Abstract
The CD8+ T cell diaspora has been analyzed after secondary challenge with an influenza A virus that replicates only in the respiratory tract. Numbers of DbNP366- and DbPA224-specific CD8+ T cells were measured by tetramer staining at the end of the recall response, then followed sequentially in the lung, lymph nodes, spleen, blood, and other organs. The extent of clonal expansion did not reflect the sizes of the preexisting memory T cell pools. Although the high-frequency CD8+ tetramer+ populations in the pneumonic lung and mediastinal lymph nodes fell rapidly from peak values, the “whole mouse” virus-specific CD8+ T cell counts decreased only 2-fold over the 4 weeks after infection, then subsided at a fairly steady rate to reach a plateau at about 2 months. The largest numbers were found throughout in the spleen, then the bone marrow. The CD8+DbNP366+ and CD8+DbPA224+ sets remained significantly enlarged for at least 4 months, declining at equivalent rates while retaining the nucleoprotein > acid polymerase immunodominance hierarchy characteristic of the earlier antigen-driven phase. Lowest levels of the CD69 “activation marker” were detected consistently on virus-specific CD8+ T cells in the blood, then the spleen. Those in the bone marrow and liver were intermediate, and CD69hi T cells were very prominent in the regional lymph nodes and the nasal-associated lymphoid tissue. Any population of “resting” CD8+ memory T cells is thus phenotypically heterogeneous, widely dispersed, and subject to broad homeostatic and local environmental effects irrespective of epitope specificity or magnitude.
The vertebrate immune system is dynamic, mobile, and anatomically dispersed. Mammalian immune responses are thought to develop predominantly in the circumscribed microenvironments of the secondary lymphoid tissue, the lymph nodes (LNs), spleen, and foci embedded in mucosal sites such as the Peyer's patches (PP). However, the cellular elements of immunity, particularly the CD4+ and CD8+ T cells, are known to move via blood and lymph through a spectrum of other organs that are not normally considered to be part of the immune system. Measuring the extent of this lymphocyte diaspora has only recently become possible with the development of tetrameric complexes of MHC class I glycoprotein + peptide (tetramers) for the direct, flow cytometric identification of antigen-specific CD8+ T cells (1−5). The tetramers allow us to quantitate both the CD8+ T cell response to pathogens and the subsequent return to homeostasis after the elimination of the invading organism.
Analysis of the diaspora effect in viral immunity should optimally distinguish between the antigen-driven dispersion of CD8+ effectors to sites of virus replication and distribution profiles reflecting the normal physiology of lymphocyte recirculation. The latter will be more characteristic of a localized infection. The influenza A viruses replicate only in respiratory epithelium, although they also can undergo a defective growth cycle in other cell types, such as dendritic cells and macrophages (6−9). The net consequence is that the site of virus-induced pathology after intranasal (i.n.) challenge is restricted to the infected lung, although antigen-presenting dendritic cells are thought to travel via the afferent lymph and blood to the regional, mediastinal LNs (MLNs) and the spleen (9, 10). However, there is no detectable viremic phase and little, if any, evidence of productive infection in these organs (8, 9, 11).
The present study quantitates the tissue diaspora for the recall CD8+ T cell response to two prominent H-2Db-restricted peptides (4, 12, 13) derived from the influenza A virus nucleoprotein (NP366–374) and acid polymerase (PA224–233). The use of secondary rather than primary challenge reflects both the need to generate large lymphocyte populations for measurement of the diaspora and the intent to explore effects related to the immunodominance hierarchy (CD8+DbNP366+ > CD8+DbPA224+) that only becomes apparent with boosting (4, 13). Diversity within these two sets of virus-specific CD8+ T cells was further assessed by staining for expression of the CD69 “early activation” marker (14, 15). The experiments provide insights into the nature of T cell expansion, homeostasis, and loss subsequent to a localized, transient infectious process.
Materials and Methods
Mice and Infection.
The 6-week-old female C57BL/6J (B6) mice were primed by i.p. inoculation with 106.9 EID50 of the A/PR8/34 (PR8, H1N1) influenza A virus. The PR8 virus replicates little, if at all, following this route of exposure, and causes no detectable pneumonia. These immune mice then were challenged i.n. 6 weeks later with 106.8 EID50 (in 30 μl) of the serologically distinct HKx31 (H3N2) virus that shares the NP and PA peptides (4, 13, 16) of the PR8 virus.
