Abstract
In this study, we examine the temporal pattern of colony appearance during cultivation experiments, and whether this pattern could inform on optimizing the process of microbial discovery. In a series of long-term cultivation experiments, we observed an expected gradual increase over time of the total number of microbial isolates, culminating in a 700-fold colony count increase at 18 months. Conventional thought suggests that long-term incubations result in a culture collection enriched with species that are slow growing or rare, may be unavailable from short-term experiments, and likely are novel. However, after we examined the phylogenetic novelty of the isolates as a function of the time of their isolation, we found no correlation between the two. The probability of discovering either a new or rare species late in the incubation matched that of species isolated earlier. These outcomes are especially notable because of their generality: observations were essentially identical for marine and soil bacteria as well as for spore formers and non-spore formers. These findings are consistent with the idea of the stochastic awakening of dormant cells, thus lending support to the scout model. The process of microbial discovery is central to the study of environmental microorganisms and the human microbiome. While long-term incubation does not appear to increase the probability of discovering novel species, the technology enabling such incubations, i.e., single-cell cultivation, may still be the method of choice. While it does not necessarily allow more species to grow from a given inoculum, it minimizes the overall isolation effort and supplies needed.
INTRODUCTION
Most microorganisms in the environment and animal microbiomes remain uncultivated and are essentially unexplored. Among them are representatives of more than half of all known microbial phyla (7, 21) and countless lower taxa. Discovering these microorganisms and bringing them into culture is essential for progress in both basic and applied microbiology. Traditional cultivation on solid media or via enrichment is thought to favor fast growers at the expense of slow growers and also mask rare species (19). The implication is that since most cultivation studies have been conducted using traditional plating or isolation from liquid enrichments, the available culture collections are disproportionally represented by fast-growing, so-called low-hanging fruit. This bears directly on the phenomenon of the great plate count anomaly (27): if incubations in the plate are by necessity short term, the anomaly may be, in part, the result of artifacts associated with the plate. Truly long-term incubations, then, have a chance of partially resolving the phenomenon by allowing access to slow-growing, rare species, and thus a new pool of microbial novelty (8, 9). Empirical evidence appears to confirm this. Indeed, recent long-term cultivation efforts led to the isolation of the first representatives of the SAR 11 clade, the phylum Verrucomicrobia, and other remarkably novel or rare microorganisms (5, 11, 12, 20, 22, 23).
While these views are logical and consistent with available observations, alternative views may have merit as well. If the majority of cells in the environment are in a nongrowing state (28), and if most of them awake into activity randomly (6 and S. Buerger, A. Spoering, E. Gavrish, C. Leslin, L. Ling, and S. S. Epstein, unpublished data), then appearing early or late in the incubation may be a result of random awakening, not an inherently low growth rate. If so, the probability of finding a novel species may not change during the length of incubation, so that a given amount of cultivation effort would bring the same amount of novelty independently of whether the effort was focused on many short-term or fewer long-term cultivation experiments. This is important for designing optimal cultivation strategies and to illuminate aspects of microbial growth behavior. The long-term cultivation experiments described here provide a unique opportunity to look at the rate of microbial discovery in relation to the length of incubation.
MATERIALS AND METHODS
Environmental microorganisms: soil microorganisms. (i) Sample collection and preparation.
Five terrestrial soils were collected at various times during 2009 and 2010. The soils represented both forest and open-field environments across the United States. Samples of ∼10 g were pulverized with mortar and pestle and air dried at room temperature, followed by heating for 15 min at 55°C to enrich for spore-forming species (14). Cell suspensions were generated by vortexing 1 g of prepared soil in a total volume of 10 ml with sterile dionized water for 30 min.
(ii) Growth conditions.
Ten-fold serial dilutions were made and plated at various densities in 2% SMS agar (0.125 g/liter casein [MP Biochemicals, San Diego, CA], 0.1 g/liter starch from potato [Sigma, St. Louis, MO], 1 g/liter Casamino Acids [Difco, Franklin Lakes, NJ], 20 g/liter Bacto agar [Difco]) in 96-well microtiter plates. Plates were sealed in Ziploc bags (1.75 mil) and incubated in a humidified chamber at room temperature. Growth was scored under a dissecting microscope (6.5× to 50× magnification range; Zeiss Stemi 2000) weekly for the first month and monthly thereafter for 3 more months. After incubation, 96-well plates containing between 1 and 3 colonies per well were chosen for microbial isolation and identification.
