Abstract
Biocides play an essential role in limiting the spread of infectious disease. The food industry is dependent on these agents, and their increasing use is a matter for concern. Specifically, the emergence of bacteria demonstrating increased tolerance to biocides, coupled with the potential for the development of a phenotype of cross-resistance to clinically important antimicrobial compounds, needs to be assessed. In this study, we investigated the tolerance of a collection of susceptible and multidrug-resistant (MDR) Salmonella enterica strains to a panel of seven commercially available food-grade biocide formulations. We explored their abilities to adapt to these formulations and their active biocidal agents, i.e., triclosan, chlorhexidine, hydrogen peroxide, and benzalkonium chloride, after sequential rounds of in vitro selection. Finally, cross-tolerance of different categories of biocidal formulations, their active agents, and the potential for coselection of resistance to clinically important antibiotics were investigated. Six of seven food-grade biocide formulations were bactericidal at their recommended working concentrations. All showed a reduced activity against both surface-dried and biofilm cultures. A stable phenotype of tolerance to biocide formulations could not be selected. Upon exposure of Salmonella strains to an active biocidal compound, a high-level of tolerance was selected for a number of Salmonella serotypes. No cross-tolerance to the different biocidal agents or food-grade biocide formulations was observed. Most tolerant isolates displayed changes in their patterns of susceptibility to antimicrobial compounds. Food industry biocides are effective against planktonic Salmonella. When exposed to sublethal concentrations of individual active biocidal agents, tolerant isolates may emerge. This emergence was associated with changes in antimicrobial susceptibilities.
INTRODUCTION
Biocides play an essential and effective role in limiting the spread of infection and disease. Concerns with regard to the overuse of these chemicals/agents and the potential selective pressure that they may confer have been expressed (21). Such overuse and selective pressure can lead to an increased tolerance to these biocides concomitant with the emergence of cross-resistance to clinically important antimicrobial compounds (7). These phenotypes may also contribute to a change in susceptibility to common food-processing stresses (6).
In the modern food industry, the scale of food production allied to consumer demands for healthy, nutritious, and minimally processed food devoid of additives, such as chemical preservatives and other antimicrobial agents, has had an important impact on the volume of biocide(s) used in this environment. In attempting to improve hygiene measures and to ensure food safety, the food industry has increased its use of biocides and chemical-based disinfectants to control the microbial ecology of the production environment (19), and thus, tolerance to biocides has been documented for most classes of agents. Exhibition of increased tolerance to biocides by zoonotic food-borne bacteria has important implications for public health. This feature would undoubtedly compromise the role of disinfectants as an effective means to control these bacterial hazards. Any failure of, or altered tolerance to, these biocidal agents could facilitate the survival of pathogenic organisms and contribute to the emergence of persistent strains.
Unlike antimicrobial compounds which are selectively toxic, most biocides do not have a distinct bacterial cell target upon which to act. Resistance to antimicrobial compounds can emerge following one or more target gene mutations. In contrast, when tolerance to one or more biocides arises, it is mediated by mechanisms that are less well characterized. Some of the modifications that can occur in a bacterial cell include upregulation of efflux pump activity or structural alterations in the cell wall, which impact permeability (33).
In the current scientific literature, researchers have raised concerns with regard to the consequences arising from the overuse of biocides and the ease with which antibiotic-resistant bacteria and isolates possessing an enhanced tolerance to food-processing stress(es) may emerge. It has been suggested that cross-resistance to antimicrobial compounds, following exposure and adaptation to a biocide, could occur in a limited number of situations. These can be summarized as follows: (i) when the biocide and an antimicrobial compound act on the same cellular target, (ii) when the biocide and the antimicrobial compound have the same transport mechanism, (iii) where a biocide and antibiotic can be accommodated by the same resistance mechanism (9), and finally, (iv) in situations where genes contributing toward biocide tolerance and antibiotic resistance are carried on the same mobile genetic element (31).
Salmonella enterica is an important zoonotic food-borne pathogen and the causative agent of gastroenteritis and typhoid fever (3). This bacterium persists in the environment (37), with enhanced survival in nonhost niches, including water, soil (40), food (8, 35), and food-processing environments (26, 39) being noted. Elimination of Salmonella through the use of effective and validated control measures, including adequate cleaning and sanitation, is essential. Improper cleaning may lead to contamination of the final food product with this pathogen, an event that can have major health and economic consequences.
The objective of this study was to investigate a large collection of well-characterized Salmonella strains for their susceptibility to a panel of commercially available food-grade biocidal formulations and their constituent active ingredients. The propensity of these bacteria to become tolerant following in vitro selection was assessed. The antimicrobial activity of the biocide formulations was subsequently reassessed under conditions that more accurately simulate relevant food production environments, including their activity against Salmonella when surface dried and enmeshed in a laboratory-induced biofilm. In the final part of this study, cross-tolerance of biocidal active agents and clinically important antimicrobial compounds was investigated.
MATERIALS AND METHODS
Biocide susceptibility testing.
Biocide susceptibility testing using a panel of seven biocide formulations of different chemical classes (Table 1) was carried out on a collection of 189 Salmonella strains, including 48 serotypes from various origins (such as clinical sources, food, the environment, and water; data not shown). All Salmonella isolates were stored on beads in cryopreservation fluid at −80°C (Technical Service Consultants Ltd., Lancashire, England). Isolates were streaked onto Mueller-Hinton (MH) agar (Oxoid, Cambridge, United Kingdom), a single isolated colony was picked and used to inoculate 10 ml of MH broth (Oxoid, Cambridge, United Kingdom) and then grown for 16 to 18 h at 37°C, with shaking at 250 rpm. The resulting culture was then used to inoculate 10 ml of fresh MH broth at a dilution of 1 in 10,000, to achieve a final cell number of approximately 105 log10 CFU/ml. One 96-well plate was used to test each isolate individually against all seven biocide formulations. All biocides tested were provided as a stock solution, and the dilution factor required to achieve the concentration recommended by the manufacturer for use was also given. A serial dilution of each formulation was made accordingly across the plate ranging from 200 to 0.2% of the recommended working concentration (i.e., twice the concentration recommended by the manufacturer for use, down to 0.2% of that concentration). Plates were then filled with 100 μl per well of the Salmonella culture, at approximately 105 log10 CFU/ml. The plates were then incubated at 37°C for 24 h in an Omnilog microplate reader (Biolog Inc., Hayward, CA). The digital imagery of this instrument tracks changes in the respiration of cultures growing in individual wells over time. The Omnilog output for a given plate consists of an optical density (OD) reading for each well, recorded every 15 min over a 24-h period. To calculate the MIC, the OD reading for each well was normalized. For normalization, the mean OD for the first hour of readings for each well, denoted the background OD, was subtracted from all the OD readings for each well, over the 24-h period. If this difference in OD above the background crossed a predetermined breakpoint, the well was considered positive for bacterial growth. We decided upon this breakpoint from calibration curve and plate count experiments; any reading giving an OD difference at or above the breakpoint was considered positive for growth when transferred to a bacterial culture medium. The MIC was calculated as the minimum concentration of the biocide formulation at which the optical density did not exceed this breakpoint.
Table 1.
