Abstract
As more whole-genome sequences become available, there is an increasing demand for high-throughput methods that link genes to phenotypes, facilitating discovery of new gene functions. In this study, we describe a new version of the Tn-seq method involving a modified EZ:Tn5 transposon for genome-wide and quantitative mapping of all insertions in a complex mutant library utilizing massively parallel Illumina sequencing. This Tn-seq method was applied to a genome-saturating Salmonella enterica serotype Typhimurium mutant library recovered from selection under 3 different in vitro growth conditions (diluted Luria-Bertani [LB] medium, LB medium plus bile acid, and LB medium at 42°C), mimicking some aspects of host stressors. We identified an overlapping set of 105 protein-coding genes in S. Typhimurium that are conditionally essential under at least one of the above selective conditions. Competition assays using 4 deletion mutants (pyrD, glnL, recD, and STM14_5307) confirmed the phenotypes predicted by Tn-seq data, validating the utility of this approach in discovering new gene functions. With continuously increasing sequencing capacity of next generation sequencing technologies, this robust Tn-seq method will aid in revealing unexplored genetic determinants and the underlying mechanisms of various biological processes in Salmonella and the other approximately 70 bacterial species for which EZ:Tn5 mutagenesis has been established.
INTRODUCTION
In recent years, an increasing number of complete genome sequences have been becoming available for numerous bacterial species, mainly due to rapidly developing DNA sequencing technologies (26). This situation has raised pressing needs for understanding the functional significance of the numerous genes identified or predicted in these complete genomes. Bioinformatics analyses of the complete genomes have brought significant insights into our understanding of the biological implications of the genomic contents (18). However, these approaches would provide little help in assigning functions to the genes with no homology to any known genes or predicting the genes responsible for the phenotypes that have never been explored yet. In this view, experimental approaches to study gene functions are essential tools in understanding the genome biology of bacteria. Particularly, high-throughput approaches to study gene functions on a genome-wide scale are especially attractive in this postgenomic era, with overwhelming amounts of bacterial genomes to be explored, each containing thousands of genes.
Several high-throughput methods to study gene functions in bacteria have been developed using transposon mutagenesis based on the same framework of negative selection, yet using different strategies to compare mutant pools before and after selection (2). More recently, the extension of these strategies has undergone dramatic improvement in their capacity to determine gene functions in terms of the accuracies in genome mapping of transposon insertion sites and quantitative measurement of each insertion by employing next generation sequencing (5, 9, 12, 14, 17, 23, 41). These methods use different procedures to capture transposon-junction sequences and sequence them by massively parallel Illumina sequencing to accomplish an in-depth profiling of transposon-junction sequences in a complex mutant library. Although distinct names have been used, all of these methods can be appropriately referred to as different variations of the “Tn-seq” method (12, 41).
Here, we describe a new version of Tn-seq conveniently tailored to the EZ:Tn5 transposon system, which has been broadly tested and used in more than 70 bacterial and archaeal species (http://www.epibio.com/transcite.asp). We applied this Tn-seq method to conduct genome-wide identification of Salmonella enterica serotype Typhimurium genes that are conditionally essential for growth or survival under the 3 selected in vitro growth conditions. These conditions include low-nutrient condition, a bile-rich environment, and body temperature of avian species (42°C), mimicking some aspects of the conditions in the host Salmonella cells are expected to encounter during infection. As a common human bacterial pathogen, Salmonella has to survive when it goes through a variety of stress conditions in the environments and the host during infection (8, 11, 33, 36). Although a wealth of information on the genetic determinants and the underlying mechanisms of stress resistance have been obtained for Salmonella, there is no doubt that many gaps still exist in our knowledge and understanding in this area. This is especially true in view of the most comprehensive phenomic profiling performed for Escherichia coli K-12 recently (30).
This study demonstrates the utility and efficiency of this Tn-seq method for the comprehensive identification of conditionally essential genes in Salmonella. The genes identified here could be an important resource for better understanding or control of Salmonella, including development of novel antimicrobials and vaccines.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The S. Typhimurium 14028 wild-type strain, isogenic deletion mutant strains, and a spontaneous mutant resistant to nalidixic acid (NA) were grown in Luria-Bertani (LB) medium or LB agar plates and stored at −80°C in 30% glycerol. The cultures were incubated at 37°C unless described otherwise. Where appropriate, the LB agar plates contained NA (25 μg/ml) or kanamycin (Km; 50 μg/ml).
Construction of transposon mutant library.
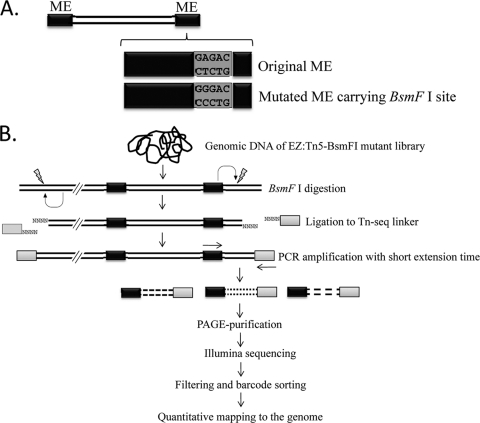
The QuikChange site-directed mutagenesis kit (Agilent Technologies La Jolla, CA) was used to change one nucleotide of pMOD-6 <KAN-2/MCS> plasmid (Epicentre BioTechnologies, Madison, WI) in one of the mosaic end (ME) sequences using the oligonucleotides shown in Table S1 in the supplemental material. This introduced the recognition sequence of type IIS enzyme BsmFI in one ME sequence (Fig. 1A). The nucleotide change A/T→G/C was confirmed by DNA sequencing, and the modified plasmid, pMOD-BsmFI, was used for transposon mutagenesis of the S. Typhimurium ATCC 14028 wild-type strain. Briefly, pMOD-BsmFI was digested with PvuII enzyme, and the 1,116-bp fragment of the modified EZ:Tn5 (EZ:Tn5-BsmFI) was extracted from agarose gel using the QIAquick gel extraction kit (Qiagen, Valencia, CA), avoiding exposure of the fragment to UV light. EZ:Tn5-BsmFI was then incubated with EZ-Tn5 transposase (Epicentre BioTechnologies) to form a transposon complex according to the instruction manual. Two microliters of the complex was subsequently used to transform electrocompetent S. Typhimurium 14028 wild-type cells by electroporation. Km-resistant (Kmr) transformants were selected on LB plates supplemented with Km. The resulting mutants were combined to form a complex library of EZ:Tn5-BsmFI mutants containing approximately 1.6 × 104 mutants. The library was stored at −80°C in 30% glycerol.
