped in OPU 9 and OPU 14. In addition, A. millepora and P. damicornis sequences were correlated strongly with OPU 7, while A. millepora and A. muricata sequences were correlated with OPU 12.
Fig 3.
PCA biplot representing relative abundances of nifH-deduced protein sequences derived from tissue clone libraries of three coral species sampled from each of three midshelf reefs. Gray lines show vectors from representative OPUs that drive the differences among clone libraries. The corresponding taxonomic affiliation and GenBank accession number of each OPU can be found in Fig. 2 and Table 2. For this analysis, only OPUs with more than 5 sequences are represented.
In contrast to patterns found for tissue samples, PCA analysis of retrieved nifH sequences derived from mucus and seawater samples showed no distinct patterns, with no evidence of mucus libraries grouping in relation to either coral species or reef location (see Fig. S2 in the supplemental material). Interestingly, the most abundant nifH sequences from seawater, which grouped in OPU 1 and were affiliated with a Trichodesmium sp., were also abundant in mucus libraries of A. millepora and A. muricata from Knife reef (Fig. 2). The presence of this OPU in these mucus samples likely represents contamination with seawater and results in these samples being separated from other mucus sequences in the PCA (see Fig. S2 in the supplemental material). The first two components of the PCA explained 40% of the variation in mucus and seawater clone library sequences.
DISCUSSION
Coral tissues are dominated by rhizobia.
Phylogenetic analysis of nifH gene clone libraries derived from tissues of three common coral species on the GBR revealed that diazotrophic communities are characterized by low diversity and a striking dominance of phylotypes from the Alphaproteobacteria class, with 71% of the total nifH sequences (n = 767 sequences) retrieved from coral tissues belonging to this class. Interestingly, all Alphaproteobacteria phylotypes were closely related to bacterial species belonging to the Rhizobiales order (between 97% and 99% amino acid identity) and were found in all three coral species investigated. Rhizobia are soil bacteria that inhabit nodules in the roots of legume plants. Rhizobia fix nitrogen, enabling plants to thrive and reproduce in nitrogen-poor environments, and in return, the rhizobia receive carbon and amino acids (19, 37). Rhizobium-like ribotypes have also been documented in 16S rRNA gene-based surveys of other coral species (29, 41, 47), as well as in functional gene arrays and metagenomic studies (26, 57), highlighting the potential importance of this group of bacteria for supplementing the nutritional requirements of the coral holobiont in nitrogen-limited oligotrophic waters. Plant rhizobia require specific signals to enter into symbiosis and for nitrogen fixation (11, 18). Similarly, close symbiotic pathways between diazotrophs and Symbiodinium and/or the coral could exist, as both are capable of ammonium assimilation (34, 55, 59). In plants, the cortex layer of nodules protects rhizobia from high concentrations of oxygen by acting as an oxygen diffusion barrier, while the plants' leghaemoglobin facilitates diazotroph respiration at low oxygen concentrations (16). It is currently unclear how rhizobia might be protected from high concentrations of oxygen arising from dinoflagellate photosynthesis in coral tissues (31). Coral microhabitats including bacterial aggregates within the gastrodermis, as reported by Ainsworth and Hoegh-Guldberg (2), or microaerophilic regions in the gastrovascular cavities of coral polyps, as observed by Agostini et al. (1), may host diazotrophic communities under oxygen-depleted conditions.