Food industry biocide formulations used in this study
| Biocide (formulation no.) | Active agent(s) (%) | Class | Recommended contact time (min) |
|---|---|---|---|
| 1 | Potassium hydroxide (5–15), hypochlorite | Alkali | 5–20 |
| 2 | Benzalkonium chloride (30–50), fatty alcohol ethoxylate (1–2), ethylene glycol (1–2) | Quaternary ammonium compound | 5–20 |
| 3 | Tetrasodium EDTA (5–15), alkyldimethylbenzyl ammonium chloride (5–15), alkyl alcohol ethoxylate (<5), sodium carbonate (<5) | Multiple classes | None specified |
| 4 | Neutral detergent (≤5), propane-1,2-diol (≤2.5), ethanol (≤2.5), coco alkyl dimethylbenzyl ammonium chloride (≤2.5), didecyldimethylammonium chloride (≤2.5), 2-bromo-2-nitropropane-1,3-diola (≤2.5) | Multiple classes | None specified |
| 5 | Propane-1,2-diol (2.5–10), ethanol (2.5–10), coco alkyl dimethylbenzyl ammonium chloride (2.5–10), didecyldimethylammonium chloride (2.5–10), 2-bromo-2-nitropropane-1,3-diola (2.5–10), polyhexamethylene biguanide polymer (≤2.5), propan-2-ol (≤2.5) | Multiple classes | 5 |
| 6 | Hydrogen peroxide (10–20), acetic acid (1–2), alkylamino oxides (2–5), peroxyacetic acid (1–2) | Acid | 15 |
| 7 | Hydrogen peroxide (27.5), peroxyacetic acid (5.8), sulfamic acid (10–20), citric acid (20–25), alkylethersulfates (1–5), 3-butoxy-2-propanol (1–5) | Oxidizing agent/acid | 20 |
Bronopol (INN).
The minimum bactericidal concentration (MBC) was also determined in each case. A microplate replicator (96 pin; Thermo Fisher Scientific, Loughborough, United Kingdom) was used to transfer 2 μl from each of the wells in the MIC plates (see above) into a fresh 96-well plate containing xylose lysine deoxycholate (XLD) agar. Inoculated plates were incubated at 37°C for 18 to 24 h, and the MBC was determined.
Antibiotic susceptibility profiling.
Antibiotic susceptibility testing was carried out on all 189 Salmonella strains against a panel of 15 clinically important antimicrobial compounds (Table 2). This assay was carried out using Sensititre Gram-negative National Antimicrobial Resistance Monitoring System (NARMS) plates (CM V1GNF; Trek Diagnostic Systems Inc., Cleveland, OH). Plates were set up and interpreted according to the manufacturer's instructions, using the same culture of Salmonella, at 105 log10 CFU/ml, as used for the biocide susceptibility. Antibiotic susceptibility profiling was repeated for selected isolates by using the Vitek 2 Gram-negative antimicrobial susceptibility cards (AST-GN) (bioMérieux, Marcy l'Etoile, France), and results were interpreted according to the manufacturer's instructions.
Table 2.
Antimicrobial compounds, the numbers of resistant Salmonella isolates detected, and the frequencies of resistance
| Antimicrobial compounda | Range tested (μg/ml) | No. of resistant isolates | Frequency of resistance | Breakpoints (μg/ml)b |
|---|---|---|---|---|
| AMI | 0.5–32 | 25 | 0.13 | ≤16, 32, ≥64 |
| AMP | 1–32 | 19 | 0.10 | ≤8, 16, ≥32 |
| AUG2 | 1–32/0.5–16 | 12 | 0.07 | ≤8/4, 16/8, ≥32/16 |
| AXO | 0.5–64 | 16 | 0.08 | ≤8, 16–32, ≥64 |
| CHL | 2–32 | 36 | 0.19 | ≤8, 16, ≥32 |
| CIP | 0.015–4 | 77 | 0.40 | ≤1, 2, ≥4 |
| FIS | 16–512 | 142 | 0.72 | ≤256, ≥512 |
| FOX | 0.5–32 | 22 | 0.12 | ≤8, 16, ≥32 |
| GEN | 0.25–16 | 28 | 0.15 | ≤2, 4, ≥8 |
| KAN | 8–64 | 25 | 0.13 | ≤16, 32, ≥64 |
| NAL | 0.5–32 | 61 | 0.32 | ≤16, ≥32 |
| STR | 32–64 | 33 | 0.17 | ≤32, ≥64 |
| SXT | 0.12–4/2.38–76 | 108 | 0.57 | ≤2/3, ≥4/76 |
| TET | 4–32 | 59 | 0.31 | ≤4, 8, ≥16 |
| XNL | 0.25–8 | 61 | 0.32 | ≤2, 4, ≥8 |
AMI, amikacin; AMP, ampicillin; AUG2, amoxicillin-clavulanic acid; AXO, ceftriaxone; CHL, chloramphenicol; CIP, ciprofloxacin; FIS, sulfisoxazole; FOX, cefoxitin; GEN, gentamicin; KAN, kanamycin; NAL, nalidixic acid; STR, streptomycin; SXT, trimethoprim-sulfamethoxazole; TET, tetracycline; XNL, ceftiofur.
The Clinical and Laboratory Standards Institute (CLSI) 2008 breakpoints were used.
Biocide stability testing.
Biocide stability was examined under two different conditions. First, stability was assessed in the presence of organic matter, in tryptic soya broth (TSB), over a period of 8 days, and second, the same assays were repeated (without organic matter) in water over the same period. Biocide solutions were made up at the recommended working concentrations in a 10-ml volume of TSB or water. A 200-μl volume was removed from all tubes each day, and the MICs and MBCs were determined (as described above) and compared with those of the type strain Salmonella enterica serotype Enteritidis NCTC 13349. All tests were carried out in duplicate.
Increasing the tolerance of Salmonella to food industry biocide formulations following in vitro selection.
A subset of Salmonella isolates were selected from the study collection. This subset consisted of isolates for which the MIC of two or more biocide formulations was above the average for the complete collection (data not shown). The isolates were grown for several generations at 37°C in MH broth, containing a specific biocide formulation at a concentration below its MIC (0.25× or 0.5× MIC). The biocide formulation concentration was increased in a stepwise manner, and cultures were grown at 37°C for 24 to 48 h. This iterative approach was continued until the biocide concentration reached 4× MIC.
As an alternative (to the approach described above), the same set of isolates was cultured on MH agar plates containing the same biocides at a concentration of 0.5× MIC. Cultures were grown at 37°C for 24 h, before being subcultured onto fresh MH plates containing an increased biocide formulation concentration. This process was continued until the concentration of biocides on agar plates reached 2× MIC.
Surviving colonies were selected at each stage and transferred to fresh MH broth; this in vitro selection strategy was repeated at increasing concentrations.
Susceptibility testing and generating tolerance to biocidal active agents.
Biocidal compounds triclosan, chlorhexidine, benzalkonium chloride, and hydrogen peroxide were chosen. These were representative of the four main classes of biocide commonly used in the food industry formulations (Table 3). Susceptibility testing was carried out as described previously, with the exception that the compounds were made up in a stock solution of 100 μg/ml for triclosan, chlorhexidine, and benzalkonium chloride and 40 mM for hydrogen peroxide. Triclosan was dissolved in ethanol, and this was used as a master solution of 10 mg/ml. This stock was then diluted to 1 mg in a solution consisting of one part ethanol–three parts water and then further diluted to 100 μg/ml in water. Chlorhexidine was made up, in water, as a stock solution of 2 mg/ml, and benzalkonium chloride was made up in water as a stock solution of 4 mg/ml; both compounds were diluted to 100 μg/ml in water. Hydrogen peroxide solutions were made up in water. These stock solutions at concentrations of 100 μg/ml or 40 mM were then diluted across the 96-well plates. In vitro selection was carried out as described above. Pulsed-field gel electrophoresis (PFGE) as described by Mullane et al. (25) was used to compare the DNA fingerprints of the 10 biocide-tolerant isolates shown in Table 5 along with their eight original susceptible wild-type isogenic parent strains (and these included S. enterica serotype Gaminara S5, S. enterica serotype Typhimurium ST24, S. enterica serotype Hvittingfoss S41, S. enterica serotype Typhimurium ST23, S. enterica serotype Senftenberg C81, S. enterica serotype Enteritidis 5408, S. enterica serotype Enteritidis NCTC 13349, and S. Typhimurium SL1344). PFGE was carried out using a 3-h XbaI digestion; DNA fragments were then resolved using a 1% agarose gel (Seakem Gold, Rockland, ME) in 0.5× Tris-borate-EDTA (TBE) buffer at 14°C for 18 h at 6.0 V cm−1 with pulse times ramped linearly from 2.16 to 54.17 s. Gels were stained with ethidium bromide (10 mg/ml) and destained for 1 h in distilled water before being visualized under UV light by using a Gel Doc 2000 system (Bio-Rad Laboratories). A strain of S. enterica serotype Braenderup was included in the PFGE analysis as a marker for molecular weight determination. DNA fingerprints were stored as tagged-image-file-format (TIFF) files and imported into the BioNumerics software program (Applied Maths, Sint-Martens-Latem, Belgium). A dendrogram was created using the Dice coefficient, with an optimization of 1% and tolerance of 1.5% and applying the unweighted-pair group method with arithmetic means (UPGMA).
