Abstract
Acinetobacter baumannii can colonize body surfaces of hospitalized patients. From these sites, invasion into the host and spread to other patients and the hospital environment may occur. The eradication of the organism from the patient's skin is an important infection control strategy during epidemic and endemic episodes. In this study, a three-dimensional (3D), air-exposed human epidermal skin equivalent was exploited to study Acinetobacter skin colonization. We characterized the adherence of A. baumannii ATCC 19606T and Acinetobacter junii RUH2228T to and biofilm formation on the skin equivalent and the responses to these bacteria. Furthermore, we assessed the ability of the disinfectant chlorhexidine to decolonize the skin equivalents. The results revealed that both strains replicated on the stratum corneum for up to 72 h but did not invade the epidermis. A. baumannii, in contrast to A. junii, formed large biofilms on the stratum corneum. Bacterial colonization did not affect keratinocyte activation, proliferation, or differentiation, nor did it induce a strong inflammatory response. Disinfection with chlorhexidine solution resulted in complete eradication of A. baumannii from the skin, without detrimental effects. This 3D model is a promising tool to study skin colonization and to evaluate the effects of novel disinfectant and antimicrobial strategies.
INTRODUCTION
Multidrug-resistant (MDR) strains of Acinetobacter baumannii are notorious for their association with outbreaks of colonization and infection worldwide (6, 23). During such outbreaks, A. baumannii can colonize body surfaces of severely ill patients, from where the organisms may invade the patient, causing infection and/or spread to other patients and their environment. Thus, the skin is thought to constitute an important reservoir for A. baumannii during outbreaks and endemic episodes (1, 17). Insight into Acinetobacter skin colonization and the microbial ecology of the skin may result in novel strategies to prevent or interfere with skin colonization and thus contribute to the eradication of the organisms from a ward.
Adherence to and biofilm formation on plastic and adherence to human cells are widely used systems to study the interactions of bacteria with abiotic and biotic surfaces. However, these systems may not adequately reflect the association of bacteria with the human skin, a process that takes place under relatively dry conditions (20). Moreover, adherence and subsequent replication are strongly influenced by environmental conditions (19), which in turn can be expected to depend on the physicochemical barrier properties of the skin surface and its nutrient availability. Once bacteria have adhered to the skin, they may invade the epidermis and trigger an inflammatory response. To our knowledge, little is known about the possible response of human skin cells to Acinetobacter.
Tissue-engineered, air-exposed human skin models are three-dimensional (3D) systems that mimic the native skin to a high degree (10, 11). Such epidermal skin equivalents are generated by culturing primary keratinocytes at the air-liquid interface on cell-free matrices (e.g., inert filters or de-epidermized dermis). The keratinocytes will proliferate, migrate, and differentiate during epidermal development, resulting in skin equivalents that contain all layers of the native epidermis (11). The skin equivalents also have barrier properties that show many similarities with the human skin (21). These systems are interesting models for studying pathogen-skin interactions.
In the present study, we exploited a 3D human epidermal skin equivalent to study the adherence of an A. baumannii strain and an Acinetobacter junii strain to the skin, the subsequent biofilm formation, and the skin's response to these bacteria. Furthermore, we explored the usefulness of this model to investigate the effects of a disinfectant on the bacteria and the human skin.
MATERIALS AND METHODS
Generation of epidermal skin equivalents.
Human keratinocytes were isolated from fresh plastic surgery surplus skin as previously described (10). Briefly, the epidermis and dermis were enzymatically and mechanically separated, and each layer was subsequently digested to obtain single-cell suspensions. Keratinocytes were cultured in Dermalife medium (Lifeline Cell Technology) supplemented with penicillin (10,000 U) and streptomycin (10 mg/ml). Epidermal skin models were generated as described previously (11, 21). In short, approximately 2 × 105 keratinocytes from a secondary culture were seeded onto a filter insert (12 mm in diameter) (Costar; Corning) in 12-well plates in Dermalife medium. Three days after seeding, cells were put air exposed by aspirating the apical medium from the keratinocytes, leaving only the filter insert in contact with the medium. The basal medium was replaced with CnT-02-3D medium (CellTec) supplemented with 2.4 × 10−2 μM bovine serum albumin, 25 μM palmitic acid, 15 μM linoleic acid, and 7 μM arachidonic acid. Prior to bacterial inoculation, the medium was replaced with keratinocyte medium without penicillin and streptomycin. Experiments were performed by using 7-day air-exposed cultures.