Rhizobium-affiliated sequences were clustered into four identified OPUs (OPUs 5, 6, 7, and 9). OPU 7 was closely related (99% amino acid similarity) to Bradyrhizobium spp., one of the most commonly occurring rhizobia that form symbioses in the nodules of legume plants. OPU 9 was closest to an uncultured bacterium (ACH99081) that is affiliated with a mesorhizobium. Interestingly, OPUs 5 and 6 were affiliated with a nifH sequence of Methylocystis echinoides, a methanotroph type II organism that also belongs to the Rhizobiales group. Moreover, OPU 5 represented the largest number of sequences retrieved and derived from all coral species sampled (215 sequences). The ability to fix nitrogen is an important phenotypic trait of most currently known methanotrophic bacteria, and nifH has been used to distinguish representatives at the clade level (4, 14). There are only a few reports of the presence of methanotrophs in corals. Using a functional gene array, Kimes et al. (26) found several genes involved in methane oxidation, with some belonging to diverse methanotrophs from type I and II and also from archaea. Siboni et al. (53) investigated archaea associated with the mucus of Acanthastrea, Favia, and Fungia species and found that 8% of the euryarchaeotal sequences were affiliated with anaerobic methanotroph type II organisms. In our study, a methanotroph type I (OPU 13) was also found, although only in P. damicornis tissue libraries. Our finding that this type of bacteria is the dominant diazotrophic group in three common coral species is an indication that methane is potentially abundant in coral tissues. Accordingly, our results highlight the need for further investigations of the fate of methane in the coral holobiont.
Diazotrophs from coral tissues are specific to coral species.
Although all coral species had high abundances of rhizobium-like sequences in their tissues, PCA of nifH sequences grouped coral samples from 3 different reefs according to coral species. We conclude that differences in the relative abundances of OPUs were responsible for grouping diazotrophic communities according to coral species and that diazotrophic communities display some coral species-specific associations. Previous studies targeting the 16S rRNA gene have reported that bacterial communities on a coral species are similar at geographically separated locations (5, 46, 47; K. B. Ritchie and G. W. Smith, presented at the 8th International Coral Reef Symposium, Panama City, Panama, 1997). These studies have contributed to the development of the coral holobiont model, in which the coral is viewed as a structured symbiotic system composed of the animal host and an array of microscopic partners, including bacteria (28, 44, 48). This study is the first to report that bacteria with a specific functional role can also be specific to a coral host, and it therefore provides additional support for the holobiont model. It is important to note that all reefs sampled in our study were mid- or outer-shelf reefs separated by distances ranging from 50 km to 200 km, and they thus experienced similar water quality parameters. Accordingly, it is possible that diazotrophic communities from coral species might differ between more distant reefs, particularly between locations with differing nutrient inputs, such as inshore versus offshore reefs. Findings that bacterial communities associated with corals differ among locations that differ in environmental characteristics (27, 44) highlight the need for further investigations of coral diazotrophic communities exposed to different water quality parameters.
The ephemeral nature of coral mucus.
In contrast to the consistent patterns found for diazotrophic communities associated with coral tissues, a variety of nifH groups were retrieved from coral mucus samples, and these belonged to five different classes of bacteria (Alphaproteobacteria, Gammaproteobacteria, Epsilonproteobacteria, Deltaproteobacteria, and Cyanobacteria). The differences in the consistency of diazotrophic communities between coral tissue and mucus samples found in our study are corroborated by previous 16S rRNA studies, which also found differences in general bacterial communities between coral tissue and mucus samples (6, 20, 21, 30, 33, 47). The overlap found between the coral mucus microbiota and that of the surrounding seawater is not surprising, as small amounts of seawater are taken up when coral mucus samples are collected in situ. Indeed, the highest number of sequences recovered from mucus grouped in OPU 1, which also possessed the highest number of seawater-derived sequences and were closely affiliated to a nifH sequence derived from Trichodesmium erythraeum. In addition, PCA analysis showed no relationship between nifH gene diversity recovered from mucus libraries and either coral species or location (see Fig. S1 in the supplemental material). The location of mucus bacterial communities at the interface between seawater and coral tissues undoubtedly promotes continuous exchange and contributes to the ephemeral, complex, and dynamic nature of mucus microbial communities (7, 25). Indeed, bacterial groups within coral mucus exhibit high spatial heterogeneity because of the many different factors that may have an impact on this habitat, such as various degrees of light exposure, nutrient availability, sedimentation, mucus age, and competition with other microbes (3, 13, 21, 49).