(iii) DNA analysis.
From these plates, all visible colonies were subsampled for subculturing, and 407 colonies were isolated into pure culture as determined by visual observation using a dissecting scope and 16S rRNA gene sequencing. For molecular identification, chromosomal DNA was isolated from approximately 106 cells after 5 min of vigorous agitation in the presence of 50 mg of glass beads (106 nm or smaller) and 100 μl of H2O in a 0.5-ml Eppendorf tube. The PCR-aided amplification of the 16S rRNA gene was carried out using chromosomal DNA, GoTaq Green master mix (Promega, Madison, WI), and universal primers 27F and 782R (Table 1) (1). PCR thermocycler parameters included 30 cycles at 95°C for 30 s, 45°C for 30 s, and 72°C for 105 s. The amplified DNA fragment (∼720 bp) was cleaned up and sequenced at Macrogen (Rockville, MD) using primer 782R and compared by BLAST alignment to the nucleotide collection in GenBank. Sequences were edited using Chromas Lite 2.01 (Technelysium Pty. Ltd., Brisbane, Australia) (A. Griekspoor and T. Groothuis) and clustered into operational taxonomic units (OTUs) based on 99, 97, and 95% sequence similarity cutoff values. This was achieved by first making all possible pairwise sequence alignments using ClustalW at default settings and calculating percent sequence similarities, followed by the clustering of the sequences into OTUs employing the mean unweighted-pair group method and using average linkages as implemented in the OC clustering program (http://www.compbio.dundee.ac.uk/Software/OC/oc.html). From each OTU, the sequence least different from the other members of the cluster was compared to the NCBI database using the BLAST search function. The top culturable and taxonomically identified hits were used to establish the identity of the OTUs. For soil samples enriched for spore formers, OTUs combining rRNA gene sequences sharing ≥99% identity were considered to belong to the same species, as is commonly accepted for actinobacteria (24, 25).
Table 1.
Sequences of primers used for genetic analysis
| Primer name | Sequence |
|---|---|
| 27F | 5′-AGA GTT TGA TCC TGG CTC AG-3′ |
| 782R | 5′-GAT TAG ATA CCC TGG TAG-3′ |
| 1492R | 5′-GGT TAC CTT GTT ACG ACT T-3′ |
| 907R | 5′-CCG TCA ATT CCT TTA AGT TT-3′ |
| 357F | 5′-CCTACG CGA GGC AGC AG-3′ |
Environmental microorganisms: marine microorganisms. (i) Sample collection and preparation.
Marine cells were obtained from a sample of intertidal sand collected on 31 October 2007 from Massachusetts Bay along the U.S. northeast seashore. The sample was vortexed for 5 cycles of 15 min each, and dislodged cells in the supernatant were counted by epifluorescence (Zeiss Axioskop 50 compound microscope at 1,000× magnification).
(ii) Growth conditions.
Counted cells were mixed with 0.1× lysogeny broth (LB) in natural seawater to a final density of 40 cells/ml. A 12-channel micropipetter was used to distribute aliquots of 50 μl containing single cells (on average) into individual wells of 31 384-well microtiter plates (designated the single-cell format), each containing a row of wells with sterile medium to control for contamination. The plates were sealed with Parafilm, separated into three groups to serve as replicates, placed into humidifying chambers, and incubated at room temperature in the dark for 18 months. Plates were scored for growth under a dissecting scope (Zeiss Stemi 2000 at 6.5× magnification) at monthly intervals. New growth was removed after 5, 6, 12, and 18 months of incubation. No contamination was observed in any of the control wells.
(iii) Genetic analysis.