Preparation of the bacterial inoculum.
A. baumannii strain ATCC 19606T and A. junii strain ATCC 17908T (RUH2228T) were used. Bacteria were preserved for prolonged periods in nutrient broth supplemented with 20% (vol/vol) glycerol at −80°C. Inocula from frozen cultures were grown overnight at 37°C on sheep blood agar plates (bioMérieux). Bacteria were cultured for 2.5 h at 37°C in Luria-Bertani (LB) medium (10 g of Bacto tryptone, 5 g of yeast extract [both from BD, Sparks, MD], and 5 g of sodium chloride [Merck] in 1 liter of distilled water) under vigorous shaking. This suspension was diluted in phosphate-buffered saline (PBS) (pH 7.4) to a concentration of approximately 3 × 105 CFU/ml, as calculated from the absorbance of the suspension at 600 nm and as verified afterwards by using standard vital counts.
Colonization of epidermal skin equivalents.
Skin equivalents were incubated with 300 μl of the bacterial suspension at 37°C in 7.3% CO2. After 1 h, the bacterial suspension was aspirated to remove nonadherent bacteria. At different intervals after inoculation, the numbers of viable detachable and adherent bacteria were assessed microbiologically. Briefly, 600 μl of PBS was applied onto the skin, and the detachable bacteria were collected, serially diluted, and plated onto diagnostic sensitivity test (DST) agar plates to determine the number of CFU. To assess the number of adherent bacteria, two biopsy specimens (each 4 mm in diameter) were taken from the skin and homogenized in PBS by using a glass Potter-Elvehjem tissue homogenizer, and the homogenates were subsequently serially diluted. The lower limits of detection of detachable and adherent bacteria were 12 and 115 CFU/skin equivalent, respectively. The number of adherent bacteria per skin equivalent (113.04 mm2) was calculated by multiplying the number of adherent bacteria in two biopsy specimens (25.12 mm2) by 4.5. The total number of bacteria per skin equivalent was calculated by adding the number of detachable bacteria per skin equivalent and the number of adherent bacteria per skin equivalent.
Microscopic analysis of bacterial replication on skin equivalents.
To visualize the bacterial colonization of the skin equivalents, one biopsy specimen of each skin equivalent was fixed in 4% formaldehyde, dehydrated, and embedded in paraffin. Next, paraffin blocks were cut into 5-μm sections, deparaffinized, rehydrated, and then stained with Alcian blue in combination with Periodic acid-Schiff (PAS) stain (both from Merck) to detect polysaccharides.
Immunohistochemical analysis of the keratinocyte response.
For the analysis of the effects of colonization of the skin equivalents, the levels of keratin 16 (K16), a marker for keratinocyte activation or stress; Ki67, a marker for keratinocyte proliferation; and K10, a marker for early keratinocyte differentiation, were determined by immunohistochemical analysis. In short, standard antigen retrieval of deparaffinized and rehydrated paraffin sections was performed by immersing sections in 0.01 M citrate buffer (pH 6.0) for 30 min at 90°C, followed by slow cooling to room temperature for at least 3 h prior to the staining of the sections. The sections were incubated overnight at 4°C with primary mouse antibodies directed against human K16 (clone LL025, 5× diluted; AbD Serotec), Ki67 (clone MIB-1, 75× diluted; Dako), and K10 (clone DE-k10, 50× diluted; Abcam). Thereafter, sections were incubated for 60 min with secondary biotinylated goat anti-mouse antibodies (Dako) and subsequently incubated for 30 min with streptavidin-biotinylated horseradish peroxidase (Dako). Chromogen 3-amino-9-ethyl-carbozole (AEC) solution was used as the substrate solution. Sections were washed three times with PBS between subsequent incubations and finally counterstained with hematoxylin.