A large number of nifH cluster III sequences (10), all closely related to sulfate reducers, including Desulfovibrio spp., represented the second largest group retrieved from mucus samples in our study and were also found in coral tissues (OPU 14). Kimes and colleagues (26) determined that 22% of dsr genes in corals belong to the Deltaproteobacteria subclass and suggested that inorganic sulfate might be another potential source of sulfur for the coral holobiont. Interestingly, nifH sequences close to Desulfovibrio and other uncultured sulfate reducers were also found in the only other study that investigated nifH in corals (43). As found by Olson and colleagues (43), our coral-derived nifH sequences were closest to Desulfovibrio salexigens and had low amino acid identity (only 87 to 88%) with all other sequences, indicating the presence of potentially novel species. OPU 14, which was the only Deltaproteobacteria nifH-affiliated sequence derived from coral tissue in this study, had a 94% similarity to a nifH sequence from the study by Olson et al. (43). Interestingly, other OPUs were observed to be closely affiliated with nifH sequences retrieved from corals from Hawaii, including OPU 12, a tissue gammaproteobacterium, and OPU 8, a mucus alphaproteobacterium. The similarity of ribotypes between this and other studies further supports that some diazotrophic communities are consistent in corals and therefore that they represent important members of the coral holobiont symbiosis.
Conclusions.
Tissues of three common coral species on the GBR displayed species-specific patterns in diazotrophic communities, suggesting that this bacterial group is likely to have a symbiotic role in the coral holobiont. The presence of consistent and dominant populations of rhizobia in all three coral species at all reefs suggests that coral rhizobia are important symbiotic members of coral tissues, although further studies are required to demonstrate that symbionts are actively fixing nitrogen. The specificity of tissue diazotrophs to each of the coral species provides further support for the holobiont model of coral symbioses. In contrast, coral mucus is a complex microbial microhabitat with an ephemeral nature, given its location at the interface between coral tissues and seawater and potential for microbial exchange, which appears to give rise to a diverse and unstructured diazotrophic community. Overall consistency in the identification of diazotrophic nifH sequences associated with corals in our and other studies suggests that these microbial groups may be essential in nitrogen cycling within the coral holobiont. Importantly, changes in diazotrophic communities could directly reflect shifts in environmental parameters, such as nutrient inputs, and could be used to detect changes in coral fitness in response to environmental change.
Supplementary Material
ACKNOWLEDGMENTS
We thank Andrew Muirhead, Jean Baptiste Raina, and Christian Pfeffer for their time and technical help in the laboratory. We also thank Jean Baptiste Raina for valuable intellectual discussions and revisions which improved earlier versions of the manuscript.
We thank the Australian Institute of Marine Sciences (AIMS), AIMS@JCU, and the ARC Centre of Excellence for Coral Reef (James Cook University) for their financial contributions to this research. We also thank CONACYT (Mexico) for giving financial support to Kimberley A. Lema.