Tissue Sampling and Treatment.
At time of sampling, the mice were anesthetized, the axillary artery was cut, and blood was collected into heparin (1,000 units/ml, Elkins-Sinn, Cherry Hill, NJ). Each mouse then was perfused via the left, then the right ventricle with 40–50 ml of heparinized PBS. Inflammatory cells were recovered by bronchoalveolar lavage (BAL), and the lungs, MLNs, liver, and spleen were removed. Pools then were made from the superficial LNs (SLNs) (cervical, axillary, brachial, popliteal) and internal LNs (ILNs) (mesenteric, inguinal, periaortic) of each mouse. Femurs and tibias were removed, and the bone marrow (BM) was flushed out with 5–10 ml of RP-10 medium (RPMI1640 with 10% FCS). Lastly, the nasal-associated lymphoid tissue (NALT) and PP were excised and, because of the small amounts of tissue, were pooled for groups of five mice. The NALT was obtained by peeling the soft palate to expose two small, elongated strips of tissue that, in situ, sit on either side of the nasal septum. These strips were removed and dispersed with needles. The PPs were excised from well-rinsed small intestine.
Peripheral blood lymphocytes (PBLs) were enriched on a 10-ml gradient of 1-STEP (Accurate Chemicals) by centrifugation for 30 min at 2,000 rpm, then collected from the interface, washed once with RP-10, lysed 2× for 1.5 min with red cell lysis buffer (Sigma) and suspended in RP-10. The BAL cells were absorbed on plastic for 60 min at 37°C to remove macrophages. Single cell suspensions were made from the MLN, SLNs, ILNs, and spleen, by grinding gently between ground glass slides or in ground glass tissue homogenizers. Lungs and livers were minced and forced through a fine stainless-steel mesh, filtered, and digested with collagenase (17). The PP also were treated with collagenase. With the exception of the BAL, NALT, PP (due to their small numbers), and BM, CD8+ T cells were enriched by using mAbs to CD4 (GK1.5) and MHC class II glycoproteins (TIB120), followed by magnetic depletion with sheep anti-mouse Ig and sheep anti-rat Ig Dynabeads (Dynal).
Flow Cytometry.
The phycoerythrin-labeled DbNP366 and DbPA224 tetramers were prepared as described (4, 13). Cell surface Fc receptors were blocked with anti-mouse CD16/CD32 Fc-RIII/II (PharMingen). The T cells were first stained with tetramer for 1 h at room temperature, followed by anti-CD8α-tricolor (Caltag, South San Francisco, CA) and anti-CD69-FITC (PharMingen), for 30 min on ice. Samples were resuspended in FACS buffer (PBS with 1% BSA and 0.1% sodium azide) and analyzed on a FACScan by using cell quest software (Becton Dickinson). Where possible, 20,000 CD8+ lymphocytes were collected. Further analysis involved gating on the tetramer-positive populations within the CD8+ gate and the CD69hi and CD69lo populations within the tetramer+ population. Data are presented as mean ± SE.
Statistical Analysis.
The experiments used 28 mice, five from days (d) 0, 10, 20, 31, 52, and three from d125. The analysis focused on CD8+ T cells from the BAL, MLN, lung, blood, BM, liver, and spleen. The SLNs and ILNs that contained LNs combined from a variety of locations were excluded, as were the pooled (because of small numbers) PP, NALT, and d0 MLN samples. Data for a variety of organs from mice at different time points were compared by using ANOVA techniques implemented in sas software (18). These immune response data sets were based on T cell frequency measurements and included the total and CD69hi CD8+DbNP366+ and CD8+DbPA224+ populations recovered from each sampling site, the cell numbers calculated from these frequencies, and the lymphocyte counts per sample. Initially, two-way ANOVA was used to determine whether (on average) the differences in the immune responses for individual organs (or groups of organs) changed significantly with time. When this was the case, one-way ANOVA was then used to analyze (on average) how organ and time point (d) affect the characteristics of the CD8+DbNP366+, CD8+DbPA224+, and CD69hi sets. Least-squares means and corresponding SEMs then were estimated for each organ and time point. Differences that were significant after Dunnet adjustment for multiple comparisons are identified in the text.