Fig 1.
Schematic diagrams for the Tn-seq method used in this study. (A) A single nucleotide was changed in one mosaic end (ME) of EZ:Tn5 to introduce the BsmFI recognition site (5′-GGGAC(N)10↓-3′/3′-CCCTG(N)14↑-5′). (B) Deep profiling of Tn5-junction sequences by Illumina sequencing.
In vitro selection of transposon mutant library.
The mutant library was subjected to selection under 4 different in vitro conditions: LB medium, 10× diluted LB medium (dLB), LB medium supplemented with 5% crude ox bile extract (Sigma; LB-bile), and LB medium incubated at 42°C (LB-42°C). To prepare the inoculum, 1 ml of the library in glycerol stock was diluted by adding 9 ml of LB medium and incubated at 37°C for 1 h with vigorous shaking (225 rpm). After being washed 3 times with phosphate-buffered saline (PBS) solution, the library was resuspended in 10 ml LB medium and diluted with LB medium to reach an optical density at 600 nm (OD600) of ≈0.9. An aliquot (2 ml) of the inoculum was used as the input pool. One milliliter of the inoculum (∼7.3 × 107 CFU/ml) of the mutant library was used to inoculate each selection medium (100 ml in a 200-ml Erlenmeyer flask) and incubated at the relevant temperature with vigorous shaking (225 rpm). After 24 h, 1 ml of the culture was transferred to the fresh selection medium of the same kind. This selection was repeated 3 times to increase the selection sensitivity of the screening. After 3 consecutive selections, 2 ml of the final culture in each selection was collected to be used as the output pools. The cell pellets from one input pool and 4 output pools were used to isolate genomic DNA using a DNA minikit (Qiagen, CA). The quantity and purity of the purified genomic DNA were measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE).
Sample preparation and Illumina sequencing.
Extracted genomic DNA was digested with BsmFI restriction enzyme (New England BioLabs, Ipswich, MA) at 65°C for 3 h. After heat inactivation at 80°C for 20 min, the digested DNA was treated with calf intestinal alkaline phosphatase (NEB) at 37°C for 1 h to prevent self-ligation in the following ligation step by incubation for additional 1 h. DNA was then phenol-chloroform extracted, ethanol precipitated, and dissolved in 10 μl H2O. DNA digests were subsequently ligated to Tn-seq linker, formed by annealing Tn-seq linker 1 and 2 oligonucleotides (see Table S1 in the supplemental material), by overnight incubation at room temperature. The Tn-seq linker-ligated samples were subsequently used as templates in PCR using cloned Pfu DNA polymerase (Agilent Technologies) with one of the 5 bar-coded Tn5 primers and Tn-seq linker primer (see Table S1). The PCR cycles consisted of initial denaturation at 94°C for 2 min, 5 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 30 s, and 20 cycles of 94°C for 30 s, 62°C for 1 min, and 72°C for 30 s, followed by final extension at 72°C for 10 min. The amplicon, which was 129 bp long, was PAGE purified and dissolved in H2O (see Fig. S1 in the supplemental material). The 5 DNA samples tagged with different bar codes were mixed with the same amount based on measurements with a NanoDrop apparatus (Thermo Fisher Scientific). The final mixed sample was analyzed using an Affymetrix BioAnalyzer to check the quality and sequenced using the Illumina genome analyzer II at the Institute for Integrative Genome Biology at the University of California at Riverside.
Analysis of Illumina sequencing data.
A computer program was written in the Python programming language to perform the following analysis. (The Python script is available upon request.) First, the sequence reads obtained from Illumina sequencing were sorted for the reads that contain a perfect 19-bp modified ME sequence (5′-CTGTCCCTTATACACATCT-3′). Next, the filtered sequences were sorted according to the 6-nucleotide (nt) bar code sequences demanding a perfect match to one of the 5 bar codes. The transposon-junction sequences were subsequently extracted from the filtered reads, and the junction sequences 11 to 13 bp long were selected for further analysis. These selected transposon-junction sequences were then mapped to the complete genome of S. Typhimurium 14028 (20) (accession numbers: chromosome, CP001363.1; plasmid, CP001362.1) to select the reads that perfectly map to the genome. Additional filtering was performed to select the reads that map to the genome only in one locus. The final output data obtained by the Python script contained the information on the transposon-junction sequence, origin (chromosome versus plasmid), genomic coordinate corresponding to EZ:Tn5-BsmFI insertion site, protein-coding gene containing the insertion within the internal 5 to 80% of the coding region, strand (positive versus negative strand), and the number of the reads in each pool for 5 mutant pools (1 input pool and 4 output pools).
The above output data were processed separately from this step for chromosome and plasmid data using JMP8 software (SAS, Cary, NC). For additional filtering, insertions with read counts in the input pool of <10 were eliminated to remove nonspecific background reads. For normalization, a normalization factor was calculated according to the formula (Ri/Ro)/(Si/So), in which the variables represent the total number of sequence reads (R) and insertion sites (S) detected in the input (i) and output (o) pools for each output pool (14). The number of sequence reads for each insertion in each output pool was multiplied by the corresponding normalization factor. After normalization, the insertions in a gene with <3 insertion sites within the internal 5 to 80% of the coding region were removed. The numbers of all normalized sequence reads within each gene were subsequently combined for each pool to obtain the total number of normalized sequence reads originated from each gene. Following this, the fitness index value was calculated for each gene by dividing the total number of sequence reads in the output pool by those in the input pool.
Construction of deletion mutants.
Single deletion mutants of S. Typhimurium 14028 carrying a deletion in pyrD, glnL, recD, and STM14_5307 were constructed with a lambda red recombination system by the method described by Cox et al. (6), using pKD13 (7) as the template for amplification of the Kmr cassette and the oligonucleotides shown in Table S1 in the supplemental material. After confirmation by DNA sequencing, the deletions were transferred to the fresh wild-type background by P22 transduction followed by selection on LB agar plates supplemented with Km. The final Kmr strains were purified on EBU plates to obtain phage-free colonies for further analysis.