Table 3.
Biocidal active agents
| Biocide active agent | Class | Mechanism(s) of action |
|---|---|---|
| Triclosan | Phenolic compound | FabIa inhibition (12, 13, 36, 38), membrane disruption (12, 38) |
| Chlorhexidine | Biguanide | Membrane damage (10, 16); inhibits membrane enzymes (23); leakage of cellular constituents (16) |
| Benzalkonium chloride | Quaternary ammonium compound | Membrane damage; leakage and coagulation of cellular proteins (10, 18) |
| Hydrogen peroxide | Oxidizing agent | Oxidative damage; cellular proteins (18); nucleic acids (14) |
FabI is an enoyl-acyl carrier protein involved in fatty acid biosysthesis.
Table 5.
Salmonella serotypes displaying a stable tolerance to biocidal active agentsa
| Serotype and lab no./strainb | Biocide | MIC (μg/ml) |
|
|---|---|---|---|
| Wild type | Mutant | ||
| Gaminara S5 | Chlorhexidine | 50 | >1,000 |
| Typhimurium ST24 | Chlorhexidine | 1.96 | >100 |
| Hvittingfoss S41 | Benzalkonium chloride | 15 | 50 |
| Typhimurium ST23 | Triclosan | 1.95 | >1,000 |
| Senftenberg C81 | Triclosan | 1 | >1,000 |
| Enteritidis 5408 | Triclosan | 0.1 | >1,000 |
| Enteritidis NCTC 13349 | Chlorhexidine | 6 | >100 |
| Triclosan | 0.1 | 100 | |
| Typhimurium SL1344 | Chlorhexidine | 4 | >100 |
| Triclosan | 0.1 | 150 | |
Stable tolerance refers to a tolerance phenotype that persists over time, on continuous exposure, or in the absence of selective pressure.
The strains are S. Gaminara S5 (R type; Fis*, Inn*, Kan*), S. Typhimurium ST24 (R type; Inn*, Tet), S. Hvittingfoss S41 (R type; Fis*), S. Typhimurium ST23 (R type; Chl, Fis, Fox*, Fur*, Inn*, Prl*, Xnl*), S. Senftenberg C81 (R type; Fox*, Inn*, Str), S. Enteritidis 5408 (R type; Amp*, Chl, Cip, Cpd*, Enr, Inn*, Mar, Xnl), S. Enteritidis NCTC 13349 (R type; Str), and S. Typhimurium SL1344 (R type; Str) (where * indicates intermediate resistance). Amp, ampicillin; Chl, chloramphenicol; Cip, ciprofloxacin; Cpd, cefpodoxime; Enr, enrofloxacin; Fis, sulfisoxazole; Fox, cefoxitin; Fur, nitrofurantoine; Inn, cefalexin; Kan, kanamycin; Mar, marbofloxacin; Prl, piperacillin; Str, streptomycin; Tet, tetracycline; Xnl, ceftiofur.
Biofilm formation assay.
Salmonella Enteritidis strains were examined under two laboratory-imposed conditions known to result in biofilm formation: a rich nutrient medium, TSB, grown at 37°C, and a minimal medium, M9, incubated at 22°C. As outlined previously, 200 μl of culture media (either TSB or M9) containing approximately 106 log10 CFU/ml Salmonella was added into fresh sterile 96-well plates. TSB-containing plates were incubated for 48 h at 37°C, and the M9-containing plates were incubated for 1 week at 22°C. We determined the concentration of Salmonella and the incubation times to result in a number of bacteria for susceptibility testing that was equivalent to or lower than that used for the planktonic susceptibility testing outlined above (giving a similar starting OD and growth curve pattern as measured by the Omnilog phenotypic microplate reader). Following incubation in each case, spent growth medium was aspirated and the plates washed; 200 μl phosphate-buffered saline (PBS) per well was added and then aspirated. A viability assay was carried out as follows. Two hundred microliters of a bacterial cell recovery medium was added to each well of the 96-well plate. The cell recovery medium used was the Biolog recovery medium GN IF-10 (Biolog Inc., Hayward, CA), which consists of a buffer containing detergent, salts, gelling agent, redox dye, and other components specifically designed for maximum recovery of Gram-negative cells and optimal detection of growth using the Omnilog microplate reader. Plates were placed in the Omnilog phenotypic microarray reader for 24 h at 37°C. The MIC for each biocide formulation was calculated from the instrument output as the lowest concentration of biocide formulation at which the well was not considered positive for growth using the same calculations outlined above.
Biofilm biocide survival assay.
Biofilms of Salmonella Enteritidis (formed as described above) were incubated in the presence of the same panel of industrial biocide formulations (using the same dilution series as outlined above) for 5 different time points: 5, 10, 15, 30, and 60 min. At the end of the incubation period, the biocides were aspirated and plates washed twice in PBS. Viability assays were performed as described above. Each assay was carried out in duplicate.
Surface-dried cell biocide survival assay.
Briefly, 106 log10 CFU/ml of overnight culture of Salmonella Enteritidis, in a volume of 10 μl TSB, was added to each well of a 96-well plate and dried in a sterile laminar airflow cabinet for a period of 4 h. Biocides were applied as described above using the same 5 time points. Plates were washed twice in PBS and a viability test was carried out as described above.
Statistical analysis.
All results were analyzed using Microsoft Excel and SPSS (PASW Statistics 18). The distribution and spread of MIC values from the biocides and antibiotic susceptibility profiling were examined using descriptive statistics. A correlation between biocide and antimicrobial compound susceptibilities was derived using a Fisher exact test and a Spearman's rank correlation.
The significance of the stability of biocide activity over time was examined by using a Student t test on the MIC values. Similarly, a Student t test was used to determine whether results from surface-dried cell and biofilm testing were significantly different from the output of the planktonic testing.
RESULTS
Food industry biocide formulation susceptibility testing.
Of the 189 strains tested, there were no surviving Salmonella strains in six of the seven food industry biocide formulations at concentrations greater than 50% of the recommended working concentration. For the remaining biocide, formulation 1, there was survival of one isolate, S. enterica serotype Fresno, when challenged at the recommended working concentration of formulation 1. This tolerant phenotype, however, was unstable, and the isolate became susceptible on repeated testing. The same phenotype was observed when the isolate was cultured in the absence of biocide or if the isolate was stored on cryoprotectant beads at −80°C.