Results
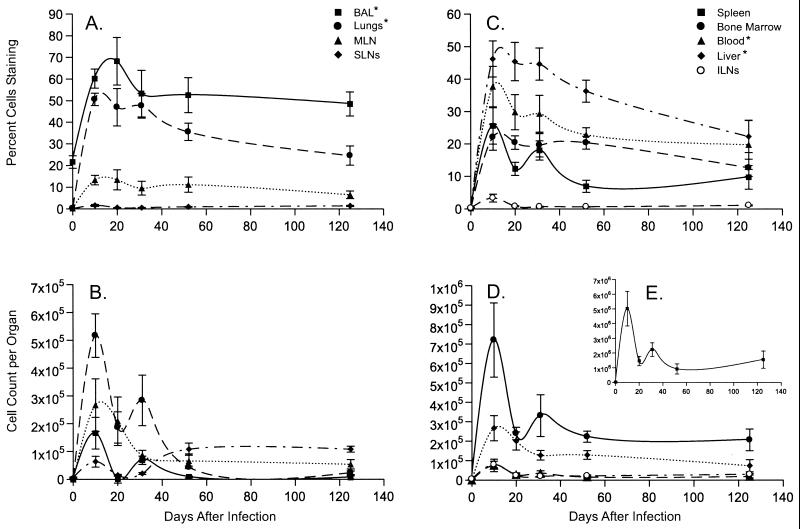
Earlier studies established that the massive proliferation of the CD8+DbNP366+ set in the MLN and spleen is over by d8 after secondary i.n. infection of PR8-primed mice with the HKx31 virus, coincident with the elimination of the virus from the lung (7). The present experiments used the same HKx31 challenge in mice given the PR8 virus i.p. 42 days previously (day 0). The high frequency of both the CD8+DbNP366+ and CD8+DbPA224+ populations (Figs. 1A and 2A) in the minimal d0 BAL set reflects that small numbers of memory T cells localize to this site, irrespective of the route of priming (19, 20).
Figure 1.
Expansion and maintenance of the immunodominant CD8+DbNP366+ T cells. It is important to note that the scales on the y axis differ for A–E. The results for lymphocyte populations recovered directly from the site of virus-induced pathology (BAL and lung) are grouped with those for the draining LNs (MLN and cervical LN, the latter included in the SLNs) in A and B, and the findings for the other sites sampled are shown in B, D, and E. Cell numbers (B, D, and E) were calculated from the frequencies (A and C) and the total CD8+ T cell counts (data not shown) for the particular organ. Epitope-specific CD8+ T cell counts for the spleen were generally 10-fold higher than for any other tissue. * denotes that the frequencies (A and C) were significantly higher in these samples than in the MLN throughout the course of the experiment.
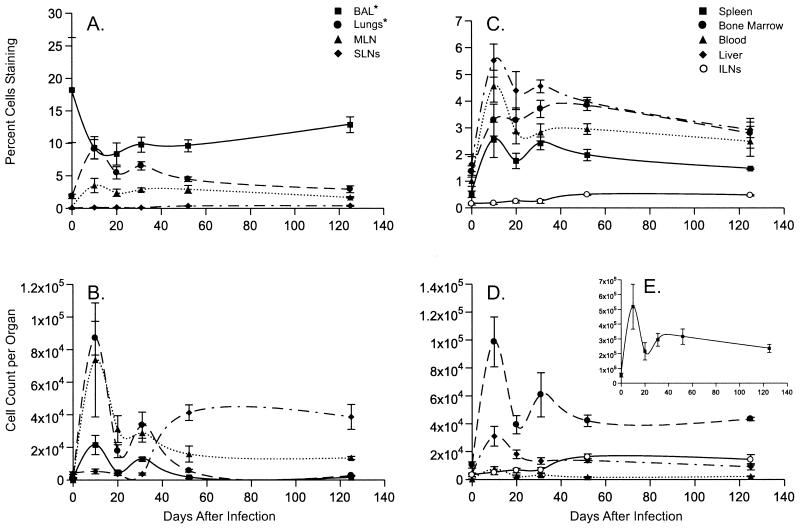
Figure 2.