Competition assays.
Each deletion mutant (ΔpyrD, ΔglnL, ΔrecD, and ΔSTM14-5307) was competed against NAr wild-type strain in the control (LB-37°C) or appropriate test conditions (dLB-37°C for ΔpyrD ΔglnL; LB-42°C for ΔrecD or ΔSTM14_5307). Briefly, 100 μl of the inoculum consisting of equal volumes of the overnight cultures of the wild type and a mutant was used to inoculate 100 ml of LB medium (control) or appropriate test medium in a 200-ml Erlenmeyer flask. The culture was incubated at the indicated temperature with vigorous shaking (225 rpm). After 24 h, 1 ml of the culture was transferred to the fresh medium and 0.1 ml of the culture was used for dilution and plating on LB agar plates (NA) and LB agar plates (Km) for selective counting of the wild-type and mutant strains, respectively. The LB agar plates were incubated overnight at 37°C, and the colonies were enumerated. This selection and counting procedure were repeated every 24 h for up to approximately 4 to 8 days for different mutants.
RESULTS
Overview of the method.
EZ:Tn5 is a Tn5 derivative with modified inverted repeat (IR) sequences of 19 bp (AGATGTGTATAAGAGACAG), which was termed mosaic ends (ME). We observed that this ME region contains DNA sequence (underlined in the above ME sequence) similar to the recognition sequence of type IIS restriction enzyme BsmFI (5′-GGGAC(N)10↓-3′/3′-CCCTG(N)14↑-5′), except for one nucleotide (Fig. 1A). Since BsmFI cuts the site 14 bp away from the recognition site, this enzyme site can be exploited to extract 12-nt sequences immediately adjacent to Tn5 insertion sites. If these 12-bp transposon-junction sequences could be selectively amplified from a mutant library and sequenced en mass in a massively parallel manner, the resulting profile would provide information on both the identity and relative quantity of each insertion in the library. Therefore, we tested if EZ:Tn5 could still be functional even after the substitution of one nucleotide (A→G) in one ME region. We found that the efficiency of mutagenesis with the modified EZ:Tn5 (EZ:Tn5-BsmFI) as measured by the number of resulting Kmr colonies was very similar to that of the original EZ:Tn5. In repeated transformation experiments, we routinely obtained approximately 1 × 104 to 3 × 104 mutants of S. Typhimurium 14028 per electroporation.
This would provide an excellent opportunity to use this EZ:Tn5-BsmFI construct as a powerful tool for deep profiling of complex insertion mutant library via Illumina sequencing, as shown in Fig. 1B. A genome-saturating library of Tn5 mutants is subjected to a selection condition of interest, and the mutant pools before and after the selection can be compared to identify all insertions exhibiting any significant changes in relative abundance after selection. This analysis will allow identification of bacterial genes important in various biological processes of interest on a genome-wide scale. However, the length of the transposon-junction sequences extracted by this method will be 12 bp, which may not be long enough for unambiguous identification of the genomic locations from which the insertions were originated. To address this issue, we performed a computer simulation using a custom Python script to extract 100,000 short sequences of different lengths (10 to 20 bp) from random locations in both strands of the complete genome of S. Typhimurium strain 14028. The short sequences were subsequently mapped back to the genome to determine the portion of the sequences that perfectly match to the genome only in one location (see Fig. S2 in the supplemental material). The result shows that approximately 47% of the 12-bp transposon-junction sequences mapped uniquely to the genome, suggesting that on average one-half of the transposon-junction sequences experimentally extracted from a mutant library should be discarded because they cannot be mapped unambiguously to one genomic locus. However, we expected that this shortcoming can be overcome by increasing the size of the mutant library and the huge number of sequence reads that can be obtained from Illumina sequencing.
Selection of the mutant library.
To test the feasibility and utility of this method, we used this method for genome-wide identification of S. Typhimurium genes conditionally essential for fitness under 3 different in vitro conditions. Three selective conditions in this study were chosen to mimic the low-nutrient condition in infected host tissues (10× diluted LB medium; dLB), bile-rich intestinal environment (LB medium plus bile acid; LB-bile), and the elevated body temperature associated with chickens and other avian species (LB medium at 42°C; LB-42°C). Our EZ:Tn5-BsmFI mutant library consists of approximately 1.6 × 104 different mutants, and the inoculum of 1 ml contained approximately 7.3 × 107 cells, indicating each mutant in the library was represented by approximately 4,600 cells. Mutants were transferred 3 times under the same selective conditions to increase the selection sensitivity. In each transfer during the selection, the inoculum of 1 ml contained approximately 108 to 109 cells, depending on the selective conditions, indicating each mutant is well represented by a sufficient number of cells in the inoculum. The final output pool of 2 ml also contained enough cells to represent all surviving mutants at the end of the final selection. We also included LB medium incubated at 37°C as a control condition to identify the genes that are essential for fitness under an optimal growth condition. Since we are interested in the genes uniquely essential under the 3 selective conditions of interest, the genes identified under the control condition were removed from those identified under each of 3 selective conditions.
Analysis of Illumina sequencing data.
The summary of the Illumina sequencing data and its analysis is shown in Table S2 in the supplemental material. Among the total of 23,141,540 sequence reads obtained from a single flow cell lane, 76% (17,490,113 reads) contained the complete 19-bp ME sequence. Among these reads, 15,893,768 reads (91%) contained the complete 6-bp bar code sequences perfectly matching one of the 5 bar codes. When these reads were sorted according to the bar code, we obtained a relatively even distribution across different bar codes: 2,374,190 (ATCACG; input), 2,859,671 (CGATGT; LB), 2,652,543 (TTAGGC; dLB), 4,382,503 (TGACCA; LB-bile), and 3,624,861 (ACAGTG; LB-42°C) reads. The transposon-junction sequences were subsequently extracted from the reads for each bar code. As expected, the majority (>99%) of the sequence reads were approximately 11 to13 bp long (see Table S2 and Fig. S3 in the supplemental material). These 11- to13-bp sequence reads for each bar code were further filtered for those that map to the genome at only one genomic locus. Finally, we obtained 1,204,021, 1,312,302, 1,367,315, 2,249,816, and 1,554,249 reads for each bar code, which corresponds to approximately 49% of the total number of the transposon-junction sequences of 11 to 13 bp for each bar code.