A range of susceptibilities was observed when Salmonella isolates were compared. Based on our experimental design, formulation 1 was the least effective of the biocides tested, with MICs ranging from 3.13 to 100% of the working concentration. Formulation 4 and formulation 5 were the most effective compounds, with limited differences being recorded in the susceptibilities of the Salmonella isolates to both. In this case, MICs ranged from <0.2 to 3.13 and 6.25%, respectively (Table 4). For the remaining four biocide formulations, MIC values ranged from 0.2% of the working concentration to 25% for formulation 2, formulation 3, and formulation 7 and from 0.2 to 50% for formulation 6.
Table 4.
Distribution of Salmonella MICs (recommended working concentrations) of food industry biocide formulations
| Formulation no. | % recommended working concna |
|||
|---|---|---|---|---|
| Range | Minimum | Maximum | Mean | |
| 1 | 96.87 | 3.13 | 100.00 | 35.2303 |
| 2 | 24.80 | 0.20 | 25.00 | 2.9562 |
| 3 | 24.80 | 0.20 | 25.00 | 3.3955 |
| 4 | 3.13 | <0.20 | 3.13 | 0.2731 |
| 5 | 6.25 | <0.20 | 6.25 | 0.3502 |
| 6 | 49.80 | 0.20 | 50.00 | 3.0092 |
| 7 | 24.80 | 0.20 | 25.00 | 2.6174 |
Shown are the percent recommended working concentrations (relating to the concentrations in which the formulations are applied in industry, where 100% is the concentration recommended by the manufacturer and 50% is half that concentration). The range statistic is defined as the difference recorded in the maximum and minimum MICs (this statistical value, expresses the variation in response of the bacterial strain in the collection to the individual biocides). The minimum and maximum MICs recorded for the entire collection are shown, as are the mean MICs for the given biocides across the strain collection.
There were no significant differences between the MICs and the MBCs (data not shown) for any of the seven food industry formulations; all were classed as bactericidal.
With the exception of formulation 1, all biocide formulations were effective when applied at their working concentration and eliminated all planktonic Salmonella strains tested.
Antibiotic susceptibility profiling.
All Salmonella strains in the collection were tested against a panel of clinically important antimicrobial compounds (Table 2). Of the 189 isolates investigated, 46 were susceptible to all compounds tested. A further 48 Salmonella strains were resistant to at least a single antimicrobial compound, whereas 95 were defined as multidrug resistant (MDR), being resistant to three or more classes of antimicrobial compounds.
Resistance to sulfisoxazole was the most frequently encountered phenotype (frequency, 0.72; n = 142), followed by resistances to trimethoprim-sulfamethoxazole (0.57; n = 108), ciprofloxacin (0.40; n = 77), ceftiofur (0.32; n = 61), and nalidixic acid (0.32; n = 61). Of the serotypes defined as MDR, S. Typhimurium and S. Braenderup were the most common. S. Typhimurium comprised 9.52% (n = 6) and S. Braenderup comprised 6.35% (n = 4) of isolates with an MDR phenotype. A summary of resistance frequencies is shown in Table 2.
When statistical evaluation was performed, there was no correlation between antimicrobial resistance and reduced susceptibility to food industry biocide formulations. In addition, no significant correlation between any antimicrobial compound and biocide formulation combination was observed.
Biocide stability and activity testing.
Biocide stability testing refers to the determination of the activity of a biocide over a defined time period. It was measured over a period of 8 days. Each biocide formulation was prepared at its working concentration and then tested. Any biocide for which the MIC/MBC remained consistent over time, when tested against the type strain, was defined as stable. Conversely, a biocide for which the MIC/MBC increased (i.e., the antimicrobial efficacy decreased) was designated unstable.
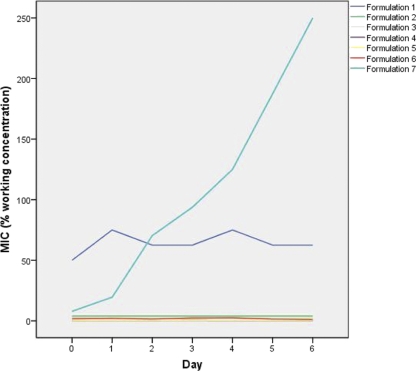
All biocides were stable, retaining their bactericidal activity, over a period of 8 days in water. With the exception of formulation 7, all biocides also retained their activity in the presence of organic matter (TSB) over the same period. Formulation 7 gradually lost its activity in TSB, with a 2-fold decrease in activity recorded over 24 h, increasing to a 60-fold decrease over 6 days (Fig. 1).
Fig 1.
Stability of biocide activity over time in the presence of organic matter.
Selecting for increased tolerance to commercial biocidal formulations.
In vitro selection for Salmonella strains displaying an increased phenotype of tolerance to the commercial formulations could not be achieved, despite several rounds of selection, either by incrementally increasing the concentration of the biocide formulation or by subculturing at the recommended concentration. A 2-fold increase in the MIC of two Salmonella strains to formulation 1 was obtained by subculturing in increasing concentrations (data not shown), but the tolerance phenotype developed was unstable, being lost following passage in the absence of the biocidal formulation or following storage on cryoprotectant beads at −80°C.
As no isolates tolerant to the biocide formulations were recovered, a subset of six Salmonella strains, of various serotypes, was selected for further study. This selection was based on the observation that these isolates demonstrated an MIC higher than the mean MIC of two or more biocide formulations (data not shown) and thus were selected for in vitro exposure to the active component of a biocide and subsequent testing.
Selecting for increased tolerance to the active component of a biocide.
After in vitro selection, bacteria exhibiting a high-level tolerance to three of four active biocidal agents were obtained; these agents included triclosan, chlorhexidine, and benzalkonium chloride (Table 5). Similarly, a biocide tolerance phenotype corresponding to triclosan and chlorhexidine could be selected in a further two Salmonella type strains, S. Typhimurium SL1344 and S. Enteritidis NCTC 13349. On this occasion, the phenotype selected was stable, being maintained after several rounds of culture in the absence of the selective agent. Rates at which tolerance developed also differed, with tolerance to triclosan emerging faster than when benzalkonium chloride was used in the selection (data not shown).
PFGE profiling confirmed that most of the bacteria displaying biocide tolerance phenotypes were indistinguishable from their wild-type parents (data not shown), with 100% similarity on dendrogram analysis (data not shown). In the case of two of the parent and mutant combinations, S. Typhimurium ST23 and its triclosan-tolerant counterpart along with S. Typhimurium ST24 and its chlorhexidine-tolerant counterpart, a single band difference was noted in the PFGE profiles; in these cases, similarities of 96% and 95.34% were obtained by dendrogram analysis (data not shown).
Differences in the ease with which a phenotype of tolerance to an active biocide compound was selected in vitro among the Salmonella serotypes were also noted. Salmonella Gaminara was the quickest to be selected; a 50-fold increase in the MIC developed following six passages over a period of 7 days. In contrast, the type strains took longer to develop a tolerance. In each case, similar fold increases in MIC were recorded following 17 passages over a period of 28 days.
Cross-tolerance of the six Salmonella strains to industrial biocide formulations, the active biocidal agents, and antimicrobial compounds.
There were no significant differences detected between the patterns of tolerance of the isogenic parent and mutant combinations to either food industry biocide formulations (data not shown) or the active biocide contained therein (Table 6).
Table 6.