Quantitative analysis of the CD8+DbPA224+ T cell response and memory. The data are presented exactly as described in the legend to Fig. 1. The y axis scales for A–E differ and also differ from those in the comparable panels in Fig. 1.
Divergence Between the Recall Response and Preexisting Memory.
Significantly more CD8+ DbPA224+ than CD8+DbNP366+ T cells were found in the BM (P < 0.006), liver (P < 0.0002), lungs (P < 0.0004), and spleen (P < 0.02) immediately before the HKx31 challenge of the PR8-primed mice (d0). However, both the total counts (P < 0.04) and the frequencies (P < 0.0001) were greater for the CD8+DbNP366+ than the CD8+DbPA224+ set throughout the experiment (Figs. 1 and 2), confirming the immunodominance of DbNP366 in the secondary response (4, 7, 13, 21). Comparing the numbers of available memory T cells does not, therefore, explain why the CD8+DbNP366+ population is so much more prominent after secondary challenge (Figs. 1 and 2, Tables 1 and 2).
Table 1.
Response in the BAL, MLN, and spleen
| Epitope and day | BAL, ×104 | MLN, ×104 | Spleen, ×106 |
|---|---|---|---|
| DbNP366+ | |||
| 9 | 21.9 ± 5.4 | 11.7 ± 5.0 | 5.0 ± 1.4 |
| 21 | 4.0 ± 0.5 | 23.0 ± 6.3 | 5.0 ± 1.1 |
| 31 | 5.2 ± 2.0 | 13.0 ± 5.1 | 4.0 ± 0.5 |
| DbPA224+ | |||
| 9 | 5.4 ± 1.8 | 5.1 ± 1.4 | 1.2 ± 0.4 |
| 21 | 0.4 ± 0.02 | 10.3 ± 1.7 | 1.1 ± 0.1 |
| 31 | 1.1 ± 0.5 | 4.1 ± 1.5 | 0.7 ± 0.1 |
Table 2.
Characteristics of NALT and PP populations
| Epitope and day | NALT
|
PP
|
||
|---|---|---|---|---|
| % Tetramer+ | % CD69Hi | % Tetramer+ | %CD69Hi | |
| DbNP366+ | ||||
| 0 | 4.1 | — | 4.0 | — |
| 10 | 48.5 | 69.5 | 4.3 | 32.3 |
| 20 | 51.0 | 85.0 | 3.0 | 25.5 |
| 31 | 59.4 | 75.0 | 4.0 | 35.4 |
| 52 | 31.0 | 74.5 | 8.2 | 96.5 |
| DbPA224+ | ||||
| 0 | 1.4 | — | 1.7 | — |
| 10 | 2.4 | 79.2 | 2.2 | 38.1 |
| 20 | 2.5 | 94.0 | 2.8 | 40.0 |
| 31 | 2.8 | 90.6 | 1.3 | 47.4 |
| 52 | 4.6 | 98.9 | 3.1 | 74.6 |
The data are derived from NALT and PP populations pooled from groups of five mice. The total numbers of epitope-specific CD8+ T cells recovered from both sites combined did not exceed 3 × 103 at any time point.
Tissue Distribution and the Kinetics of Attrition.
The largest numbers of tetramer+ CD8+ T cells were detected in the spleen at every time point (Figs. 1 and 2 B, D, and E). The next highest values were recorded from the BM, followed by the lung, then the liver or MLN, with the patterns generally, being consistent for both epitopes (Figs. 1 and 2 B and D). Maximum counts for both tetramer+ populations were generally found on d10 (Figs. 1 and 2 B and D) as were peak frequencies for the CD8+DbNP366 (P < 0.002 for d10 vs. d0; Fig. 1 A and C) and CD8+DbPA224+ (P < 0.02, Fig. 2 A and C) sets. The exception was the % values for the CD8+DbPA224+ T cells in the BAL (Fig. 2A), although the d10 BAL counts were greatly increased (Fig. 2B).
The estimates of cell number for both the CD8+DbNP366+ and CD8+DbPA224+ populations looked to be smaller on d20 than d31 (Figs. 1 and 2 B, D, and E). However, a limited, repeat study did not show this pattern for the BAL, MLN, and spleen (Table 1), although cell counts for the BAL fell rapidly between d10 and d20 in both experiments. Comparing the cumulated data from all seven sites for d10 and d31 indicated that the number of CD8+DbNP366+ and CD8+DbPA224+ T cells had fallen by a factor of 2.1 and 1.9 times, respectively. Despite the difference in magnitude (Figs. 1 and 2), the rates of cell loss for these two CD8+ T cell populations are similar for the first month after secondary challenge.