To determine the proliferation index, the number of Ki67-positive nuclei among 100 basal cells was counted in sections of three different samples at a magnification of ×200.
Determination of cytokine and chemokine levels.
The levels of interleukin-1α (IL-1α), IL-1β, IL-6, IL-8, IL-10, and tumor necrosis factor alpha (TNF-α) in culture media were determined by an enzyme-linked immunosorbent assay (ELISA) (all from Biosource, Invitrogen) according to the manufacturer's instructions. The lower limits of detection were 1 pg/ml for IL-1α and IL-1β, 15 pg/ml for IL-6, 7 pg/ml for IL-8, and 25 pg/ml for IL-10 and TNF-α.
Determination of gene expression levels.
Total RNA was extracted from homogenized skin biopsy specimens by using the RNeasy minikit (Qiagen), followed by a treatment with DNase I (Qiagen). cDNA synthesis was performed on 300 ng of total RNA by using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. For each sample, a control for genomic DNA contamination was included by adding sterile water instead of reverse transcriptase. Real-time quantitative PCR was performed by use of an ICycler IQ instrument (Bio-Rad), with a final volume of 25 μl comprising 1× IQ SYBR green Supermix (Bio-Rad), 10 pmol each primer (Table 1), and 5 μl of 10×-diluted cDNA. PCR conditions consisted of an initial denaturation step at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15 s; annealing at 56.5°C (for Ki67), 57.8°C (for LL-37), 60.8°C (for glyceraldehyde-3-phosphate dehydrogenase [GAPDH] and IL-1β), 61.7°C (for TNF-α), or 62°C (for beta-2 microglobulin [β2 M], IL-1α, IL-6, IL-8, human beta-defensin 2 [hBD-2], hBD-3, K16, and K10) for 15 s; and elongation at 72°C for 20 s. GAPDH and β2 M were used for standardization. Experiments were performed in duplicate. Data were acquired and analyzed by using ICycler IQ Optical System software (Bio-Rad), with an automatic adjustment of the baseline and threshold parameters.
Table 1.
Sequences of primers used for reverse transcription-PCR
| Primer target | Sequence |
|
|---|---|---|
| Forward | Reverse | |
| β2 M | TGCTGTCTCCATGTTTGATGTATCT | TCTCTGCTCCCCACCTCTAAGT |
| GAPDH | AAGGTCGGAGTCAACGGATTT | ACCAGAGTTAAAAGCAGCCCTG |
| IL-1α | CGCCAATGACTCAGAGGAAGA | AGGGCGTCATTCAGGATGAA |
| IL-1β | ACGAATCTCCGACCACCACT | CCATGGCCACAACAACTGAC |
| IL-6 | GGTACATCCTCGACGGCATCT | GTGCCTCTTTGCTGCTTTCAC |
| IL-8 | GCCAGGAAGAAACCACCGGAAGG | GGCTGCCAAGAGAGCCACGG |
| TNF-α | CCTGTGAGGAGGACGAACAT | GGTTGAGGGTGTCTGAAGGA |
| hBD-2 | TGATGCCTCTTCCAGGTGTTT | GGATGACATATGGCTCCACTCTTA |
| hBD-3 | TTATTGCAGAGTCAGAGGCGG | CGAGCACTTGCCGATCTGTT |
| LL-37 | ATTTCTCAGAGCCCAGAAGC | CGGAATCTTGTACCCAGGAC |
| K16 | GAGATGCGTGACCAGTACGA | TTGTTCAGCTCCTCGGTCTT |
| K10 | AGCATGGCAACTCACATCAG | TGTCGATCTGAAGCAGGATG |
| Ki67 | AATTCAGACTCCATGTGCCTGAG | CTTGACACACACATTGTCCTCAGC |
Decontamination of colonized skin equivalents.