Susceptibility profiles of biocide-tolerant Salmonella serotypes to biocidal active agents and food industry biocide formulations
| Biocidal agent/formulation | MIC (μg/ml)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gaminara S5 |
Typhimurium ST24 |
Hvittingfoss S41 |
Typhimurium ST23 |
Senftenberg C81 |
Enteritidis 5408 |
|||||||
| WT | Chlorhexidine tolerant | WT | Chlorhexidine tolerant | WT | Benzalkonium chloride tolerant | WT | Triclosan tolerant | WT | Triclosan tolerant | WT | Triclosan tolerant | |
| Chlorhexidine | 37.5 | >1,000 | 1.96 | >100 | 62.5 | 7.81 | 3.91 | 7.81 | 15.6 | 15.6 | 7.8 | 15.6 |
| Benzalkonium chloride | 3.91 | 3.91 | 3.91 | 1.95 | 23.43 | 93.75 | 3.91 | 1.95 | 31.25 | 31.25 | 31.25 | 31.25 |
| Triclosan | 0.78 | >10 | 2.5 | 5 | >10 | 10 | 0.1 | 1,000 | 1 | >1,000 | 0.1 | >1,000 |
| H2O2 | 1.25 | 0.32 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 |
WT, wild-type isogenic parent strain. The MICs of the wild-type parent isolates and biocide-tolerant mutants are shown. Significant alterations in susceptibility patterns are displayed in bold.
However, four of the six biocide-tolerant isolates exhibited significant changes in their patterns of susceptibility to a panel of antimicrobial agents compared to their wild-type counterparts. These differences were not drug class specific (Table 7). The benzalkonium chloride-tolerant S41 had the largest number of alterations in its susceptibility profile, with a significantly decreased susceptibility to 12 of the 24 antimicrobial compounds evaluated. In this case, changes in susceptibility occurred in 8 of 11 classes of compound tested. The triclosan-tolerant C81 displayed a decreased susceptibility to seven antimicrobials of six classes, and the chlorhexidine-tolerant S5 similarly showed a decreased susceptibility to three compounds from three drug classes. The triclosan-tolerant isolate 5408 had one change in its susceptibility profile; in this case, the mutant was more susceptible to cefpodoxime.
Table 7.
Antimicrobial susceptibility profiles of susceptible and resistant/tolerant Salmonella serotypes
| Class | Antimicrobial compound | MIC (μg/ml)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gaminara S5 |
Typhimurium ST24 |
Hvittingfoss S41 |
Typhimurium ST23 |
Senftenberg C81 |
Enteritidis 5408 |
||||||||
| WT | Chlorhexidine tolerant | WT | Chlorhexidine tolerant | WT | Benzalkonium chloride tolerant | WT | Triclosan tolerant | WT | Triclosan tolerant | WT | Triclosan tolerant | ||
| β-Lactam | Ampicillin | <2 | <2 | <2 | <2 | <2* | 16* | <2 | <2 | <2 | <2 | 16‡ | <2‡ |
| Amoxacillin-clavulanic acid | 4 | <2 | <2 | <2 | <2 | 4 | <2 | <2 | <2 | <2 | 4 | <2 | |
| Piperacillin | <4 | <4 | <4 | 8 | <4* | 64* | 16‡ | <4‡ | <4* | 16* | 8* | 16* | |
| Imipenim | 2 | <2 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | |
| Cephalosporin | Cephalexin | 16‡ | 8‡ | 16‡ | 8‡ | <4* | 16* | 16‡ | 8‡ | 16 | 16 | 16 | 16 |
| Cefpodoxime | 0.5 | 0.5 | <0.25 | <0.25 | <0.25* | 2* | 0.5 | <0.25 | <0.25 | 0.5 | 4‡ | 2‡ | |
| Ceftiofur | 2 | 2 | 2* | 4* | <1* | >8* | 4‡ | 2‡ | 2† | >8† | >8 | >8 | |
| Cefpirome | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | |
| Ceftriaxone | <0.25 | <0.25 | <0.25 | ND | <0.25† | 2† | <0.25 | ND | 0.5 | 1 | ND | ND | |
| Aminoglycoside | Amikacin | <0.5 | <0.5 | <2 | <2 | <2 | 2 | <2 | <2 | 4* | 16* | <2* | 16* |
| Gentamicin | 2 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1* | 4* | <1 | 2 | |
| Tobramycin | 2 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | 3 | <1 | 2 | |
| Kanamycin | 16‡ | <8‡ | <8 | ND | <8 | <8 | <8 | ND | <8* | 32* | ND | ND | |
| Streptomycin | <32 | <32 | <32 | ND | <32 | <32 | ND | ND | 64 | >64 | ND | ND | |
| Polyketide | Tetracycline | <1§ | >16§ | >16 | >16 | <1* | 8* | >16 | >16 | <1 | 2 | 4 | 4 |
| Fluoroquinolone | Enrofloxacin | <0.12 | 0.25 | <0.12 | 0.25 | <0.12 | 0.5 | 0.25 | <0.12 | <0.12 | <0.12 | >4 | 0.5 |
| Marbofloxacin | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | >4 | <0.5 | |
| Ciprofloxacin | 0.06 | 0.06 | 0.03 | ND | 0.06* | 0.5* | 0.06 | ND | 0.03 | 0.25 | ND | ND | |
| Nitrofuran | Nitrofurantoin | <16 | 32 | 32* | 64* | 32 | 32 | 64‡ | <16‡ | <16 | <16 | 32 | <16 |
| Chloramphenicol | Chloramphenicol | 4* | 8* | 8 | 16 | 4* | 16* | 8‡ | <2‡ | 4* | 16* | 32 | 16 |
| Diaminopyrimidines/sulfonamide | Trimethprim-sulfamethoxazole | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| Cephamycin | Cefoxitin | <0.5 | <0.5 | 2 | ND | 8 | >32 | 16 | ND | 16§ | 32§ | ND | ND |
| Quinolone | Nalidixic acid | 4* | 16* | 4 | ND | 4† | 32† | 4 | ND | 8† | 32† | ND | ND |
| Sulfonamide | Sulfisoxazole | >256 | 256 | 128 | ND | >256 | 128 | >256 | ND | 32* | >256* | ND | ND |
WT, wild-type isogenic parent strain. Significant alterations in susceptibility patterns are displayed in bold.
, alteration in CLSI classification from sensitive to intermediate;
, alteration in CLSI classification from sensitive to resistant;
, alteration in CLSI classification from intermediate to sensitive;
, alteration in CLSI classification from intermediate to resistant; ND, not determined.
Neither the biocide-tolerant S. Typhimurium strains, ST23 and ST24, nor the two type strains S. Typhimurium SL1344 and S. Enteritidis ATCC 13349 demonstrated any significant alterations in their biocide susceptibilities (data not shown).
Efficacy of biocide formulations against Salmonella contained within a biofilm.
All seven biocide formulations were assessed for their efficacy to eliminate in vitro-generated biofilm of one strain, an S. Enteritidis strain; this isolate was chosen for study because it displayed the strongest biofilm formation across all conditions tested. This assay aimed to evaluate any differences in the efficacy of the various biocide formulations against the bacterium grown in a biofilm compared to the same strain planktonically cultured. Biofilms were produced under two growth conditions: in M9 minimal medium cultured at 22°C and in a rich nutrient medium (TSB) cultured at 37°C. It was noted that there was an increase in the MIC for Salmonella Enteritidis bacteria contained within the biofilm relative to the same bacteria grown as planktonic cells (Table 8). Biocide formulations differed greatly in their abilities to eliminate Salmonella when it was contained in a biofilm relative to Salmonella in plantonkic cultures.
Table 8.