The general profile was then of steady attrition until 52 days after infection. The d52 results showed increased prevalence (Figs. 1 and 2 C and D) of both epitope-specific CD8+ sets in the SLNs and ILNs, although not in the MLNs, which peak earlier and are clearly involved in the acute response. The size of the virus-specific CD8+ T cell populations then seemed to stabilize through d125 (Figs. 1 and 2 B, D, and E), when the numbers present were still significantly higher than those detected before secondary challenge (P < 0.02 and P < 0.04 for the CD8+DbNP366+ and CD8+DbPA224+ sets, respectively).
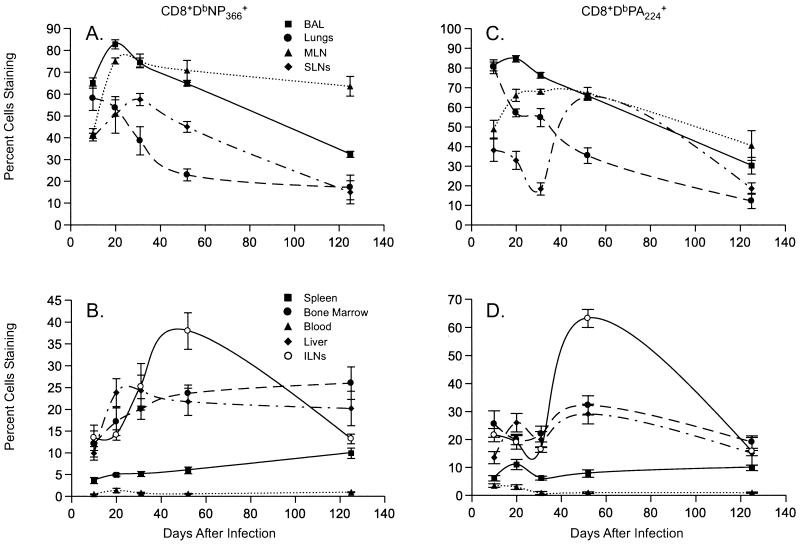
The Blood and Site-Related CD69 Expression.
The frequencies of the CD8+DbNP366+ and CD8+DbPA224+ sets in the PBL remained in excess of 20% and 2%, respectively (Figs. 1C and 2C) throughout the 4-month sampling period. Although the mice were perfused, it is still possible that some of the organ profiles (Figs. 1 and 2, Table 2) could be influenced by the continued presence of contaminating CD8+ T cells in the vasculature. However, virus-specific CD69hi T cells were invariably at minimal frequency in the PBL (Fig. 3 B and D), whereas the majority of the CD8+DbNP366+ and CD8+DbPA224+ T cells from “responding sites” (BAL, lung, and MLN) were CD69hi at d10 and d20 after the i.n. challenge (Fig. 3 A and C). Thereafter, tetramer+CD69hi T cells were at substantially greater frequency in the BAL and MLN than in the lung. The CD69hi CD8+DbNP366+ and CD8+DbPA224+ sets were also much more prevalent in the BM and liver than in the PBL at all time points (Fig. 3 B and D). The patterns for the spleen were most like those for the PBL, although the spleen and peripheral LN profiles tended to converge with time (Fig. 3 B and D). The CD69 staining profiles thus illustrate very clearly that memory CD8+ T cells are heterogeneous, and that this diversity is related to the anatomical environment rather than to epitope specificity.
Figure 3.
Distribution profiles of CD69hi virus-specific CD8+ T cells. The frequency analysis for the CD69hiCD8+DbNP366+ populations is presented in A and B and for the CD69hiCD8+DbPA224+ sets in C and D. As described in the legend to Fig. 1, the results for sites associated directly with the pneumonic lung are shown in A and C, and the findings for the blood, BM, and other solid tissues are in B and D. The scales on the y axis differ for A–D.
Mucosal-Associated Lymphoid Tissue.