Two days after inoculation with A. baumannii ATCC 19606T, 0.5% chlorhexidine solution in 70% ethanol or, as a control, PBS was applied onto the skin equivalents using a sterile cotton swab. The numbers of detachable and adherent bacteria were assessed, as described above, 24 h after the application of the disinfectant.
Statistical analysis.
Results are expressed as means ± standard errors of the means unless stated otherwise. Data were analyzed for statistical significance by using the Wilcoxon rank sum test (SPSS 17.0). P values of ≤0.05 were considered significant.
RESULTS
Replication of Acinetobacter on skin equivalents.
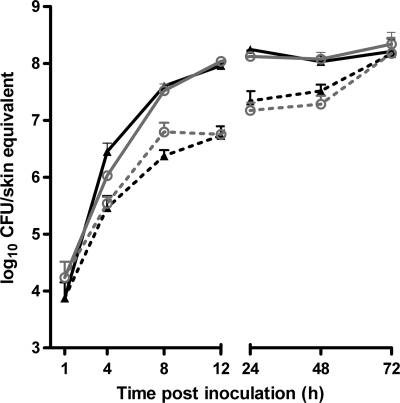
To compare the persistences of A. baumannii ATCC 19606T and A. junii RUH2228T on the skin, human epidermal skin equivalents were inoculated with approximately 1 × 105 CFU suspended in 300 μl of PBS. The suspensions were aspirated from the skin equivalents after 1 h to allow air exposure again. The total numbers of bacteria on the skin equivalent increased during the first 12 h, from 7 × 103 ± 6 × 103 to 9 × 107 ± 3 × 106 CFU for the A. baumannii strain and from 2 × 104 ± 1 × 104 to 1 × 108 ± 1 × 107 CFU for the A. junii strain, and thereafter remained stable for the duration of the experiment (Fig. 1).
Fig 1.
A. baumannii and A. junii replication on skin equivalents. Human epidermal skin equivalents were inoculated with approximately 1 × 105 CFU of A. baumannii ATCC 19606T (triangles) or A. junii RUH2228T (circles). After 1 h, the nonadhered bacteria were removed. At different intervals thereafter, the numbers of detachable and adherent bacteria were determined microbiologically. The results are expressed as mean numbers of adherent (dotted lines) and total (solid lines) CFU per skin equivalent ± standard errors of the means from three independent experiments.
At 48 and 72 h after infection, the number of adhered A. baumannii ATCC 19606T was significantly higher than the number of detachable bacteria, indicating that the proportion of bacteria that adhered increased over time (Fig. 1). For the A. junii strain, the number of adherent bacteria exceeded that of detachable bacteria at 72 h after infection.
Acinetobacter biofilm formation on skin equivalents.
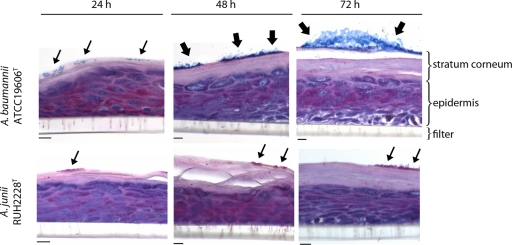
Microscopic analysis revealed that bacteria of both strains persisted on the stratum corneum and did not invade the epidermis (Fig. 2). Although there was no significant difference in the numbers of adherent bacteria between A. baumannii ATCC 19606T and A. junii RUH2228T (Fig. 1), Alcian blue-PAS staining revealed large biofilm structures formed by the A. baumannii strain but not by the A. junii strain on the stratum corneum at 48 and 72 h (Fig. 2).
Fig 2.
Biofilm formation on skin equivalents. Skin equivalents were exposed to A. baumannii ATCC 19606T and A. junii RUH2228T for up to 72 h. Thereafter, 5-μm sections of paraffin-embedded skin were stained with Alcian blue-PAS, and biofilm formation was analyzed by light microscopy. Small arrows indicate clusters of bacteria, and large arrows indicate bacteria within a biofilm matrix. Scale bars, 20 μm.
Response of skin equivalents to Acinetobacter.