MICs of food industry formulations against planktonic, biofilm, and surface-dried cultures of Salmonella Enteritidis
| Culture | Exposure time (min) | MIC (μg/ml) of formulation: |
||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| Planktonic | 100 | 6.25 | 6.25 | 0.2 | 0.2 | 12.5 | 3.13 | |
| Low-nutrient (M9) biofilm | 5 | 200 | 100 | 50 | 3.13 | 1.56 | 100 | 100 |
| 10 | 200 | 200 | 200 | 0.78 | 0.78 | 200 | 200 | |
| 15 | 200 | 25 | 100 | 1.56 | 0.78 | 100 | 100 | |
| 30 | 200 | 6.25 | 25 | 0.78 | 0.78 | 25 | 25 | |
| 60 | 50 | 1.56 | 6.25 | 0.39 | 0.39 | 12.5 | 6.25 | |
| High-nutrient (TSB) biofilm | 5 | >200 | >200 | 100 | 50 | 25 | >200 | >200 |
| 10 | >200 | >200 | 50 | 25 | 25 | 200 | 200 | |
| 15 | >200 | >200 | 100 | 12.5 | 6.25 | >200 | >200 | |
| 30 | >200 | >200 | 100 | 25 | 6.25 | >200 | >200 | |
| 60 | >200 | >200 | 50 | 3.13 | 3.13 | >200 | >200 | |
| Surface dried | 5 | 200 | 50 | 6.25 | 6.25 | 6.25 | 50 | 100 |
| 10 | 100 | 100 | 6.25 | 1.56 | 0.78 | 50 | 25 | |
| 15 | 100 | 25 | 6.25 | 12.5 | 3.13 | 50 | 12.5 | |
| 30 | 25 | 50 | 6.25 | 0.78 | 0.78 | 12.5 | 12.5 | |
| 60 | 25 | 12.5 | 3.13 | 0.39 | 0.39 | 12.5 | 12.5 | |
We examined both high- and low-nutrient-biofilm-derived matrices in order to assess whether the biocides had similar effects on both. Low-nutrient-derived biofilm is associated with greater exopolysaccharide (EPS) production and thickness (5). Both of these features are known to contribute to biofilm-associated tolerance when such a treatment is used (20). The high-nutrient-derived biofilm was associated with reduced EPS production but an increase in cell density.
Under high-nutrient culture conditions, three biocide formulations remained effective and were capable of eliminating Salmonella contained within the biofilm; these included formulation 3, formulation 4, and formulation 5. Within formulation 5's recommended contact time of 5 min, its MIC was 25% of the working concentration. Neither formulation 3 nor formulation 4 had a recommended contact time, but after 5 min, MICs of 100 and 50% of the recommended working concentrations, respectively, were recorded. The remaining biocide formulations were ineffective against Salmonella contained in the high-nutrient biofilm, at a level of two times the recommended working concentration, even after 1-h contact time, three times the recommended contact time for formulations 1, 2, and 7 and four times the recommended contact time for formulation 6.
Salmonella Enteritidis biofilms formed following growth in M9 minimal medium demonstrated an increased tolerance to the biocide formulations compared to that of their planktonic counterparts, despite having a smaller inoculum of Salmonella. However, these bacteria were less tolerant than Salmonella contained within biofilms formed in high-nutrient culture media. After 5-min contact time, MICs of 100, 3.13, and 1.56% and were recorded for formulation 3, formulation 4, and formulation 5, respectively. These three formulations remained the most effective. Of the remaining biocides, MICs of 100% of the working concentration were recorded for formulation 2 and formulation 7, within their recommended maximum contact time of 20 min. Formulation 6 inhibited the growth of Salmonella Enteritidis after 30 min, twice its recommended contact time, and formulation 1 required 60-min application time, four times that which was recommended, to kill all viable cells at its recommended working concentration.
Efficacy of biocide formulations against surface-dried Salmonella.
Surface-dried efficacy testing was carried out with the same Salmonella Enteritidis strain for all seven biocide formulations described above. An increase in MIC was observed, at all time points, for five of the formulations relative to planktonically grown cells (Table 8). However, all but one of the formulations remained effective at its recommended working concentration and recommended contact time. Formulation 3 retained the same activity against surface-dried Salmonella as it did against free-growing Salmonella; an MIC of 6.25% of the working concentration after 5 min of contact was observed. Formulation 4 and formulation 5 were the most effective biocides, with an MIC after 5-min contact of 6.25% of the working concentration. Formulation 7 and formulation 6 inhibited surface-dried Salmonella after 5 min at their recommended working concentration. Formulation 2 had an MIC of 100% of the working concentration, after 15-min contact time. Formulation 1 was the least effective, having an MIC of 100% of the working concentration but requiring 30-min contact time, 10 min longer than the manufacturer's recommended contact time.
DISCUSSION
Results of this study showed that, with the exception of one formulation, commercially available biocides used in the food industry are effective against several serotypes of planktonically grown Salmonella. Our results support findings reported earlier (24). Interestingly, after several rounds of in vitro selection with increasing concentrations of the industry biocide formulations, we could not detect any stable increases in the biocide tolerance of any Salmonella strain.
When biocide formulations are used along the food production chain, they are normally applied at concentrations that typically far exceed those used for antibiotics, without the same toxicity concerns (9). It is hypothesized that at these higher concentrations, biocides target several bacterial cell structures (34), resulting in cell death and making it difficult for tolerance to emerge. For example, oxygen-releasing biocides facilitate the indiscriminate oxidation of a range of membrane and cytoplasmic thiol groups and other chemical groups leading to the inhibition of multiple key enzymes and modification of structural proteins (9, 18). The industrial biocides tested in this study comprised specific formulations containing multiple active biocidal agents, targeting the bacterial cell at several levels. The absence of intrinsic and extrinsic tolerance within our Salmonella collection and the inability to select any increased tolerance among these isolates is, most likely, due to the indiscriminate effects of biocide action.
No correlation between reduced susceptibility to food industry biocide formulations and resistance to clinically relevant antimicrobial compounds was detected. The 95 MDR isolates studied exhibited a susceptibility phenotype indistinguishable from the phenotype of the 46 isolates classed as antibiotic susceptible, when measured with biocide formulations. Previous studies have also found that antibiotic-resistant bacteria are no more tolerant to biocides, including disinfectants and antiseptics, than susceptible strains (1, 4, 17). In addition, multidrug-resistant nosocomial isolates of Enterococcus spp., Staphylococcus aureus, Staphylococcus epidermidis, Pseudomonas aeruginosa, Escherichia coli, and Klebsiella pneumoniae have all been found to have disinfectant susceptibility patterns similar to those of their antibiotic-sensitive counterparts (1).
Commercially available biocide formulations used in the food industry contain several active agents, some of which may include triclosan, chlorhexidine, and benzalkonium chloride. Sublethal concentrations of an active ingredient may have the potential to select for tolerant bacteria from an exposed population of cells. To determine whether or not increased tolerance to these agents could be selected for, after in vitro culture, Salmonella isolates were individually exposed to subinhibitory concentrations (0.25× MIC) of triclosan, chlorhexidine, and benzalkonium chloride. Bacterial isolates recovered demonstrated an increased tolerance to each active agent, with a high degree of tolerance being developed by several isolates and linked with an increase in the corresponding MIC which was measured at 1,000-fold for triclosan, between 25- and 50-fold for chlorhexidine, and 4-fold for benzalkonium chloride. Differences in the ease with which a phenotype of tolerance to an active biocide compound was selected in vitro from the Salmonella serotypes studied was noted. The laboratory type strains were slower to develop a tolerance than those recovered from the environment. It may be reasonable to suggest that this phenomenon may arise due to the fact that laboratory-adapted type strains, unlike the environmental strains, have not been exposed to selective conditions such as would be encountered in the food-processing or host-associated environments.
In this case, bacterial isolates demonstrating an increased tolerance to a specific biocide active agent displayed alterations in their profiles of susceptibility to a panel of antimicrobial compounds. The magnitudes and numbers of changes differed for the different biocides and the Salmonella serotypes. Different serotypes displayed a variation in the number of alterations recorded. Braoudaki and Hilton previously reported that variation in the phenotype between individual serotypes was observed, i.e., a biocide-tolerant S. Virchow strain exhibited cross-tolerance to antimicrobial compounds, which contrasted with S. Typhimurium exhibiting little cross-tolerance and S. Enteritidis exhibiting none (2). A similar trend was noted in this study, with serotypes Typhimurium and Enteritidis, the type strains and nontype strains, exhibiting little or no significant changes, as measured by MIC, compared with serotypes Gaminara, Hvittingfoss, and Senftenberg.