Frequency analysis of the NALT indicated that there was a strong, secondary CD8+DbNP366+ response but, for the small numbers of cells that were analyzed, we did not detect any increase in the CD8+DbPA224+ set (Table 2). No evidence was found for the antigen-driven expansion of either population in the PP (Table 2), indicating that swallowed virus does not survive passage through the stomach to the gut. The CD8+DbNP366+ and CD8+DbPA224+ T cells recovered from the PP and the NALT were predominantly CD69hi at d52, an effect that was also seen for the ILNs and the SLNs but not for other solid tissues (Fig. 3 B and D). Otherwise, relatively more CD8+DbNP366+ than CD8+DbPA224+ T cells were CD69hi in both the NALT and the PP from d10 to d31. This effect thus seems to be unrelated to whether or not an immune response occurred at the particular site (Table 2).
Discussion
The present analysis provides insight into the nature of the return to homeostasis (22) after a secondary CD8+ T cell response to a localized pathogen. The effect of the local microenvironment on the activation status of CD8+ memory T cells is profound and persistent (23). The patterns for the immunodominant CD8+DbNP366+ and less prominent CD8+DbPA224+ populations are similar in almost every respect. Most experimental immunologists study lymphocytes derived from mouse spleen, a site shown here to contain at least 50% of the total virus-specific CD8+ memory T cells. Those who analyze humans are often limited to the PBL. The fact that the % CD8+ tetramer+ T cells was consistently higher in the blood than the spleen, whereas the CD69 staining profiles were most similar for blood and spleen, provides support for the validity of comparing results from mice and humans. The situation modeled here presumably occurs regularly in people primed naturally with one influenza A virus, then challenged a year or two later with a serologically different variant sharing epitopes from conserved, internal proteins.
The type II transmembrane protein CD69 generally is regarded as a marker of early activation (14, 15). High-level expression is rapidly induced by a variety of stimuli on a range of cells types, including T lymphocytes, macrophages, platelets, eosinophils, and B cells. The presence of CD69 on T cells usually is considered to reflect triggering via the T cell receptor, although cytokines also may have this effect (15, 24). Few of the CD8+ T cells in the blood are normally CD69hi, but the relative prevalence is increased during chronic HIV infection and allograft rejection (15, 25), situations where the antigen load is likely to be massive. The CD69lo characteristic of the PBL and spleen CD8+ tetramer+ populations analyzed in the present experiments is thus in accord with the general perception that “resting” CD8+ memory T cells do not express this glycoprotein.
The relatively small CD8+DbNP366+ and CD8+DbPA224+ T cell populations recovered from the NALT, the MLN, and the BAL of the respiratory tract remained predominantly CD69hi for more than 6 weeks after virus challenge. A similar observation has been made for CD8+ T cells isolated from the brains of mice injected intracerebrally with an influenza A virus (26). The reason for this during the early, antigen-driven phase of the response is obvious, but why should the pattern be maintained? Antigen persistence is difficult to exclude. However, no evidence for the continued presence of the influenza genome has been found from reisolation or PCR studies (7, 8) and antigen-presenting cells have not been recovered after the acute phase of the host response to the influenza A viruses or to other negative-strand RNA viruses (9, 10). Also, the massive CD8+DbNP366+ T cell proliferation characteristic of this secondary response ceases abruptly coincident with virus elimination from the lung and, in the long-term, CD8+DbNP366+ memory T cells cycle much more frequently in the spleen than in the MLN (7).
Is the persistent expression of CD69 characteristic of terminally differentiated, sessile CD8+ T cells that are destined for destruction? Although CD69hiCD8+tetramer+ T cells are at equivalent prevalence in the BAL and lung populations recovered on day 10 after i.n. challenge, they then decline much more rapidly in the set recovered by collagenase digestion of the lung. Perhaps lymphocytes that can be lavaged from the airways in the long term have been ejected from the former site of virus-induced pathology as part of a process of elimination and repair. However, apoptotic CD8+DbNP366+ effectors are not found at high frequency in the BAL during the resolution phase of influenza pneumonia (17), though the kinetic data suggests that many die and are removed by phagocytosis. Some CD69hiCD8+tetramer+ T cells may relocate to previously uninvolved LNs and other organs, but most of the high-frequency CD8+DbNP366+ and CD8+DbPA224+ sets in the PBL are CD69low. Any that exit into the circulation from the BAL and MLN must either down-regulate CD69 or be rapidly removed elsewhere. Others may be selectively eliminated in the liver (27), where relatively more (than in BAL or MLN) CD8+DbNP366+ T cells show evidence of progression to apoptosis (17).