Footnotes
Published ahead of print 17 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Agostini S, et al. 2009. Coral symbiotic complex: hypothesis through vitamin B12 for a new evaluation. Galaxea (Tokyo) 11:1–11 [Google Scholar]
- 2. Ainsworth TD, Hoegh-Guldberg O. 2009. Bacterial communities closely associated with coral tissues vary under experimental and natural reef conditions and thermal stress. Aquat. Biol. 4:289–296 [Google Scholar]
- 3. Ainsworth TD, Vega Thurber R, Gates RD. 2010. The future of coral reefs: a microbial perspective. Trends Ecol. Evol. 25:233–240 [DOI] [PubMed] [Google Scholar]
- 4. Auman AJ, Speake CC, Lidstrom ME. 2001. nifH sequences and nitrogen fixation in type I and type II methanotrophs. Appl. Environ. Microbiol. 67:4009–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bourne DG, Iida Y, Uthicke S, Smith-Keune C. 2008. Changes in coral-associated microbial communities during a bleaching event. ISME J. 2:350–363 [DOI] [PubMed] [Google Scholar]
- 6. Bourne DG, Munn CB. 2005. Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ. Microbiol. 7:1162–1174 [DOI] [PubMed] [Google Scholar]
- 7. Brown BE, Bythell JC. 2005. Perspectives on mucus secretion in reef corals. Mar. Ecol. Prog. Ser. 296:291–309 [Google Scholar]
- 8. Ceh J, Van Keulen M, Bourne DG. 2011. Coral-associated bacterial communities on Ningaloo Reef, Western Australia. FEMS Microbiol. Ecol. 75:134–144 [DOI] [PubMed] [Google Scholar]
- 9. Chao A. 1984. Nonparametric estimation of the number of classes in a population. Scand. Stat. 11:265–270 [Google Scholar]
- 10. Chien Y, Zinder SH. 1996. Cloning, functional organization, transcript studies, and phylogenetic analysis of the complete nitrogenase structural genes (nifHDK2) and associated genes in the Archaeon Methanosarcina barkeri 227. J. Bacteriol. 178:143–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cullimore J, Dénarié J. 2003. How legumes select their sweet talking symbionts. Science 302:575–578 [DOI] [PubMed] [Google Scholar]
- 12. Dang H, Luan X, Zhao J, Li J. 2009. Diverse and novel nifH and nifH-like gene sequences in the deep-sea methane seep sediments of the Okhotsk Sea. Appl. Environ. Microbiol. 75:2238–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daniels CA, et al. 2011. Spatial heterogeneity of bacterial communities in the mucus of Montastraea annularis. Mar. Ecol. Prog. Ser. 426:29–40 [Google Scholar]
- 14. Dedysh SN, Ricke P, Liesack W. 2004. NifH and NifD phylogenies: an evolutionary basis for understanding nitrogen fixation capabilities of methanotrophic bacteria. Microbiology 150:1301–1313 [DOI] [PubMed] [Google Scholar]
- 15. D'Elia CF, Wiebe WJ. 1990. Biogeochemical cycles in coral reef ecosystems, p 49–74 In Dubinsky Z. (ed), Coral reefs: ecosystems of the world, vol 25 Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 16. Dixon R, Kahn D. 2004. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2:621–631 [DOI] [PubMed] [Google Scholar]
- 17. Falkowski PG, Dubinsky Z, Muscatine L, McCloskey L. 1993. Population control in symbiotic corals. Bioscience 43:606–611 [Google Scholar]
- 18. Fischer H. 1994. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 58:352–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fred EB, Baldwin IL, McCoy E. 1932. Root nodule bacteria and leguminous plants, vol 5 Parallel Press, Madison, WI [Google Scholar]
- 20. Frias-Lopez J, Zerkle AL, Bonheyo GT, Fouke BW. 2002. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl. Environ. Microbiol. 68:2214–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guppy R, Bythell JC. 2006. Environmental effects on bacterial diversity in the surface mucus layer of the reef coral Montastraea faveolata. Mar. Ecol. Prog. Ser. 328:133–142 [Google Scholar]
- 22. Hammer O, Harper DAT, Ryan P. 2001. Past: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4:1–9 [Google Scholar]