For most isolates, the patterns of alteration in the antibiotic susceptibility profiles were distributed among different classes of antibiotic. Results indicated that the mechanisms of tolerance to biocidal agents (triclosan, chlorhexidine, and benzalkonium chloride) may be broad spectrum, imparting a reduced susceptibility to compounds with diverse chemical structures. These data suggest a contribution by efflux pumps through their upregulation and/or permeability alterations in these strains. Antimicrobial compounds, including amikacin, ampicillin, cephalosporins, cefoxitin, chloramphenicol, ciprofloxacin, kanamycin, nalidixic acid, piperacillin, and tetracycline, which demonstrated a reduced effect on one or more of the biocide-tolerant Salmonella strains, are known efflux pump substrates (11, 15, 27, 29, 32) of the resistance nodulation and cell division (RND) family (32). In some instances, changes in antibiotic susceptibility profiles were sufficiently large that these could be defined according to CLSI guidelines, whereas others were more modest. The latter, nonetheless, could still be of importance, as such changes may impart a growth advantage to the bacteria (2), particularly if efflux mechanisms are involved. Researchers have extensively reported on the contribution of active efflux to antimicrobial resistance, therapeutic failure, and virulence (28, 30, 32). Results show that sublethal exposure to common food industry biocides induced an increased tolerance to multiple antibiotics. The fact that this exposure may potentially select for strains that overexpress their efflux pumps could result in Salmonella strains that are more invasive and subsequently more resistant to antimicrobial chemotherapy, a development that would be of concern to public health. In our laboratory, further work designed to address this issue and attempt to identify those mechanisms contributing to this tolerance is under way. More recent studies focused on examining the proteomes of two of the biocide-tolerant Salmonella strains reported on here. We are additionally in the process of undertaking single-nucleotide-polymorphism (SNP) analysis on two of the isogenic/mutant isolate combinations, namely, ST23 and ST24.
Although significant changes in the patterns of susceptibility to several antimicrobial compounds were detected in four of the biocide-tolerant mutants, none gave rise to significant changes in MIC when retested against the biocide active agents. Mechanisms in those isolates exhibiting a phenotype of high-level tolerance to one class of biocidal agent did not cause a significant cross-tolerance to other biocidal agents from a different class. Similarly, there was no correlation between tolerance to any of the three individual biocidal active agents, triclosan, chlorhexidine, and benzalkonium chloride, and a reduced susceptibility to the food industry biocide formulations, even if that formulation contained that same active agent. As an example of this, formulation 3 contained benzalkonium chloride as an active agent, yet the in vitro-selected benzalkonium chloride-tolerant isolate and its susceptible isogenic parent were both inhibited at 6.25% of the working concentration. This may be due to the fact that the tolerant isolate remained susceptible to other active ingredients contained in the biocide formulation. These data showed that although isolates displaying a tolerance to individual biocidal agents may correlate with an increased tolerance to clinically important antibiotics, they are no more tolerant to industrial biocide formulations than their susceptible isogenic parents.
In our studies, Salmonella Enteritidis was chosen for the biofilm and surface-dried assays to test the efficacy of the various biocide formulations. This strain showed typical susceptibility profiles when cultured in a planktonic state. In contrast, when the same bacterium was reevaluated as part of a laboratory-generated biofilm (under low- and high-nutrient conditions) or when dried onto an abiotic surface, it demonstrated a marked increase in tolerance. The biocide formulations examined in this study differed in their activities against surface-dried and biofilm communities. Some biocide formulations, including formulation 3, formulation 4, and formulation 5, remained effective against sessile communities formed under high- and low-nutrient conditions. Conversely, others, such as formulation 2, formulation 6, formulation 7, and formulation 1, were unable to inhibit biofilm containing Salmonella even at twice their recommended working concentrations and for a contact period 4 times that recommended by the manufacturer. Similarly, formulations 1 and 6 were unable to eliminate Salmonella contained in a biofilm formed under low-nutrient conditions within their recommended contact times. These observations are in agreement with earlier work, which reported that biocides used in the food industry vary in their activity, from almost total reduction of surface-attached Salmonella to little or no effect (24). In this study, biofilms formed under high-nutrient availability were associated with an increased tolerance compared with those formed under low-nutrient conditions. The increased EPS and biofilm thicknesses associated with low-nutrient biofilms did not impart an enhanced tolerance to the biocide formulations tested, compared with the situation in which high-nutrient media was used. However, the increased tolerance of the high-nutrient biofilm may be attributed to a higher cell density in the nutrient-rich biofilm.
These data highlight the importance of implementing control strategies to prevent the development of biofilm and surface-dried communities in the food-processing environment. Cleaning frequencies and regimes should be carefully designed, monitored, and modified appropriately. A cleaning plan that ensures a sufficient surface disinfectant contact time, using biocides at a concentration sufficient to kill cells present in a biofilm and avoiding the dilution effect of an applied disinfectant with rinsing water, should prevent the persistence of adapted strains (22). Furthermore, these findings demonstrate the importance of preventing the build-up of organic matter in the production environment. Organic matter is known to reduce the activity of the biocides (1). Of the biocides included in this study, one lost its stability in the presence of organic matter. Furthermore, Salmonella strains contained in the biofilms formed in the presence of organic matter were significantly more tolerant to all biocides tested, surviving twice the concentration, and may therefore be more difficult to eradicate in the food factory environment.
Given the broad-spectrum activity demonstrated by biocide formulations against bacteria associated with their high in-use concentration, the development of persistent colonies is not likely to arise. It is arguable whether or not biocide tolerance could develop outside the laboratory setting when biocides are used at recommended concentrations. It is possible that tolerance to a biocide may arise following incorrect use or misuse of the formulation. Some examples of the latter include when the biocide is used at concentrations below that which is recommended by the manufacturer, if the biocide is applied in the presence of organic matter which can often interfere with and disrupt its activity, when used at a suboptimal temperature or pH, or if an initial high concentration of the applied biocide is reduced by unintended dilution. Furthermore, even if a biocide is used according to the recommended guidelines and biofilm formation has been allowed to develop or there are surface-dried bacteria which remain, then biocide treatments may also fail to eliminate bacteria.
Biocides play an essential and effective role in limiting the spread of infectious agents and are important to the food industry, as an aid to control the microbial contamination of the production environment. It has been demonstrated that sublethal exposure to biocidal agents can lead to the development of tolerant isolates. Studies focusing on an improved understanding of the mechanisms involved in biocide tolerance and research into the epidemiology of any emerging biocide-tolerant strains will be of increasing importance in the future.
ACKNOWLEDGMENTS
We acknowledge the financial support provided by the Food Institutional Research Measure (FIRM), administered by the Department of Agriculture, Fisheries and Food, Ireland (grant no. 08/RD/TAFRC/616).
There are no conflicts of interest to report. None of the authors is funded by commercial companies with a stake in this research.