Microbial biomass was subcultured in the same medium for identification via 16S rRNA gene sequencing. Universal bacterial primers 27F and 1492R were used to perform PCR-aided amplification. In some cases, seminested PCR-aided amplification was performed using 27F and 1492R, followed by using 27F and 907R or 357F and 907R (Table 1). Amplicon sequencing was performed commercially, and the resulting sequences were imported into the ARB database (16). Marine species were identified as the smallest clusters of sequences on a phylogenetic tree produced within the ARB database. These were comprised of sequences sharing ≥97% sequence similarity cutoff values and were considered species as accepted for most bacteria (26). Representative sequences from each cluster were entered into the GenBank database using the BLAST search tool, and the closest cultured and taxonomically identified relative was recorded.
(iv) Subculture.
Out of the total of 496 wells showing growth, 406 were successfully subcultured, and the time interval required for regrowth was noted. Twenty randomly selected isolates were also subcultured in a single-cell format. Cells grown in the original isolation experiment were enumerated as described above, diluted in fresh medium (0.1× LB in seawater), and administered to one 96-well plate per isolate in the single-cell format. A row of wells in each plate was inoculated with sterile medium to control for contamination. The plates were observed and the growth scored for at least 1 month.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of soil microorganisms have been deposited in the NCBI database under accession numbers JQ419503 to JQ419717. The 16S rRNA gene sequences of marine microorganisms have been deposited in the NCBI database under accession numbers HQ446854 to HQ446856, GQ262723, and JQ660963 to JQ661255.
RESULTS
Recovery of environmental cells.
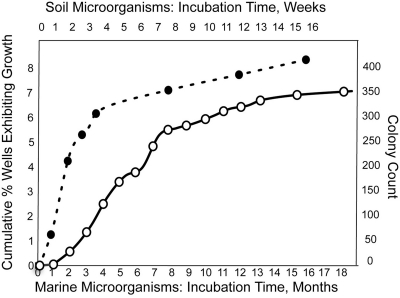
In the growth experiment involving marine sediment microorganisms, a cell suspension containing 11,000 cells was placed into individual wells of microtiter plates such that each well received, on average, a single cell. Most cells became isolated from each other and grew independently from their natural neighbors. The accumulation of new cases of visible turbidity is shown in Fig. 1. During the first month of incubation, we observed 0.01% of wells forming visible growth, indicating that very few inoculated cells grew within the first few weeks. The plates were observed during a time period of 18 months, in which the proportion of wells exhibiting growth reached 7%, an increase of more than 700-fold after the first month.
Fig 1.
Accumulation of growth events over time in long-term cultivation experiments using microorganisms from marine (○) and soil (●) samples.
In a series of growth experiments involving soil microorganisms, we first enriched the samples for spore formers by drying and preheating the samples. In these experiments, the total number of plated cells was not determined, and we cannot calculate the percentage of recovery, but the overall dynamic of the cumulative colony number showed a general trend similar to that in marine samples (Fig. 1).
Appearance of rare and novel species.
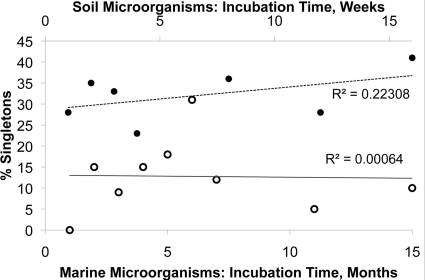
We examined the dynamics of the appearance of rare species, defined as species represented by the fewest number of isolates in the culture collection obtained here. The rarest of all are singletons, or species met only once during the incubation. Figure 2 shows the proportion of singletons among the total number of species isolated at each particular time point, and this proportion did not vary over time.
Fig 2.
Proportion of singletons (species met once study-wide) from marine microorganisms (○) and soil microorganisms (●) at each time point.