The normalization factors for the output pools were 0.94 (LB), 0.89 (dLB), 0.57 (LB-bile), and 0.80 (LB-42°C) for chromosome and 0.55 (LB), 0.61 (dLB), 0.23 (LB-bile), and 0.49 (LB-42°C) for plasmid. When the reads for all insertions in each gene were combined, we obtained the data set for 3,806 and 90 protein-coding genes for chromosome and plasmid, respectively. To obtain a more robust and reliable result, the genes that contained less than 3 insertions were removed, resulting in totals of 1,879 and 52 coding genes for the chromosome and plasmid, respectively. For all of the genes included in the final data set, each gene contained 8.8 and 9.1 authentic insertion sites on average on the chromosome and plasmid, respectively. We also determined the reproducibility of Tn-seq profiling using two biological replicates. When the data were processed separately for the chromosome and plasmid, we obtained very high levels of reproducibility for both the chromosome (R2 = 0.99) and plasmid (R2 = 0.99) (see Fig. S4 in the supplemental material). As the level of genome saturation by insertions in this final data set was not sufficiently high, we did not investigate in vitro essential genes (16, 19) but focused on conditionally essential genes in this study.
Identification of conditionally essential genes.
Among the 1,931 (1,879 + 52) genes, the genes conditionally essential for each selective condition were first selected by a cutoff fitness index of ≤0.2, which indicates at least a 5-fold reduction in relative abundance during selection. The genes selected by this criterion are expected to exhibit a strong fitness defect under each condition. When the mutants were selected in LB medium, which was used as a reference condition for optimal growth, a total of 32 genes were identified, as required for optimal in vitro fitness. These genes should be distinguished from essential genes because the mutants with insertions in the essential genes cannot be recovered by definition of the essential genes. However, the insertion mutants in these 32 genes were recovered and were well represented in the input pool. Therefore, these genes should be considered dispensable for growth or survival, yet contribute to optimal growth at 37°C in LB medium.
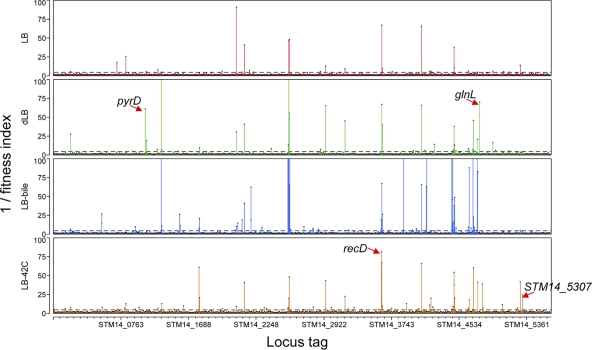
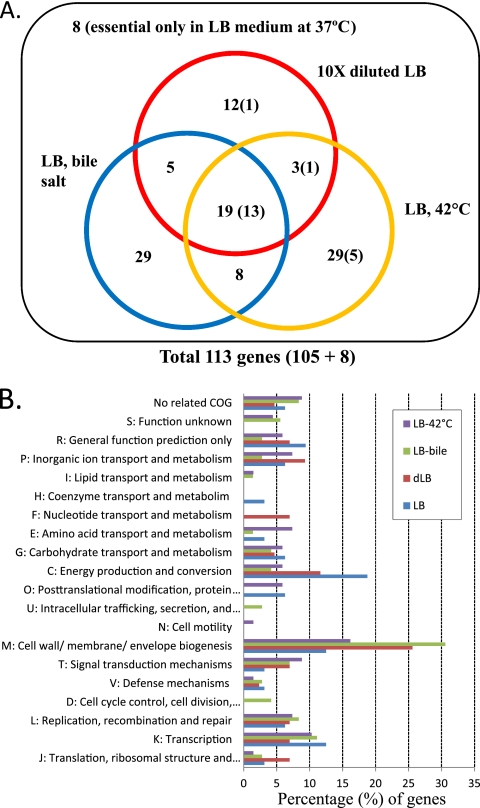
The result in Fig. 2 gives a clear overview of the genome-wide fitness profile of Salmonella mutants. We obtained 39, 61, and 56 genes required for fitness in under the dLB, LB-bile, and LB-42°C conditions, respectively (Fig. 3A). There were many genes essential for fitness under more than one condition, and many genes were also shown to be essential for general fitness under the optimal growth condition (LB at 37°C; numbers shown in parentheses in Fig. 3A). In total, 105 genes conditionally essential under either one of the selective conditions were identified (Fig. 3A; for the list of the genes, see Table S3 in the supplemental material). All of the 105 genes identified were chromosomal genes, and none of them was located on the plasmid.
Fig 2.
Identification of the genes conditionally essential for fitness under 3 different selective conditions: dLB, LB-bile, and LB-42°C. Genome-wide view of the fitness index−1 (= total read counts in the input pool/total read counts in the normalized output pool) is shown for each gene identified under the optimal growth condition (LB medium; control) and the 3 different selective conditions. The fitness index−1 was used for the y axis (instead of the fitness index) for better visualization of mutants with a fitness defect, as indicated by the high peaks. The cutoff line of 5 (= 0.2−1) used in this study to determine the fitness defect is shown (dashed lines). The genes selected in this study for further mutant characterization are shown by arrows.
Fig 3.
Conditionally essential genes. (A) The genes identified as conditionally essential for fitness under each of the three selective conditions. The numbers of genes that are also essential for optimal fitness in LB medium are shown in parentheses. There were 8 additional genes essential for fitness in LB medium, but dispensable for fitness under all 3 selective conditions. (B) Functional classification of the identified genes.
To obtain insights into functional trends associated with each condition, we assigned the identified genes to functional (cluster of orthologous groups [COG]) categories (38) using the BLAST on Orthologous Groups (BLASTO) algorithm (44) (Fig. 3B). Not surprisingly, the in vitro essential genes (LB medium) were most prominently enriched in COG categories K (transcription), M (cell wall/membrane/envelope biogenesis), and C (energy production and conversion). The genes essential for fitness in dLB exhibited a similar trend, except that category K was not enriched while category M was further enriched to represent 25% of all genes identified. In LB-bile, the genes required for fitness were significantly biased toward cell wall/membrane/envelope biogenesis (category M). This result corroborates well with previous findings that lipopolysaccharide (LPS) biosynthesis and membrane integrity are critical in bile resistance of Salmonella (23, 31).