Footnotes
Published ahead of print 24 February 2012
REFERENCES
- 1. Anderson RL, Carr JH, Bond WW, Favero MS. 1997. Susceptibility of vancomycin-resistant Enterococci to environmental disinfectants. Infect. Control Hosp. Epidemiol. 18:195–199 [DOI] [PubMed] [Google Scholar]
- 2. Braoudaki M, Hilton AC. 2004. Adaptive resistance to biocides in Salmonella enterica and Escherichia coli O157 and cross-resistance to antimicrobial agents. J. Clin. Microbiol. 42:73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coburn B, Grassl GA, Finlay BB. 2007. Salmonella, the host and disease: a brief review. Immunol. Cell Biol. 85:112–118 [DOI] [PubMed] [Google Scholar]
- 4. Cookson BD, Bolton MC, Platt JH. 1991. Chlorhexidine resistance in methicillin-resistant Staphylococcus aureus or just an elevated MIC? An in vitro and in vivo assessment. Antimicrob. Agents Chemother. 35:1997–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dewanti R, Wong AL. 1995. Influence of culture conditions on biofilm formation by Escherichia coli O157:H7. Int. J. Food Microbiol. 26:147–164 [DOI] [PubMed] [Google Scholar]
- 6. Duffy G, Walsh C. 2005. Antibiotic resistance in food borne pathogens. Teagasc, Oak Park, Carlow, Ireland: Accessed 11 October 2011 http://www.teagasc.ie/research/reports/foodprocessing/5036/eopr5036.pdf [Google Scholar]
- 7. Fraise AP. 2002. Biocide abuse and antimicrobial resistance—a cause for concern? J. Antimicrob. Chemother. 49:11–12 [DOI] [PubMed] [Google Scholar]
- 8. Gast RK, Holt PS. 1998. Persistence of Salmonella enteritidis from one day of age until maturity in experimentally infected layer chickens sampling for persistent. Poult. Sci. 77:1759–1762 [DOI] [PubMed] [Google Scholar]
- 9. Gilbert P, McBain AJ. 2003. Potential impact of increased use of biocides in consumer products on prevalence of antibiotic resistance. Clin. Microbiol. Rev. 16:189–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gilbert P, Moore LE. 2005. Cationic antiseptics: diversity of action under a common epithet. J. Appl. Microbiol. 99:703–715 [DOI] [PubMed] [Google Scholar]
- 11. Giraud E, Cloeckaert A, Kerboeuf D, Chaslus-Dancla E. 2000. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 44:1223–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heath RJ, Rock CO. 2000. A triclosan-resistant bacterial enzyme. Nature 406:145–146 [DOI] [PubMed] [Google Scholar]
- 13. Heath RJ, et al. 1999. Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J. Biol. Chem. 274:11110–11114 [DOI] [PubMed] [Google Scholar]
- 14. Imlay JA, Chin SM, Linn S. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640–642 [DOI] [PubMed] [Google Scholar]
- 15. Islam S, Jalal S, Wretlind B. 2004. Expression of the MexXY efflux pump in amikacin-resistant isolates of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 10:877–883 [DOI] [PubMed] [Google Scholar]
- 16. Jones G. 1997. Chlorhexidine: is it still the gold standard? Periodontol. 2000 15:55–62 [DOI] [PubMed] [Google Scholar]
- 17. Karatzas KAG, et al. 2008. Phenotypic and proteomic characterization of multiply antibiotic-resistant variants of Salmonella enterica serovar Typhimurium selected following exposure to disinfectants. Appl. Environ. Microbiol. 74:08–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lambert P. 2004. Mechanism of action of biocides, p 139–153 In Fraise A, Lambert PA, Maillard J-Y. (ed), Russell, Hugo & Ayliffe's principles and practice of disinfection, preservation and sterilization, 4th ed Blackwell Publishing Ltd., Malden, MA [Google Scholar]
- 19. Langsrud S, Sidhu M, Heir E, Holck A. 2003. Bacterial disinfectant resistance—a challenge for the food industry. Int. Biodeter. Biodegr. 51:283–290 [Google Scholar]
- 20. Mah T-FC, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34–39 [DOI] [PubMed] [Google Scholar]
- 21. McBain AJ, Rickard AH, Gilbert P. 2002. Possible implications of biocide accumulation in the environment on the prevalence of bacterial antibiotic resistance. J. Ind. Microbiol. 29:326–330 [DOI] [PubMed] [Google Scholar]
- 22. Meyer B. 2006. Does microbial resistance to biocides create a hazard to food hygiene? Int. J. Food Microbiol. 112:275–279 [DOI] [PubMed] [Google Scholar]
- 23. Moore SL, Payne DN. 2004. Types of antimicrobial agents. In Fraise A, Lambert PA, Maillard J-Y. (ed), Russell, Huge & Ayliffe's principles and practice of disinfection, preservation and sterilization, 4th ed Blackwell Publishing Ltd., Malden, MA [Google Scholar]
- 24. Møretrø T, et al. 2009. Evaluation of efficacy of disinfectants against Salmonella from the feed industry. Appl. Environ. Microbiol. 106:1005–1012 [DOI] [PubMed] [Google Scholar]
- 25. Mullane NR, Whyte P, Wall PG, Quinn T, Fanning S. 2007. Application of pulsed-field gel electrophoresis to characterise and trace the prevalence of Enterobacter sakazakii in an infant formula processing facility. Int. J. Food Microbiol. 116:73–81 [DOI] [PubMed] [Google Scholar]
- 26. Nesse LL, et al. 2003. Molecular analyses of Salmonella enterica isolates from fish feed factories and fish feed ingredients. Appl. Environ. Microbiol. 69:1075–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nikaido H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishino K, Latifi T, Groisman EA. 2006. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59:126–141 [DOI] [PubMed] [Google Scholar]
- 29. Nishino K, Nikaido E, Yamaguchi A. 2009. Regulation and physiological function of multidrug efflux pumps in Escherichia coli and Salmonella. Biochim. Biophys. Acta 1794:834–843 [DOI] [PubMed] [Google Scholar]
- 30. Pagès J-M, Masi M, Barbe J. 2005. Inhibitors of efflux pumps in Gram-negative bacteria. Trends Mol. Med. 11:382–389 [DOI] [PubMed] [Google Scholar]
- 31. Poole K. 2004. Acquired resistance, p 154–169 In Fraise A, Lambert PA, Maillard J-Y. (ed), Russell, Hugo & Ayliffe's principles and practice of disinfection, preservation and sterilization, 4th ed Blackwell Publishing Ltd., Malden, MA [Google Scholar]
- 32. Poole K. 2004. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 10:12–26 [DOI] [PubMed] [Google Scholar]
- 33. Poole K. 2002. Mechanisms of bacterial biocide and antibiotic resistance. Symp. Ser. Soc. Appl. Microbiol. 92:55S–64S [PubMed] [Google Scholar]
- 34. Russell AD. 2003. Similarities and differences in the responses of microorganisms to biocides. J. Antimicrob. Chemother. 52:750–763 [DOI] [PubMed] [Google Scholar]
- 35. Shi X, Namvar A, Kostrzynska M, Hora R, Warriner K. 2007. Persistence and growth of different Salmonella serovars on pre- and postharvest tomatoes. J. Food Prot. 70:2725–2731 [DOI] [PubMed] [Google Scholar]
- 36. Sivaraman S, et al. 2004. Inhibition of the bacterial enoyl reductase FabI by triclosan: a structure-reactivity analysis of FabI inhibition by triclosan analogues. J. Med. Chem. 47:509–518 [DOI] [PubMed] [Google Scholar]
- 37. Su L-H, Chiu C-H. 2006. Salmonella: clinical importance and evolution of nomenclature. Chang Gung Med. J. 30:210–219 [PubMed] [Google Scholar]
- 38. Tabak M, et al. 2007. Effect of triclosan on Salmonella Typhimurium at different growth stages and in biofilms. FEMS Microbiol. Lett. 267:200–206 [DOI] [PubMed] [Google Scholar]
- 39. Vestby LK, Møretrø T, Langsrud S, Heir E, Nesse LL. 2009. Biofilm forming abilities of Salmonella are correlated with persistence in fish meal and feed factories. BMC Vet. Res. 5:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Winfield MD, Groisman EA. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3679–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]