The expression of CD69 on a subset of virus-specific CD8+ T cells in the BM and “remote” lymphoid tissue (ILNs, PP) may be a consequence of local interactions in the particular organ. The BM is not a site of immune response in normal mice infected with the HKx31 influenza A virus (11). Memory CD8+ T cells must enter the BM from the blood. Perhaps the molecular interactions that facilitate CD8+ T cell trafficking between the intravascular, solid tissue and lymph compartments induce the expression of CD69. A further possibility is that a proportion of the CD8+ memory T cells in the LNs and BM may be receiving some inductive signal. A primary characteristic of the BM, which also provides a long-term “home” for antibody-producing plasma cells (28), is the constitutive production of hemopoietic growth factors. “Bystander” activation as a consequence of type 1 IFN and IL-15 production (29, 30) during the course of other infectious processes also may induce CD69 expression. Such bystander effects would be expected to occur more in the organized lymphoid tissue. Although the relative prevalence of CD69hi CD8+DbNP366+ and CD8+DbPA224+ T cells is low in the spleen, the numbers tend to increase with time and more closely resemble the profiles in the LNs.
Although this may be the most comprehensive numerical data set ever assembled for a CD8+ T cell response, the analysis did not include the lamina propria of the gut (31) or the BM of the forelimbs and sternum. Both sites will contain substantial memory T cell populations, some of which may be highly activated (32). Thus, although the extent of clonal expansion in the secondary lymphoid tissue has been measured accurately, these results must underestimate the size of the subsequent diaspora. The calculation that the total virus-specific CD8+ T cell numbers fall only 2-fold between d10 and d30 after infection is, therefore, still too high. The idea that most immune T cells are eliminated very rapidly after the resolution of the antigen-driven phase (24, 33) may apply to the minority population associated directly with the site of pathology but, at least in localized infections, the resolution of the majority response proceeds much more slowly.
Acknowledgments
We thank Vicki Henderson for help with the manuscript. These experiments were supported by National Institutes of Health Grants AI29579 and CA21765 and by the American Syrian Lebanese Associated Charities.
Abbreviations
- LN
lymph node
- PP
Peyer's patches
- i.n.
intranasal
- MLN
mediastinal LN
- NP
nucleoprotein
- PA
acid polymerase
- BAL
bronchoalveolar lavage
- SLN
superficial LN
- ILN
internal LN
- BM
bone marrow
- NALT
nasal-associated lymphoid tissue
- PBL
peripheral blood lymphocyte
- d(n)
day n
References
- 1.McMichael A J, O'Callaghan C A. J Exp Med. 1998;187:1367–1371. doi: 10.1084/jem.187.9.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 3.Murali-Krishna K, Altman J D, Suresh M, Sourdive D J, Zajac A J, Miller J D, Slansky J, Ahmed R. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 4.Flynn K J, Belz G T, Altman J D, Ahmed R, Woodland D L, Doherty P C. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- 5.Doherty P C, Christensen J P. Annu Rev Immunol. 2000;18:561–592. doi: 10.1146/annurev.immunol.18.1.561. [DOI] [PubMed] [Google Scholar]
- 6.Horimoto T, Kawaoka Y. Virology. 1995;210:466–470. doi: 10.1006/viro.1995.1363. [DOI] [PubMed] [Google Scholar]