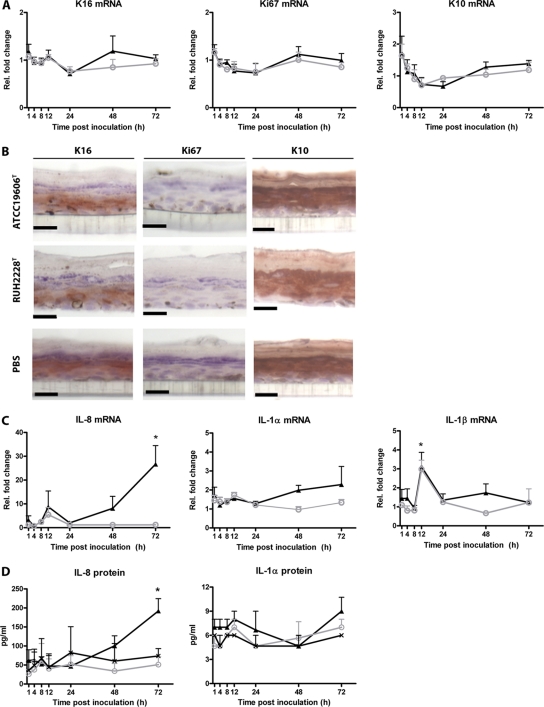
Subsequently, the effect of bacterial colonization on keratinocyte differentiation and proliferation and on cytokine and antimicrobial peptide production was evaluated. The results revealed that the exposure of the skin equivalents to A. baumannii ATCC 19606T and A. junii RUH2228T did not affect the mRNA expression levels of Ki67, K10, and K16 in keratinocytes (Fig. 3A). In agreement with this finding, no difference was seen in protein expression levels of the activation/stress marker K16 between colonized and PBS-exposed skin equivalents (Fig. 3B). Moreover, the expression of K10 in the suprabasal viable cell layers of the exposed and nonexposed skin equivalents confirms that the early differentiation program was not altered by the Acinetobacter strains. In addition, the number of proliferating cells in the basal layers was not influenced by the bacteria; i.e., 20 ± 2 versus 18 ± 1 cells out of 100 basal cells were positive for Ki67 in A. baumannii-colonized skin versus PBS-exposed skin.
Fig 3.
Response of keratinocytes to Acinetobacter. Skin equivalents were exposed to A. baumannii ATCC 19606T (triangles), A. junii RUH2228T (circles), or, as a control, PBS (crosses). (A and B) mRNA expression at various intervals thereafter (A) and protein levels at 48 h thereafter (B) of the keratinocyte activation/stress marker K16, the proliferation marker Ki67, and the differentiation marker K10 were determined by using quantitative PCR (qPCR) (A) and immunohistochemistry (B). Scale bars, 20 μm. (C and D) In addition, the mRNA expression (C) and protein (D) levels of the chemokine IL-8 and the proinflammatory cytokines interleukin-1α (IL-1α) and IL-1β at different intervals were determined by using qPCR and ELISA, respectively. Results are expressed as mean fold changes in mRNA expression levels relative to those of PBS-exposed skin equivalents (A and C) or mean cytokine levels in pg/ml (D) ± standard errors of the means from three independent experiments. *, significantly different from PBS.
Furthermore, we assessed the production of the proinflammatory cytokines TNF-α, IL-1α, IL-1β, IL-6, and IL-8 and that of the anti-inflammatory cytokine IL-10 by the skin equivalent in response to both Acinetobacter strains. During the first 48 h of infection, skin equivalents expressed levels of IL-1α and IL-8 after exposure to the two Acinetobacter strains that were similar to those expressed by skin equivalents exposed to PBS. However, after 72 h, mRNA and protein expression levels of IL-8, but not of IL-1α, were slightly but significantly (P < 0.05) higher in skin equivalents colonized by the A. baumannii strain than in those colonized by the A. junii strain and/or exposed to PBS (Fig. 3C and D). In addition to IL-8 and IL-1α, IL-1β mRNA expression levels were significantly (P < 0.05) higher in skin equivalents exposed to the Acinetobacter strains than in PBS-exposed skin equivalents but only at 12 h of infection (Fig. 3C). The IL-1β protein, however, was not detectable. Moreover, TNF-α, IL-6, and IL-10 mRNAs and proteins were not detectable in skin equivalents.