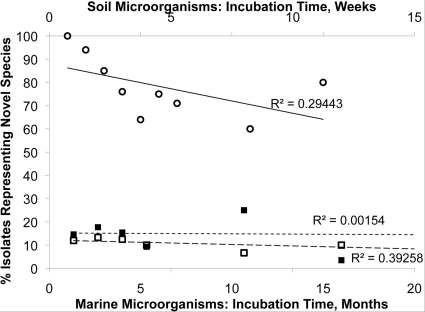
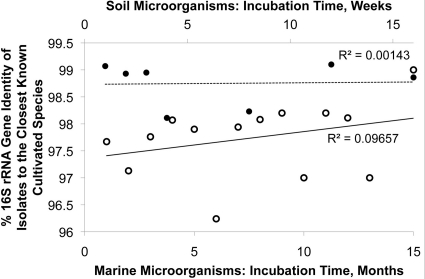
We also examined the dynamics of the appearance of novel species, defined as OTUs sharing >97 and >99% 16S rRNA gene identity for marine and soil bacteria, respectively (24–26). For each time point, we plotted the number of isolates representing novel species as a fraction of all isolates obtained at this time point (Fig. 3). This figure shows that the probability of discovering a novel species among the isolates seems to be the same at any given time point in both marine sediments and soils.
Fig 3.
Proportion of novel isolates among all isolates, separately for each time point for marine microorganisms (○), non-spore-forming soil microorganisms (■), and spore-forming soil microorganisms (□). In all cases, the isolate was considered novel if its 16S rRNA gene sequence shared less than 99% identity with previously reported isolates. Note the low value of R2, indicating that the percentage of novel isolates does not depend on the time required to form a visible biomass.
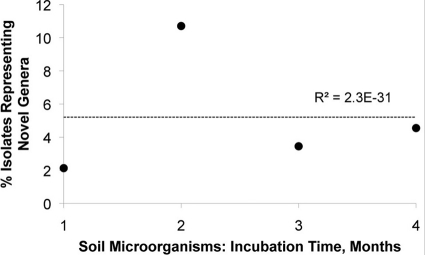
We also checked, in two different ways, if the degree of phylogenetic novelty of new isolates varied depending on the time of incubation. First, we considered OTUs sharing <95% 16S rRNA gene sequence identity as a proxy for microbial genera and plotted the number of isolates representing novel genera as a proportion of all isolates obtained at the same time (Fig. 4). As was the case with the species-level taxa, we did not observe any particular connection between the proportion of isolates representing novel genera and the time it took to grow them. Second, we plotted the average phylogenetic novelty of all microbial isolates obtained at a given time point against the time at which their growth was first observed. Should isolates detected at later stages of incubation be more novel that those that appeared first, the regression would have a positive slope. This is not what we observed in either marine or soil samples (Fig. 5). These tendencies were characteristic of both spore-forming and non-spore-forming microorganisms (Fig. 6).
Fig 4.
Proportion of soil isolates (●) representing novel genera among all soil isolates. Data on weekly observations were pooled by month to minimize scatter. Scatter was due to the relatively low number of substantially novel isolates observed on a weekly basis. Regarding the low R2 value, see the legend to Fig. 3.
Fig 5.
Average phylogenetic novelty of isolates depending on the time of the appearance of marine microorganisms (○) and soil microorganisms (●). The low value of R2 indicates that the novelty of an isolate does not depend on the time of its isolation.
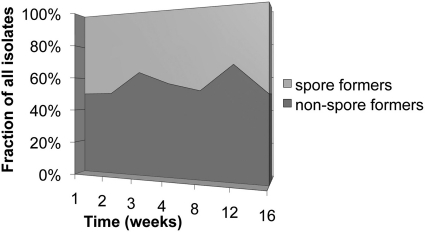
Fig 6.
Proportion of sporulating and nonsporulating soil microorganisms isolated in long-term cultivation experiments.