- 23. Hennecke H, et al. 1985. Concurrent evolution of nitrogenase genes and 16S rRNA in Rhizobium species and other nitrogen fixing bacteria. Arch. Microbiol. 142:342–348 [Google Scholar]
- 24. Hewson I, Moisander PH, Morrison AE, Zehr JP. 2007. Diazotrophic bacterioplankton in a coral reef lagoon: phylogeny, diel nitrogenase expression and response to phosphate enrichment. ISME J. 1:78–91 [DOI] [PubMed] [Google Scholar]
- 25. Johnston IS, Rohwer F. 2007. Microbial landscapes on the outer tissue surfaces of the reef-building coral Porites compressa. Coral Reefs 26:375–383 [Google Scholar]
- 26. Kimes NE, Van Nostrand JD, Weil E, Zhou J, Morris PJ. 2010. Microbial functional structure of Montastraea faveolata, an important Caribbean reef-building coral, differs between healthy and yellow-band diseased colonies. Environ. Microbiol. 12:541–556 [DOI] [PubMed] [Google Scholar]
- 27. Klaus JS, Janse I, Heikoop JM, Sanford RA, Fouke BW. 2007. Coral microbial communities, zooxanthellae and mucus along gradients of seawater depth and coastal pollution. Environ. Microbiol. 9:1291–1305 [DOI] [PubMed] [Google Scholar]
- 28. Knowlton N, Rohwer F. 2003. Multispecies microbial mutualisms on coral reefs: the host as a habitat. Am. Nat. 162:S51–S62 [DOI] [PubMed] [Google Scholar]
- 29. Kooperman N, Ben-Dov E, Kramarsky-Winter E, Barak Z, Kushmaro A. 2007. Coral mucus-associated bacterial communities from natural and aquarium environments. FEMS Microbiol. Lett. 276:106–113 [DOI] [PubMed] [Google Scholar]
- 30. Koren O, Rosenberg E. 2006. Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Appl. Environ. Microbiol. 72:5254–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kühl M, Cohen Y, Dalsgaard T, Jorgensen BB, Revsbech NP. 1995. Microenvironment and photosynthesis of zooxanthellae in scleractinian corals studied with microsensors for O2, pH and light. Mar. Ecol. Prog. Ser. 117:159–172 [Google Scholar]
- 32. Kvennefors ECE, Roff G. 2009. Evidence of cyanobacteria-like endosymbionts in Acroporid corals from the Great Barrier Reef. Coral Reefs 28:547 [Google Scholar]
- 33. Lampert Y, et al. 2008. Phylogenetic diversity of bacteria associated with the mucus of Red Sea corals. FEMS Microbiol. Ecol. 64:187–198 [DOI] [PubMed] [Google Scholar]
- 34. Leggat W, Hoegh-Guldberg O, Dove S. 2007. Analysis of an EST library from the dinoflagellate (Symbiodinium sp.) symbiont of reef-building corals. J. Phycol. 43:1010–1021 [Google Scholar]
- 35. Lesser MP, et al. 2007. Nitrogen fixation by symbiotic cyanobacteria provides a source of nitrogen for the scleractinian coral Montastraea cavernosa. Mar. Ecol. Prog. Ser. 346:143–152 [Google Scholar]
- 36. Lesser MP, Mazel CH, Gorbunov MY, Falkowski PG. 2004. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305:997–1000 [DOI] [PubMed] [Google Scholar]
- 37. Lodwig EM, et al. 2003. Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature 422:722–726 [DOI] [PubMed] [Google Scholar]
- 38. Lovell CR, Piceno YM, Quattro JM, Bagwell CE. 2000. Molecular analysis of diazotroph diversity in the rhizosphere of the smooth cordgrass, Spartina alterniflora. Appl. Environ. Microbiol. 66:3814–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mohamed NM, Colman AS, Tal Y, Hill RT. 2008. Diversity and expression of nitrogen fixation genes in bacterial symbionts of marine sponges. Environ. Microbiol. 10:2910–2921 [DOI] [PubMed] [Google Scholar]
- 40. Moisander PH, Beinart RA, Voss M, Zehr JP. 2008. Diversity and abundance of diazotrophic microorganisms in the South China Sea during intermonsoon. ISME J. 2:954–967 [DOI] [PubMed] [Google Scholar]
- 41. Mouchka ME, Hewson I, Harvell CD. 2010. Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 50:662–674 [DOI] [PubMed] [Google Scholar]