The skin equivalents constitutively expressed the antimicrobial peptides human beta-defensin 2 (hBD-2) and hBD-3 but not LL-37. The infection of skin equivalents with the Acinetobacter strains did not induce an enhanced expression of these peptides (data not shown).
Eradication of A. baumannii from skin equivalents.
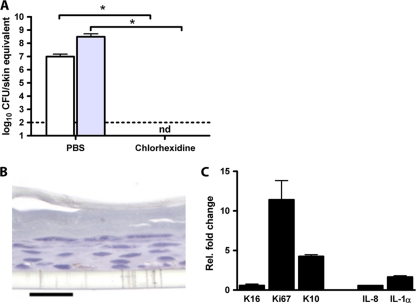
Finally, we examined the effect of chlorhexidine on A. baumannii ATCC 19606T colonization and on the epidermis. The swapping of skin equivalents colonized by the A. baumannii strain for 48 h with 0.5% chlorhexidine solution in 70% ethanol resulted in undetectable levels of A. baumannii on the skin (Fig. 4A). In addition, this chlorhexidine solution did not affect skin morphology (Fig. 4B) and mRNA expression levels of K16, IL-1α, and IL-8 but enhanced the expression levels of Ki67 and K10 11-fold and 4-fold, respectively (Fig. 4C). This finding indicates that chlorhexidine enhanced keratinocyte proliferation and differentiation but did not induce epidermal activation/stress or inflammation in the skin (Fig. 4C).
Fig 4.
Treatment of infected skin equivalents with chlorhexidine. Skin equivalents were infected with A. baumannii ATCC 19606T for 48 h. Thereafter, chlorhexidine or PBS was applied onto the skin equivalents. (A) Twenty-four hours after treatment, the numbers of detachable and adherent bacteria were determined microbiologically. Results are expressed as mean numbers of adherent (white bars) and total (gray bars) CFU per skin equivalent ± standard errors of the means from three independent experiments. The dotted line represents the lower limit of detection. nd, not detectable; *, significant (P < 0.05) difference. (B) Light micrograph of a section of chlorhexidine-treated skin stained with Alcian blue-PAS stain. Scale bar, 20 μm. (C) Twenty-four hours after treatment, mRNA expression levels of the keratinocyte activation/stress (K16), proliferation (Ki67), and differentiation (K10) markers as well as the inflammatory mediators IL-8 and IL-1α were determined by using qPCR. The results are expressed as mean mRNA expression levels relative to those of PBS-treated skin ± standard deviations from four measurements.
DISCUSSION
The main conclusion from the present findings is that the 3D human skin equivalent is a promising model to study skin colonization by Acinetobacter strains and to evaluate the effects of disinfectants and other antimicrobial agents. This is of importance, as A. baumannii is able to colonize the skin (2, 7, 8, 17, 24), which can be a source of infection and spread to other patients and the environment. Recent papers have emphasized the benefits of skin disinfection as a tool to reduce colonization pressure in a ward (2, 12, 22). Our main conclusion is based on the following findings. First, both A. baumannii ATCC 19606T and strain RUH2228T, which belongs to the species A. junii, a species that occurs on the human skin, adhered to and replicated on the stratum corneum for up to 72 h without invading the epidermis. An important difference between these two strains was the ability of the A. baumannii strain, but not the A. junii strain, to form a large biofilm on the skin equivalents. Second, bacterial colonization did not affect keratinocyte activation, proliferation, or differentiation, nor did it induce a strong inflammatory response. Third, disinfection with chlorhexidine solution resulted in complete eradication of the A. baumannii strain from the skin, without detrimental effects.