DISCUSSION
It was established more than a century ago that most environmental cells do not form colonies on standard media (30). This uncultivated microbial majority offers exciting opportunities for microbial discovery, and the nature of uncultivated species has been extensively discussed (6, 7, 21). One idea is that some uncultivated species are simply so slow growing that standard cultivation approaches select against such species, and there are a number of reports pointing to the novelty of slow-growing isolates (5, 8, 9, 11, 12, 20, 22, 23). Here, we check if long-term cultivation is indeed a promising tool for microbial discovery. In the experiment involving marine sediment cells, we saw the initial recovery at the low end of the range of values typical of marine sediments (3, 13, 15, 23). Unlike conventional growth experiments that employ mixes of microbial species cocultivated in, e.g., petri dishes, cells in our experiment could grow unimpeded by those showing growth early (31). This provided an opportunity to conduct a uniquely long incubation that lasted for 1.5 years. At the end of the experiment, the proportion of wells showing growth reached 7%, indicating a 700-fold increase in cultivability during the incubation period. Soil samples showed a similar trend. One interesting observation is that while the accumulation of new growth events slowed down over time, it did not reach zero even after very long incubations (Fig. 1). Rather, it exhibited signs that the rate of the increase stabilized at a certain measurable level. We expected that the likeliest candidates for being slow-growing, rare, and possibly novel would be the organisms that did not appear until after the initial spike of new colonies was over (18). These expectations were not met. Most microorganisms isolated late in the incubation process were not inherently slow growing (S. Buerger et al., unpublished), and they were neither particularly rare nor especially novel. In other words, while the culture collection obtained in the first half of incubation was larger than in the second (Fig. 1), the proportion of novel species was the same in each and in both (marine and soil) samples (Fig. 3).
We conclude that neither rare nor more abundant species isolated at later stages of long-term incubations are significantly different in their novelty from microorganisms isolated in short-term experiments. This may sound somewhat counterintuitive, but in fact it is consistent with the idea of the random awakening of dormant cells, if indeed such cells dominate in environmental samples. If the scout model has validity, one should expect most species, both known and novel, to appear randomly during cultivation, and our data strongly favor this inference.
One rather unexpected finding is that spore formers and non-spore formers exhibited an overall similarity in the temporal pattern of their appearance during cultivation. Spore-forming isolates that emerged late are no more and no less novel than those detected early. Their proportion among all isolates obtained at a given time point stayed essentially constant throughout the incubation, similarly to non-spore formers. In both groups and in species that were detected multiple times, the appearance of isolates appeared evenly spread out over long periods of time, so that the notion of a species-specific lag phase does not seem to be applicable (S. Buerger et al., unpublished). This suggests that the growth medium did not contain germination factors; in their absence, the spore revival seems to be stochastic. Note that a low, apparently random level of background germination was reported for Bacillus subtilis (17). It is therefore tempting to hypothesize that spore formers and non-spore formers alike have specialized cell types, spores and dormant cells, respectively, and these cell types share a certain similarity in both how deep their dormancy is and how they exit this state into activity. There may be less of a gap between these cell types than is suggested by their clear morphological differences.
In conclusion, the results of our cultivation experiments strongly suggest that, given the same growth medium, the success in discovering novel species depends on the amount of cultivation effort rather than the length of incubation. Environmental microorganisms in our study appear to behave according to the scout model: being mostly in a nongrowing state at the time of inoculation, they wake into activity and start forming colonies in a random fashion and over long periods of time. Chance then plays a very significant role in capturing novel species. A strain may be perfectly cultivable on the given medium, but if it is rare in most environments, its scarce scouts may elude cultivation for years, making an impression of the strain's uncultivability. In the end, the increased effort may bring such a strain into culture as if by accident, even without the manipulation of the media formulation. This may be applicable to both marine and soil species and both spore formers and non-spore formers. Cultivation experiences are consistent with this view. For example, representatives of the phylum Verrucomicrobia had eluded researchers for decades, yet their eventual cultivation was achieved using conventional, time-honored techniques (2, 4, 10, 15, 23, 29).
Note that these conclusions do not bear on the merits of single-cell cultivation. By preventing overgrowth by a few species that quickly colonize a petri dish, the single-cell approach allows for the recovery of many more colonies from the same inoculum, and so does cultivation by dilution to extinction (2, 4). It also allows for longer incubation times, and it is logistically easier to keep a few plates longer than establish a large number of short-living ones. Even if such a method does not bring qualitatively richer diversity into culture, it minimizes the overall labor and supplies required for the isolation of species that are, in principle, cultivable on the given medium.
Footnotes
Published ahead of print 24 February 2012
This article is contribution 282 of the Marine Science Center, Northeastern University, Nahant, Massachusetts, USA.