There are 48 (61 − 13) genes identified as conditionally essential for fitness in the presence of bile salts. Bile resistance has been studied in depth, and extensive mutant screenings have revealed a comprehensive list of genes required for bile resistance in S. Typhimurium (25, 31, 42), S. enterica serotype Typhi (23, 42), and Escherichia coli (30). Notably, 38 out of the 48 genes identified in this study were previously implicated in bile resistance in Salmonella species or E. coli. This result validates our experimental and bioinformatics approaches to identify conditionally essential genes. The remaining 10 genes newly identified in this study may reflect the differences in the sensitivities of the screening and selection conditions between the experiments.
Phenotypic characterization of deletion mutants.
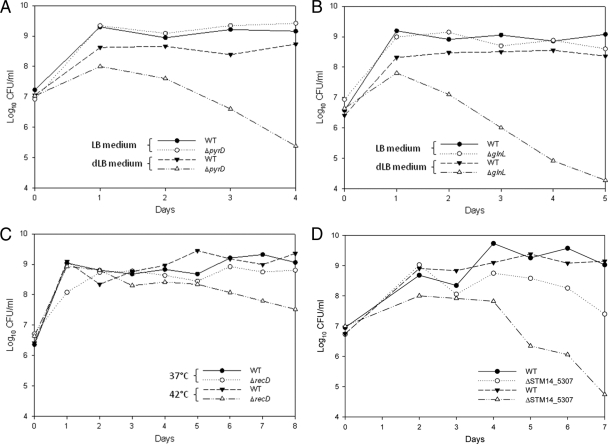
To further verify the results of the Tn-seq screening, we sought to characterize the functions of additional genes that were identified in this study, which have not been previously linked to the phenotypes. We chose 2 genes, pyrD and glnL, encoding dihydroorotate dehydrogenase and nitrogen regulation protein, respectively, among the 39 genes identified in dLB medium. The deletion mutants were analyzed for growth patterns in LB medium (control condition) and dLB medium. Unexpectedly, the ΔpyrD mutant demonstrated a slight growth defect in both LB and dLB media compared to the wild type (see Fig. S5 in the supplemental material). Conversely, ΔglnL exhibited a slight growth defect only in LB medium (see Fig. S5). The reason why the growth phenotypes did not accurately reflect those predicted from Tn-seq data could be due to the differences in the assay conditions. Initially the negative selection of mutants was performed through competition within the complex mutant library. Therefore, in order to more closely mimic the conditions under which the mutants were selected, we characterized the 2 mutants using competition assays in which each mutant was competed against the wild-type strain. As expected, both ΔpyrD and ΔglnL mutants exhibited a severe competitive disadvantage in dLB medium, but not in LB medium (Fig. 4A and B, respectively).
Fig 4.
Competition between the wild type and each of the ΔpyrD, ΔglnL, ΔrecD, and ΔSTM14_5307 mutants. For the competition assay, the wild type and each deletion mutant were mixed in a 1:1 ratio and inoculated into 2 different culture conditions (control versus the relevant test condition indicated). The cultures were diluted into respective selective conditions every 24 h for up to 4 to 8 days. The cell numbers were determined for the wild-type (NAr) and mutant (Kmr) cells each day using LB plates supplemented with the appropriate antibiotics. (A) ΔpyrD (LB versus dLB); (B) ΔglnL (LB versus dLB); (C) ΔrecD (LB at 37°C versus 42°C); and (D) ΔSTM14_5307 (LB at 37°C versus 42°C).
Additionally, two genes, recD and STM14_5307, encoding the exonuclease V subunit and putative transcriptional regulator, respectively, were chosen from the genes identified in LB-42°C, and the deletion mutants were subjected to competition assays. The ΔrecD mutant showed a slight competitive disadvantage at 37°C but a more obvious disadvantage during competition at 42°C (Fig. 4C). In the case of the ΔSTM14_5307 mutant, the mutant demonstrated a clear competitive disadvantage even at 37°C (Fig. 4D), yet a more severe competitive disadvantage was observed at 42°C (day 7 in Fig. 4D).
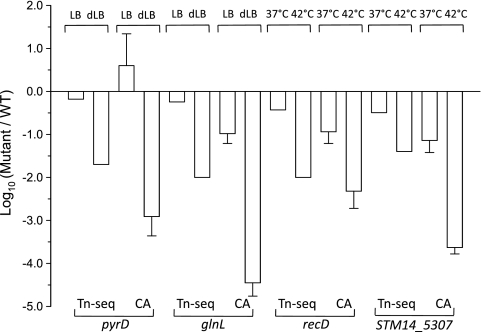
To determine the accuracy of fitness measurement inferred by the Tn-seq profiles, we compared the fitness indices obtained by Tn-seq data and competition assay for each gene. As shown in Fig. 5, Tn-seq data were able to predict the fitness of each mutant strain with high levels of accuracy.
Fig 5.
Comparison of the fitness indices obtained by Tn-seq data (see Table S3 in the supplemental material) and a competition assay (CA) for the ΔpyrD, ΔglnL, ΔrecD, and ΔSTM14_5307 mutants. The comparison was made for 2 different culture conditions (control versus relevant test condition indicated) for each mutant. The results of the competition assay (n = 3) were obtained at day 4 (ΔpyrD, ΔglnL, and ΔrecD) or 7 (ΔSTM14-5307).
Insertions in the conditionally essential genes are not lethal.