We have previously shown that both A. baumannii ATCC 19606T and A. junii RUH2228T were able to form large biofilms on plastic (5). On the human skin model reported here, however, only A. baumannii ATCC 19606T formed large biofilms. A possible explanation for the latter observation could be that A. baumannii, but not A. junii, can utilize the nutrients of the stratum corneum for biofilm formation. In agreement with this suggestion, it was shown previously that the metabolic versatility of A. baumannii is considerably greater than that of A. junii (3, 18). The ability of Acinetobacter to form biofilms appears to depend on the surface to which the bacteria adhere, as suggested previously by Gaddy and Actis (13). Although more strains should be examined before a definitive conclusion can be drawn, we hypothesize that the ability of A. baumannii to form a biofilm on human skin plays an important role in its persistence on the skin.
We have previously shown that human bronchial epithelial cells and cultured human macrophages produce considerable amounts of inflammatory mediators in response to these Acinetobacter strains (5). Using this skin equivalent model, however, A. baumannii ATCC 19606T and A. junii RUH2228T induced a poor inflammatory response. This may be explained as follows. In our previous study, the cells were cultured in monolayers and infected with bacteria, enabling direct contact between the bacteria and the cells. However, in the current experiments, the bacteria did not invade the epidermis and, thus, did not come into direct contact with live keratinocytes. These results underline the importance of the stratum corneum as a protective barrier against infections, as also shown previously by Duckney et al., who demonstrated that Staphylococcus epidermidis and Propionibacterium acnes induced an inflammatory response in a reconstructed human epidermis model only when applied subcutaneously in the culture medium and not when applied topically onto the stratum corneum (9). Moreover, this finding emphasizes that results from studies of pathogen-host interactions using keratinocyte monolayers (lacking a stratum corneum) will not reflect the in vivo situation.
With the emergence of antibiotic resistance, there has been a reappraisal of the use of antiseptics for skin disinfection (12, 15). Chlorhexidine is widely used as an antiseptic against many microorganisms, including A. baumannii (12). However, several studies have shown that chlorhexidine may have toxic effects on human fibroblasts (14) and be detrimental to wound healing (14, 16), making its use controversial in particular situations. Although the A. baumannii type strain used in this study was fully susceptible to treatment with chlorhexidine, an MDR A. baumannii isolate that was resistant to chlorhexidine concentrations of up to 1% has already been reported (4). In this light, it is necessary to develop novel disinfectant strategies to eradicate A. baumannii from the colonized skin.
In summary, we have described a novel model for Acinetobacter skin colonization using 3D human epidermal skin equivalents. This model will be beneficial in characterizing bacterial growth kinetics and the interactions of different bacterial species on a biotic surface. Moreover, this model may also be advantageous for identifying and evaluating new targets for disinfection and antimicrobial strategies.
ACKNOWLEDGMENT
This work was supported by grant 10.106 from the Dutch Burns Foundation.
Footnotes
Published ahead of print 30 January 2012
REFERENCES
- 1. Bergogne-Berezin E, Towner KJ. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Borer A, et al. 2007. Impact of 4% chlorhexidine whole-body washing on multidrug-resistant Acinetobacter baumannii skin colonisation among patients in a medical intensive care unit. J. Hosp. Infect. 67:149–155 [DOI] [PubMed] [Google Scholar]
- 3. Bouvet PJ, Grimont PA. 1987. Identification and biotyping of clinical isolates of Acinetobacter. Ann. Inst. Pasteur Microbiol. 138:569–578 [DOI] [PubMed] [Google Scholar]