- 7.Flynn K J, Riberdy J M, Christensen J P, Altman J D, Doherty P C. Proc Natl Acad Sci USA. 1999;96:8597–8602. doi: 10.1073/pnas.96.15.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichelberger M C, Wang M, Allan W, Webster R G, Doherty P C. J Gen Virol. 1991;72:1695–1698. doi: 10.1099/0022-1317-72-7-1695. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton-Easton A, Eichelberger M. J Virol. 1995;69:6359–6366. doi: 10.1128/jvi.69.10.6359-6366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Usherwood E J, Hogg T L, Woodland D L. J Immunol. 1999;162:3350–3355. [PubMed] [Google Scholar]
- 11.Tripp R A, Topham D J, Watson S R, Doherty P C. J Immunol. 1997;158:3716–3720. [PubMed] [Google Scholar]
- 12.Townsend A R, Rothbard J, Gotch F M, Bahadur G, Wraith D, McMichael A J. Cell. 1986;44:959–968. doi: 10.1016/0092-8674(86)90019-x. [DOI] [PubMed] [Google Scholar]
- 13.Belz G T, Xie W, Altman J D, Doherty P C. J Virol. 2000;74:3486–3493. doi: 10.1128/jvi.74.8.3486-3493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor-Fishwick D A, Siegel J N. Eur J Immunol. 1995;25:3215–3221. doi: 10.1002/eji.1830251203. [DOI] [PubMed] [Google Scholar]
- 15.Marzio R, Mauel J, Betz-Corradin S. Immunopharmacol Immunotoxicol. 1999;21:565–582. doi: 10.3109/08923979909007126. [DOI] [PubMed] [Google Scholar]
- 16.Christensen J P, Doherty P C, Branum K C, Riberdy J M. J Virol. 2000;74:11690–11696. doi: 10.1128/jvi.74.24.11690-11696.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belz G T, Altman J D, Doherty P C. Proc Natl Acad Sci USA. 1998;95:13812–13817. doi: 10.1073/pnas.95.23.13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.SAS Institute. SAS/STAT User's Guide. Cary, NC: SAS Institute; 1990. , Version 6, 4th Ed., Vol. 2. [Google Scholar]
- 19.Cerwenka A, Morgan T M, Dutton R W. J Immunol. 1999;163:5535–5543. [PubMed] [Google Scholar]
- 20.Hogan R J, Usherwood E J, Zhong W, Roberts A D, Dutton R W, Harmsen A G, Woodland D L. J Immunol. 2001;166:1813–1822. doi: 10.4049/jimmunol.166.3.1813. [DOI] [PubMed] [Google Scholar]
- 21.Belz, G. T., Xie, W. & Doherty, P. C. (2001) J. Immunol., in press. [DOI] [PubMed]
- 22.Freitas A A, Rocha B. Annu Rev Immunol. 2000;18:83–111. doi: 10.1146/annurev.immunol.18.1.83. [DOI] [PubMed] [Google Scholar]
- 23.Doherty P C. J Immunol. 1995;155:1023–1027. [PubMed] [Google Scholar]
- 24.Tough D F, Sun S, Zhang X, Sprent J. Immunol Rev. 1999;170:39–47. doi: 10.1111/j.1600-065x.1999.tb01327.x. [DOI] [PubMed] [Google Scholar]
- 25.De Martino M, Rossi M E, Azzari C, Gelli M G, Chiarelli F, Galli L, Vierucci A. Clin Exp Immunol. 1999;117:513–516. doi: 10.1046/j.1365-2249.1999.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hawke S, Stevenson P G, Freeman S, Bangham C R. J Exp Med. 1998;187:1575–1582. doi: 10.1084/jem.187.10.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehal W Z, Juedes A E, Crispe I N. J Immunol. 1999;163:3202–3210. [PubMed] [Google Scholar]
- 28.Slifka M K, Ahmed R. Curr Opin Immunol. 1998;10:252–258. doi: 10.1016/s0952-7915(98)80162-3. [DOI] [PubMed] [Google Scholar]
- 29.Sprent J, Zhang X, Sun S, Tough D. Philos Trans R Soc London B. 2000;355:317–322. doi: 10.1098/rstb.2000.0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ku C C, Murakami M, Sakamoto A, Kappler J, Marrack P. Science. 2000;288:675–678. doi: 10.1126/science.288.5466.675. [DOI] [PubMed] [Google Scholar]
- 31.Masopust D, Shen H, Lefrancois L. J Immunol. 2001;166:2348–2356. doi: 10.4049/jimmunol.166.4.2348. [DOI] [PubMed] [Google Scholar]
- 32.Masopust D, Vezys V, Marzo A L, Lefrancois L. Science. 2001;291:2413–2417. [PubMed] [Google Scholar]
- 33.Marrack P, Hugo P, McCormack J, Kappler J. Immunol Rev. 1993;133:119–129. doi: 10.1111/j.1600-065x.1993.tb01513.x. [DOI] [PubMed] [Google Scholar]