We analyzed our data in comparison with those from in vitro metabolic reconstruction (MR) modeling (10). A metabolic reconstruction breaks down all known metabolic pathways in the cell into their respective reactions and enzymes and analyzes them within the perspective of the entire system. One such MR recently reported on Salmonella Typhimurium LT2 (39) predicted 144 genes lethal for growth or survival in LB media. We found that the 21 genes out of the 105 genes identified in our study were also listed as lethal genes. However, construction of deletion mutants has been reported previously for at least 12 of those 21 genes (13, 21, 22, 37, 43), suggesting that the prediction of lethal genes by this MR is not accurate. The nonessential nature of the 105 conditionally essential genes identified in this study is also supported by additional experimental evidence by Langridge et al. (23), in which 356 essential genes were discovered in S. Typhi with high confidence using 1.1 million random Tn5 insertions in the genome. Among the 105 genes identified in our study, only two genes (icdA and pssA) were reported as essential, and 29 genes were classified as advantageous for growth in LB medium (23).
Biological roles of the identified conditionally essential genes during host infection.
As an important bacterial pathogen with a battery of genetic tools available, S. Typhimurium has been used commonly as a model organism to study bacterial pathogenesis. Consequently, a large quantity of “omics” (genomics, transcriptomics, and proteomics) data has been accumulated for S. Typhimurium and other related serotypes (15). Particularly, a large portion of those studies have been conducted in the context of host-pathogen interactions. In order to gain deeper insights into the biological implications of the genes identified in this study, we analyzed them in light of those data. We found the results obtained from comprehensive functional profiling of mutant libraries (random insertions or targeted deletions) obtained using animal infection models would be particularly useful in shedding light on the roles of the genes and their products during host infection. These large-scale high-throughput screenings of Salmonella mutants have provided almost saturating lists of the Salmonella genes essential for in vivo infection using different mouse infection models (3, 4, 24, 34). For a large portion of these in vivo essential genes, however, the biochemical bases for attenuation of the mutants are unknown. Our study revealed comprehensive sets of conditionally essential genes with relevance to stress resistance during host infection. Therefore, the comparison of our data with the existing functional profiling data would provide the biological basis for the requirement of at least a subset of the genes essential for in vivo infection.
For each of the 105 genes identified as conditionally essential in this study, we determined if they have been previously identified as essential for in vivo fitness during infection in animal models (mouse or chicken). One requirement for a gene to have a role for in vivo fitness is that the protein encoded by the gene should be expressed in vivo. Therefore, we also used the comprehensive in vivo proteomics data of S. Typhimurium obtained using both systemic and enteric infection models of mice to determine if protein encoded by each of the 105 genes was expressed in vivo during infection in the mouse (1). However, no in vivo protein expression data are available for chickens. The summary of the functional information and proteome data is shown in Fig. 6 (see Tables S3 to S6 for more details).
Fig 6.
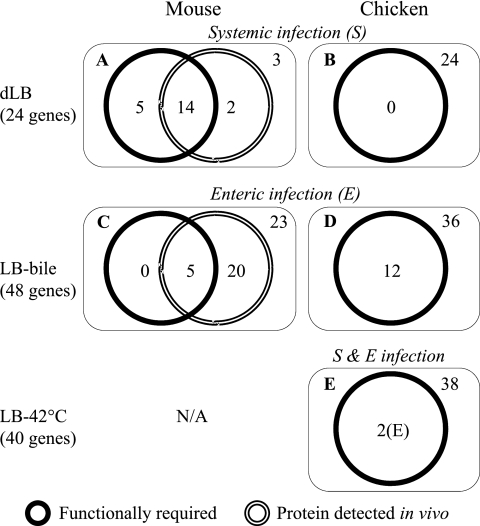
In vivo functions of the gene products during animal infection. Among the genes conditionally required for fitness under each selective condition, the numbers of the genes required for in vivo fitness during animal infection (mouse versus chicken) through 2 different infection routes (systemic versus enteric infection) are indicated. The numbers of the genes that produce proteins in vivo are also indicated for the mouse infection model using the 2 different routes of infection. The genes also essential for fitness in LB medium were not included in the numbers of the conditionally essential genes.
The low-nutrient condition reflected in dLB medium is encountered by Salmonella cells when they are present inside macrophage vacuoles during systemic infection (11). Out of 24 genes essential for fitness in dLB, 19 genes were shown to be essential for in vivo fitness during a systemic infection in the mouse, among which in vivo protein expression during systemic infection was detected for 14 genes (Fig. 6A; see Table S4 in the supplemental material). Five proteins previously shown to be required for in vivo survival during systemic infection in mice (RecG, GlnL, LepA, RfaQ, and ZntA) were not detected in vivo, probably due to the limited amount and high complexity of the sample (1). Two proteins among the 16 proteins detected in vivo (Pnp and OmpA) are not required for fitness during systemic infection. This analysis suggests that the 19 genes were required for in vivo fitness during systemic infection in mice due to their requirements for fitness under the low-nutrient condition. However, none of the 24 genes was shown to be important during systemic infection in chickens (Fig. 6B). This reflects the fact that all large-scale mutant screenings have been performed with the mouse systemic infection model, and the number of mutants screened in chickens is very limited (35, 40).
In case of LB-bile, Salmonella should encounter bile stress in the intestinal tract of the host during enteric infection (11). In the mouse enteric infection model, 25 genes out of the total of 48 genes required for bile resistance in vitro were shown to be expressed from the cecal samples of the infected mouse (Fig. 6C; see Table S5 in the supplemental material). Among the 25 gene's proteins, 5 (RfaL, RfaJ, RfaI, Rfc, and RfbP) were shown to be important for in vivo fitness. Interestingly, all of these 5 proteins are involved in biosynthesis of lipopolysaccharide core and O-antigen (22). The remaining 20 genes have not been linked to enteric infection in the mouse, which probably reflects the lack of comprehensive screening conducted with enteric infection model of the mouse due to the technical difficulty associated with the bottleneck existing in the enteric infection model. However, the fact that these 20 genes are both expressed in vivo and required for bile resistance in vitro strongly suggests that these genes play roles in efficient colonization in the intestinal tract via conferring resistance to bile. Interestingly, 12 of the 48 genes were shown to be important for cecal colonization during chicken infection (Fig. 6D). Nine of the 12 genes (acrB, rfbN, rfbD, rfbB, tolC, rfbI, rfbK, rbsK, and rfbP) were shown to be expressed in vivo in the mouse enteric infection model, yet only one of them (rfbP) has been functionally linked to the mouse enteric infection model (see Table S5).