- 42. Muscatine L, Porter JW. 1977. Reef corals: mutualistic symbioses adapted to nutrient-poor environements. Bioscience 27:454–460 [Google Scholar]
- 43. Olson ND, Ainsworth TD, Gates RD, Takabayashi M. 2009. Diazotrophic bacteria associated with Hawaiian Montipora corals: diversity and abundance in correlation with symbiotic dinoflagellates. J. Exp. Mar. Biol. Ecol. 371:140–146 [Google Scholar]
- 44. Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E. 2006. The coral probiotic hypothesis. Environ. Microbiol. 8:2068–2073 [DOI] [PubMed] [Google Scholar]
- 45. Ritchie KB, Smith GW. 2004. Microbial communities of coral surface mucopolysaccharide layers, p 259–264 In Rosenberg E, Loya Y. (ed), Coral health and disease. Springer, Berlin, Germany [Google Scholar]
- 46. Rohwer F, Breitbart M, Jara J, Azam F, Knowlton N. 2001. Diversity of bacteria associated with the Carbbean coral Montastraea franksi. Coral Reefs 20:85–91 [Google Scholar]
- 47. Rohwer F, Seguritan V, Azam F, Knowlton N. 2002. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243:1–10 [Google Scholar]
- 48. Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. 2007. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5:355–362 [DOI] [PubMed] [Google Scholar]
- 49. Rypien KL, Ward JR, Azam F. 2010. Antagonistic interactions among coral-associated bacteria. Environ. Microbiol. 12:28–39 [DOI] [PubMed] [Google Scholar]
- 50. Schauer M, Massana R, Pedros-Alio C. 2000. Spatial differences in bacterioplankton composition along the Catalan coast (NW Mediterranean) assessed by molecular fingerprinting. FEMS Microbiol. Lett. 33:51–59 [DOI] [PubMed] [Google Scholar]
- 51. Shannon CE, Weaver W. 1949. The mathematical theory of communication. University of Illinois Press, Champaign, IL [Google Scholar]
- 52. Shashar N, Cohen Y, Loya Y, Sar N. 1994. Nitrogen fixation (acetylene reduction) in stony corals: evidence for coral-bacteria interactions. Mar. Ecol. Prog. Ser. 111:259–264 [Google Scholar]
- 53. Siboni N, Ben-Dov E, Sivan A, Kushmaro A. 2008. Global distribution and diversity of coral-associated Archaea and their possible role in the coral holobiont nitrogen cycle. Environ. Microbiol. 10:2979–2990 [DOI] [PubMed] [Google Scholar]
- 54. Simpson EH. 1949. Measurment of diversity. Nature 163:688 [Google Scholar]
- 55. Stambler N. 2011. Zooxanthellae: the yellow symbionts inside animals, part 3, p 87–106 In Dubinsky Z, Stambler N. (ed), Coral reefs: an ecosystem in transition. Springer, New York, NY [Google Scholar]
- 56. Ueda T, Suga Y, Yahiro N, Matsuguchi T. 1995. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J. Bacteriol. 177:1414–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wegley L, Edwards RA, Rodriguez-Brito B, Liu H, Rohwer F. 2007. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9:2707–2719 [DOI] [PubMed] [Google Scholar]
- 58. Williams WM, Viner AB, Broughton WJ. 1987. Nitrogen fixation (acetylene reduction) associated with the living coral Acropora variabilis. Mar. Biol. 94:531–535 [Google Scholar]
- 59. Yellowlees D, Alwyn T, Rees V, Leggat W. 2008. Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ. 31:679–694 [DOI] [PubMed] [Google Scholar]
- 60. Young JPW. 1992. Phylogenetic classification of nitrogen fixing organisms, p 43–86 In Stacey G, Evans HJ, Burris RH. (ed), Biological nitrogen fixation. Chapman and Hall, New York, NY [Google Scholar]
- 61. Zehr JP, Jenkins BD, Short SM, Steward GF. 2003. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5:539–554 [DOI] [PubMed] [Google Scholar]
- 62. Zehr JP, McReynolds LA. 1989. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl. Environ. Microbiol. 55:2522–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zehr JP, Mellon MT, Zani S. 1998. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl. Environ. Microbiol. 64:3444–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.