REFERENCES
- 1. Baker GC, Smith JJ, Cowan DA. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541–555 [DOI] [PubMed] [Google Scholar]
- 2. Button DK, Schut F, Quang P, Martin R, Robertson BR. 1993. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl. Environ. Microbiol. 59:881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cifuentes A, et al. 2000. Prokaryotic diversity in Zostera noltii-colonized marine sediments. Appl. Environ. Microbiol. 66:1715–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Connon SA, Giovannoni SJ. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davis KER, Joseph SJ, Janssen PH. 2005. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71:826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Epstein SS. 2009. Microbial awakenings. Nature 457:1083. [DOI] [PubMed] [Google Scholar]
- 7. Handelsman J. 2004. Metagenomics: application of genomics to non-cultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hattori T. 1976. Plate count of bacteria in soil on a diluted broth as a culture medium. Rep. Inst. Agr. Res. Tohoku Univ. 27:23–30 [Google Scholar]
- 9. Hattori T, et al. 1997. Advances in soil microbial ecology and the biodiversity. Antonie Van Leeuwenhoek 72:21–28 [DOI] [PubMed] [Google Scholar]
- 10. Janssen PH, Schuhmann A, Mörschel E, Rainey FA. 1997. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl. Environ. Microbiol. 63:1382–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Janssen PH, Yates PS, Grinto BE, Taylor PM, Sait M. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Joseph SJ, Hugenholtz P, Sangwan P, Osborne CA, Janssen PH. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210–7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Keller M, Zengler K. 2004. Tapping into microbial diversity. Nat. Rev. Microbiol. 2:141–150 [DOI] [PubMed] [Google Scholar]
- 14. Labeda DP, Shearer MC. 1990. Isolation of actinomycetes for biotechnological applications, p 1–19 In Labeda DP. (ed), Isolation of biotechnological organisms from nature. McGraw-Hill Publishing Co., New York, NY [Google Scholar]
- 15. Llobet-Brossa E, Rossello-Mora R, Amann R. 1998. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64:2691–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ludwig W, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paidhungat M, Setlow P. 2000. Role of ger proteins in nutrient and non-nutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182:2513–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pedrós-Alió C. 2006. Marine microbial diversity: can it be determined? Trends Microbiol. 14:257–263 [DOI] [PubMed] [Google Scholar]
- 19. Poindexter JS. 1981. Oligotrophy: feast and famine existence. Adv. Microb. Ecol. 5:63–89 [Google Scholar]
- 20. Rappe MS, Connon SA, Vergin KL, Giovannoni SJ. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630–633 [DOI] [PubMed] [Google Scholar]
- 21. Rappe MS, Giovannoni SJ. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369–394 [DOI] [PubMed] [Google Scholar]
- 22. Sait M, Hugenholtz P, Janssen PH. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654–666 [DOI] [PubMed] [Google Scholar]
- 23. Sangwan P, Kovac S, Davis KER, Sait M, Janssen PH. 2005. Detection and cultivation of soil verrucomicrobia. Appl. Environ. Microbiol. 71:8402–8410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stach JE, Bull AT. 2005. Estimating and comparing the diversity of marine actinobacteria. Antonie Van Leeuwenhoek 87:3–9 [DOI] [PubMed] [Google Scholar]
- 25. Stach JE, et al. 2003. Statistical approaches for estimating actinobacterial diversity in marine sediments. Appl. Environ. Microbiol. 69:6189–6200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stackebrandt E, Goebel BM. 1994. Taxonomic note: a place for DNA:DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteria. Int. J. Syst. Bacteriol. 44:846–849 [Google Scholar]
- 27. Staley JT, Konopka A. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321–346 [DOI] [PubMed] [Google Scholar]
- 28. Stevenson LH. 1978. A case for bacterial dormancy in aquatic systems. Microb. Ecol. 4:127–133 [DOI] [PubMed] [Google Scholar]
- 29. Ward N, Rainey FA, Stackebarndt E, Schlesner H. 1995. Unraveling the extent of diversity within the order Planctomycetales. Appl. Environ. Microbiol. 61:2270–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Winterburg H. 1898. Zur Methodik der Bakterienzahlung. Z. Hyg. 29:75–93 [Google Scholar]
- 31. Zengler K. 2009. Central role of the cell in microbial ecology. Microbiol. Mol. Biol. Rev. 73:712–729 [DOI] [PMC free article] [PubMed] [Google Scholar]