The elevated temperature of 42°C is the body temperature of avian species, including chickens. Therefore, any mutant with a fitness defect at 42°C is very likely to be attenuated during infection in a chicken. Among the 40 genes identified as essential for fitness at 42°C in this study, only 2 genes were previously implicated with an in vivo fitness defect during chicken infection (Fig. 6E; see Table S6 in the supplemental material). The result suggests that these 2 genes, rfaY and rfbP, are required for infection in chicken through their requirements for fitness at 42°C. However, one of the proteins (RfbP) is also required for both systemic and enteric infection in the mouse, whose body temperature is 37°C, which indicates that there is another mechanism(s) underlying the attenuation of the mutants during host infection (see Table S6). For example, the fitness indices for the rfbP gene were 0.45 and 0.01 for dLB and LB-bile, respectively, indicating that this gene is also required for fitness under low-nutrient conditions (although the fitness index was >0.2) and bile-rich environments.
DISCUSSION
In this study, we described a new version of Tn-seq method and used it with stringent cutoffs to obtain robust fitness profiling of the entire genome of S. Typhimurium strain 14028 under 3 different in vitro stress conditions, including low-nutrient, bile-rich, and high-temperature (42°C) environments. The phenotype characterization of the 4 deletion mutants along with previous studies on bile acid resistance of Salmonella demonstrated that our Tn-seq method and bioinformatics analysis assessed gene functions accurately.
This method as a variation on existing Tn-seq methods is based on the use of the modified EZ:Tn5 transposon that carries the recognition site of type IIS enzyme BsmFI on one ME sequence to allow straightforward and robust extraction of Tn5-junction sequences of the identical length of 12 nt from a complex mutant library. We used our Tn-seq method in conjunction with a bar coding strategy and Illumina sequencing to allow high-resolution functional genome scanning for multiple selective conditions of interest.
There are other existing methods for comprehensive functional screening of a transposon mutant library with the aid of next generation sequencing (5, 9, 12, 14, 17, 23, 41). Our method is distinct from other methods in that we employed the EZ:Tn5 mutagenesis system. The proven broad host range of this mutagenesis system along with a commercially available EZ-Tn5 transposase (Epicentre BioTechnologies) will make this method easily accessible and applicable to variety of bacterial species with appropriate modifications on an antibiotic cassette and its promoter. The EZ:Tn5 system was also used in the method developed by Langridge et al. (23), and the strategies developed by Gawronski et al. (14), Gallagher et al. (12), Eckert et al. (9), and Christen et al. (5) should be applicable to the EZ:Tn5 system with appropriate modifications. However, our Tn-seq method is technically more simple and straightforward than other methods involving mechanical shearing and fractionation of DNA fragments (9, 12, 14, 23). In addition, the uniform length of PCR amplicons obtained by our protocol provides an efficient means to remove fragments resulting from a possible aberrant PCR.
One drawback of our method is the relatively short length of the transposon-junction sequences, which made it necessary to discard approximately one-half of the transposon-junction sequences mapped to the genome. However, increasing the number of mutants in the library along with more sequencing reads would eventually overcome this problem. Particularly with rapidly increasing sequencing capacity of next generation sequencing technologies, this will become a negligible issue (29).
We also demonstrate for the first time that this Tn-seq approach could be used in conjunction with bar codes to analyze multiple samples simultaneously. This approach will allow high-resolution functional screening of a bacterial genome for multiple selective conditions of interest, opening the door for multidimensional comprehensive understanding of bacterial gene functions.
In this study, we modified the DNA sequence of plasmid pMOD-6 <KAN-2/MCS> to obtain the EZ:Tn5-BsmFI fragment. Alternatively, EZ:Tn5-BsmFI could be prepared by amplifying the template plasmid pMOD-6 <KAN-2/MCS> or any other derivative plasmids using a pair of primers corresponding to the ME sequences where one of the primers contains one nucleotide change to introduce the BsmFI site into the ME region (unpublished data). This can simplify the procedure by eliminating the step for site-directed mutagenesis.
We have identified 105 genes conditionally essential for fitness under 3 different conditions reflecting stressors Salmonella would encounter during survival in the environments and infected hosts. We examined the biological significance of these genes during host infection by analyzing the data in light of currently available mutant fitness data and proteomics data obtained from animal infection studies. Through this analysis, we assigned biological bases for in vivo requirements of the proteins for all 58 genes among the 105 genes identified in this study. This process resembles virulence-attenuated pool (VAP) screening using signature-tagged mutagenesis (STM), which is performed with a subset of the original mutants shown to be attenuated in vivo to reveal roles that the identified factors play in the infection process (27, 28). For VAP screening based on STM, the need to reduce the pool for further screening came from the limited (∼96) number of mutants that can be screened simultaneously by STM. However, the global scale and cost-effectiveness of the Tn-seq method eliminate the need to prepare a smaller size of VAP for secondary screening under various stress conditions. Instead, a complex library of transposon mutants could be screened simultaneously or one at a time under multiple conditions, including animal infection and other host-associated stressors. If this approach is used in an animal infection model in conjunction with screenings for a variety of virulence-associated phenotypes representative of all known host barriers to overcome for successful infection, it is expected to provide a wealth of functional information for most of the in vivo essential factors.
The genes identified in this study include many putative or hypothetical genes or genes with unknown functions. Understanding the functions of these genes and products is expected to reveal unknown mechanisms of Salmonella survival and persistence during its life cycle. In addition, these genes have great potentials to be used as good candidate targets for development of vaccines and novel antimicrobials. The mutants with deletions in some of those genes that would still allow in vivo survival, yet at appropriately reduced levels, could be very effective in eliciting an adaptive immune response in the host, while they are likely be cleared from the host faster than the wild-type strain. It would be interesting to test the S. Typhimurium mutant with reduced fitness at 42°C as an attenuated live vaccine for poultry.
With the rapidly increasing sequencing capacity of the 2nd generation sequencing technologies (29) and the emergence of 3rd generation sequencing methods with even greater potential (32), our Tn-seq method will allow exploration and comprehensive understanding of the functional implications of genetic elements (both coding and noncoding genes) at increasingly higher resolutions for a variety of biological contexts (5).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant R21 AI063137 and a USDA Food Safety Consortium grant.