- 4. Brooks SE, Walczak MA, Hameed R, Coonan P. 2002. Chlorhexidine resistance in antibiotic-resistant bacteria isolated from the surfaces of dispensers of soap containing chlorhexidine. Infect. Control Hosp. Epidemiol. 23:692–695 [DOI] [PubMed] [Google Scholar]
- 5. de Breij A, et al. 2010. Do biofilm formation and interactions with human cells explain the clinical success of Acinetobacter baumannii? PLoS One 5:e10732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939–951 [DOI] [PubMed] [Google Scholar]
- 7. Dijkshoorn L, Van Vianen W, Degener JE, Michel MF. 1987. Typing of Acinetobacter calcoaceticus strains isolated from hospital patients by cell envelope protein profiles. Epidemiol. Infect. 99:659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dijkshoorn L, et al. 1989. Use of protein profiles to identify Acinetobacter calcoaceticus in a respiratory care unit. J. Clin. Pathol. 42:853–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duckney P, Wong HK, Serrano J, Oddos T, Stamatas G. 2011. Inflammatory effects of common skin microbial species on cultured keratinocytes. J. Invest. Dermatol. 131(Suppl):S12 [Google Scholar]
- 10. El Ghalbzouri A, Commandeur S, Rietveld MH, Mulder AA, Willemze R. 2009. Replacement of animal-derived collagen matrix by human fibroblast-derived dermal matrix for human skin equivalent products. Biomaterials 30:71–78 [DOI] [PubMed] [Google Scholar]
- 11. El Ghalbzouri A, Siamari R, Willemze R, Ponec M. 2008. Leiden reconstructed human epidermal model as a tool for the evaluation of the skin corrosion and irritation potential according to the ECVAM guidelines. Toxicol. In Vitro 22:1311–1320 [DOI] [PubMed] [Google Scholar]
- 12. Evans HL, et al. 2010. Effect of chlorhexidine whole-body bathing on hospital-acquired infections among trauma patients. Arch. Surg. 145:240–246 [DOI] [PubMed] [Google Scholar]
- 13. Gaddy JA, Actis LA. 2009. Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol. 4:273–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hidalgo E, Dominguez C. 2001. Mechanisms underlying chlorhexidine-induced cytotoxicity. Toxicol. In Vitro 15:271–276 [DOI] [PubMed] [Google Scholar]
- 15. Khan MN, Naqvi AH. 2006. Antiseptics, iodine, povidone iodine and traumatic wound cleansing. J. Tissue Viability 16:6–10 [DOI] [PubMed] [Google Scholar]
- 16. Lucarotti ME, White H, Deas J, Silver IA, Leaper DJ. 1990. Antiseptic toxicity to breast carcinoma in tissue culture: an adjuvant to conservation therapy? Ann. R. Coll. Surg. Engl. 72:388–392 [PMC free article] [PubMed] [Google Scholar]
- 17. Marchaim D, et al. 2007. Surveillance cultures and duration of carriage of multidrug-resistant Acinetobacter baumannii. J. Clin. Microbiol. 45:1551–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nemec A, et al. 2011. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res. Microbiol. 162:393–404 [DOI] [PubMed] [Google Scholar]
- 19. O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49–79 [DOI] [PubMed] [Google Scholar]
- 20. Shepherd J, Douglas I, Rimmer S, Swanson L, MacNeil S. 2009. Development of three-dimensional tissue-engineered models of bacterial infected human skin wounds. Tissue Eng. Part C Methods 15:475–484 [DOI] [PubMed] [Google Scholar]
- 21. Thakoersing VS, et al. 18 October 2011, posting date Unravelling barrier properties of three different in-house human skin equivalents. Tissue Eng. Part C Methods. [Epub ahead of print.] doi:10.1089/ten.TEC.2011.0175 [DOI] [PubMed] [Google Scholar]
- 22. Urban C, Segal-Maurer S, Rahal JJ. 2003. Considerations in control and treatment of nosocomial infections due to multidrug-resistant Acinetobacter baumannii. Clin. Infect. Dis. 36:1268–1274 [DOI] [PubMed] [Google Scholar]
- 23. Visca P, Seifert H, Towner KJ. 18 October 2011, posting date Acinetobacter infection—an emerging threat to human health. IUBMB Life. [Epub ahead of print.] doi:10.1002/iub.534 [DOI] [PubMed] [Google Scholar]
- 24. Zeana C, et al. 2003. The epidemiology of multidrug-resistant Acinetobacter baumannii: does the community represent a reservoir? Infect. Control Hosp. Epidemiol. 24:275–279 [DOI] [PubMed] [Google Scholar]