Footnotes
Published ahead of print 24 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Becker D, et al. 2006. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440:303–307 [DOI] [PubMed] [Google Scholar]
- 2. Bossé JT, Zhou L, Kroll JS, Langford PR. 2006. High-throughput identification of conditionally essential genes in bacteria: from STM to TSM. Infect. Disord. Drug Targets 6:241–262 [DOI] [PubMed] [Google Scholar]
- 3. Chan K, Kim CC, Falkow S. 2005. Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect. Immun. 73:5438–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chaudhuri RR, et al. 2009. Comprehensive identification of Salmonella enterica serovar typhimurium genes required for infection of BALB/c mice. PLoS Pathog. 5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Christen B, et al. 2011. The essential genome of a bacterium. Mol. Syst. Biol. 7:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cox MM, et al. 2007. Scarless and site-directed mutagenesis in Salmonella enteritidis chromosome. BMC Biotechnol. 7:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Durant JA, Corrier DE, Byrd JA, Stanker LH, Ricke SC. 1999. Feed deprivation affects crop environment and modulates Salmonella enteritidis colonization and invasion of leghorn hens. Appl. Environ. Microbiol. 65:1919–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eckert SE, et al. 2011. Retrospective application of transposon-directed insertion site sequencing to a library of signature-tagged mini-Tn5Km2 mutants of Escherichia coli O157:H7 screened in cattle. J. Bacteriol. 193:1771–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feist AM, Herrgård MJ, Thiele I, Reed JL, Palsson B. 2009. Reconstruction of biochemical networks in microorganisms. Nat. Rev. Microbiol. 7:129–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Foster JW, Spector MP. 1995. How Salmonella survive against the odds. Annu. Rev. Microbiol. 49:145–174 [DOI] [PubMed] [Google Scholar]
- 12. Gallagher LA, Shendure J, Manoil C. 2011. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2(1):e00315-e00310 doi:10.1128/mBio.00315-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Van Immerseel F. 2009. The Salmonella Enteritidis lipopolysaccharide biosynthesis gene rfbH is required for survival in egg albumen. Zoonoses Public Health 56:145–149 [DOI] [PubMed] [Google Scholar]
- 14. Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. 2009. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc. Natl. Acad. Sci. U. S. A. 106:16422–16427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gillespie JJ, et al. 2011. PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infect. Immun. 79:4286–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glass JI, et al. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U. S. A. 103:425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goodman AL, et al. 2009. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6:279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Horner DS, et al. 2010. Bioinformatics approaches for genomics and post genomics applications of next-generation sequencing. Brief. Bioinform. 11:181–197 [DOI] [PubMed] [Google Scholar]
- 19. Hutchison CA, et al. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165–2169 [DOI] [PubMed] [Google Scholar]
- 20. Jarvik T, Smillie C, Groisman EA, Ochman H. 2010. Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J. Bacteriol. 192:560–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karasova D, et al. 2009. Comparative analysis of Salmonella enterica serovar Enteritidis mutants with a vaccine potential. Vaccine 27:5265–5270 [DOI] [PubMed] [Google Scholar]
- 22. Kong Q, et al. 2011. Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar Typhimurium. Infect. Immun. 79:4227–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Langridge GC, et al. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 19:2308–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lawley TD, et al. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. López-Garrido J, Cheng N, García-Quintanilla F, García-del Portillo F, Casadesús J. 2010. Identification of the Salmonella enterica damX gene product, an inner membrane protein involved in bile resistance. J. Bacteriol. 192:893–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. MacLean D, Jones JD, Studholme DJ. 2009. Application of ‘next-generation’ sequencing technologies to microbial genetics. Nat. Rev. Microbiol. 7:287–296 [DOI] [PubMed] [Google Scholar]
- 27. Merrell DS, Camilli A. 2002. Information overload: assigning genetic functionality in the age of genomics and large-scale screening. Trends Microbiol. 10:571–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Merrell DS, Hava DL, Camilli A. 2002. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol. Microbiol. 43:1471–1491 [DOI] [PubMed] [Google Scholar]
- 29. Metzker ML. 2010. Sequencing technologies—the next generation. Nat. Rev. Genet. 11:31–46 [DOI] [PubMed] [Google Scholar]
- 30. Nichols RJ, et al. 2011. Phenotypic landscape of a bacterial cell. Cell 144:143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Prouty AM, Van Velkinburgh JC, Gunn JS. 2002. Salmonella enterica serovar Typhimurium resistance to bile: identification and characterization of the tolQRA cluster. J. Bacteriol. 184:1270–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rothberg JM, et al. 2011. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475:348–352 [DOI] [PubMed] [Google Scholar]
- 33. Rychlik I, Barrow PA. 2005. Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol. Rev. 29:1021–1040 [DOI] [PubMed] [Google Scholar]
- 34. Santiviago CA, et al. 2009. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 5:e1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shah DH, et al. 2005. Identification of Salmonella gallinarum virulence genes in a chicken infection model using PCR-based signature-tagged mutagenesis. Microbiology 151:3957–3968 [DOI] [PubMed] [Google Scholar]
- 36. Slauch J, Taylor R, Maloy S. 1997. Survival in a cruel world: how Vibrio cholerae and Salmonella respond to an unwilling host. Genes Dev. 11:1761–1774 [DOI] [PubMed] [Google Scholar]
- 37. Su J, et al. 2009. RfaB, a galactosyltransferase, contributes to the resistance to detergent and the virulence of Salmonella enterica serovar Enteritidis. Med. Microbiol. Immunol. 198:185–194 [DOI] [PubMed] [Google Scholar]
- 38. Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637 [DOI] [PubMed] [Google Scholar]
- 39. Thiele I, et al. 2011. A community effort towards a knowledge-base and mathematical model of the human pathogen Salmonella Typhimurium LT2. BMC Syst. Biol. 5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Turner AK, Lovell MA, Hulme SD, Zhang-Barber L, Barrow PA. 1998. Identification of Salmonella typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect. Immun. 66:2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6:767–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van Velkinburgh JC, Gunn JS. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 67:1614–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yethon JA, et al. 2000. Salmonella enterica serovar Typhimurium waaP mutants show increased susceptibility to polymyxin and loss of virulence in vivo. Infect. Immun. 68:4485–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou Y, Landweber LF. 2007. BLASTO: a tool for searching orthologous groups. Nucleic Acids Res. 35:W678–W682 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.