Abstract
Antimicrobial proteins and peptides (AMPs) are essential effectors of innate immunity, acting as a first line of defense against bacterial infections. Many AMPs exhibit high affinity for cell wall structures such as lipopolysaccharide (LPS), a potent endotoxin able to induce sepsis. Hence, understanding how AMPs can interact with and neutralize LPS endotoxin is of special relevance for human health. Eosinophil cationic protein (ECP) is an eosinophil secreted protein with high activity against both Gram-negative and Gram-positive bacteria. ECP has a remarkable affinity for LPS and a distinctive agglutinating activity. By using a battery of LPS-truncated E. coli mutant strains, we demonstrate that the polysaccharide moiety of LPS is essential for ECP-mediated bacterial agglutination, thereby modulating its antimicrobial action. The mechanism of action of ECP at the bacterial surface is drastically affected by the LPS structure and in particular by its polysaccharide moiety. We have also analyzed an N-terminal fragment that retains the whole protein activity and displays similar cell agglutination behavior. Conversely, a fragment with further minimization of the antimicrobial domain, though retaining the antimicrobial capacity, significantly loses its agglutinating activity, exhibiting a different mechanism of action which is not dependent on the LPS composition. The results highlight the correlation between the protein's antimicrobial activity and its ability to interact with the LPS outer layer and promote bacterial agglutination.
INTRODUCTION
Lipopolysaccharide (LPS) is the main outer membrane component of Gram-negative bacteria. The LPS molecule is a phosphorylated glycolipid composed of lipid A, also known as endotoxin, and a polysaccharide moiety. In turn, the latter is composed of the inner core (comprising three heptose residues and a 3-deoxy-d-manno-octulosonic acid residue), the outer core (comprising two glucose residues, two galactose residues, and one N-acetylglucosamine residue), and the antigenic “O” region composed of several repeats of different oligosaccharide units (Fig. 1A and B) (2, 9, 25, 27).
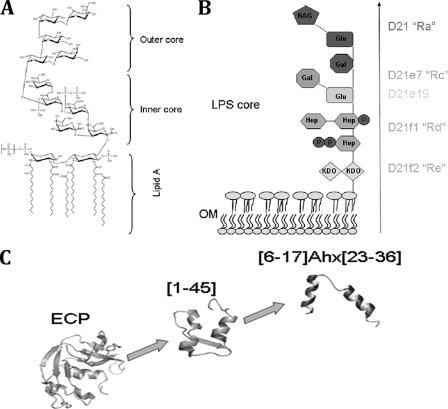
Fig 1.
(A and B) Chemical structure of LPS core (A) and chemotype scheme for the strains used (B). The Ra chemotype (strain D21) possesses the entire LPS core, the Rc chemotype includes the heptose chain and a Glu residue (strain D21e7) and an additional Gal residue (strain D21e19), the Rd chemotype contains the heptose and Kdo (strain D21f1), and the Re chemotype (strain D21f2) contains uniquely the Kdo portion. Only the D21 strain contains the phosphate residues associated with the heptose residues. (C) Structure scheme for ECP and derived peptides showing [6-17]-Ahx-[23-36] structure based on nuclear magnetic resonance (NMR) information (16, 21, 35).
LPS serves as a first barrier against antibiotics by hampering the diffusion of chemicals through the outer membrane (12). In fact, some modifications targeting the LPS structure in Gram-negative bacteria have been reported to confer resistant phenotypes against antibiotics and particularly against antimicrobial proteins and peptides (AMPs) (29). LPS, and in particular the lipid A portion, is released during bacterial infection, inducing inflammation and ultimately septic shock (4, 11). Unraveling the structural determinants for LPS interaction can assist in the design of new antibiotics with endotoxin-neutralizing properties (5, 15, 17).
AMPs interact with LPS competitively, displacing the divalent cations, breaking LPS compactness, and thus furthering the transport of peptides across the outer membrane (18). This process is modulated by the LPS charge, structure, and packing of lipid acyl chains in the outer layer (6, 24, 26). Several studies have determined that many AMPs require crossing of the outer membrane in order to reach the inner phospholipid bilayer, where they exert their action (7, 8, 23).
Eosinophil cationic protein (ECP) is an antimicrobial protein stored in the secondary granules of eosinophils that has an active role in infection control, immunomodulation, and tissue-remodelling processes (3, 37). ECP displays high antimicrobial activity and is able to specifically agglutinate Gram-negative bacteria before cell permeation occurs (30). Interestingly, in model membrane studies, ECP also promotes vesicle aggregation previous to any leakage event (36). Our previous results highlighted the correlation between ECP activity and its affinity to LPS (33).
Further experimental work evidenced that the antimicrobial domain of the protein lies in its N terminus (10, 28, 31). Indeed, a peptide containing the first 45 N-terminal residues (peptide [1-45]) (Fig. 1C) has been shown to retain the antimicrobial activity of the whole protein (32). A refinement of the antimicrobial domain by rational structure minimization led us to the synthesis of a shorter peptide ([6-17]-Ahx-[23-36]) that links the two first α-helical segments (Fig. 1C) and embodies the protein pharmacophore (35). However, though the shortest construct can agglutinate bacterial cells, its affinity for LPS is significantly reduced compared with that of the whole protein (35).
In order to study the role of LPS in the bactericidal properties of ECP and its derivative peptides, a wide battery of Escherichia coli K-12 mutant strains have been tested. These mutant strains, presenting distinctive chemotypes, are defective in genes involved in the biosynthesis and assembly of LPS (20, 29). From the results presented here, we conclude that for ECP and its N-terminal domain, both bacterial agglutination and antimicrobial activities are correlated with the LPS structure. A peptide with a further trimming of the N-terminal domain, though retaining the antimicrobial action, displays a different mechanism of action that is not dependent on the LPS layer. Therefore, the protein and the N-terminal antimicrobial domain can induce bacterial cell death by action at the cell outer layer without any active transport into the cytoplasm.
MATERIALS AND METHODS
Strains and chemicals.
Escherichia coli K-12 mutants deficient in lipopolysaccharide (LPS) synthesis were obtained from the E. coli Genetic Stock Center (Department of Biology, Yale University, New Haven, CT). The Alexa Fluor 488 protein labeling kit and the LIVE/DEAD bacterial viability kit were purchased from Molecular Probes (Eugene, OR). The BacTiter-Glo assay kit was from Promega (Madison, WI).
Expression and purification of recombinant ECP.
Wild-type ECP was expressed using a synthetic gene for the human ECP-coding sequence. Protein expression in E. coli strain BL21(DE3) (Novagen, Madison, WI), folding of the protein from inclusion bodies, and purification were carried out as previously described (1).
Peptides.
Peptides [1-45] and [6-17]-Ahx-[23-36] were prepared by fluoren-9-ylmethoxycarbonyl (Fmoc) solid-phase peptide synthesis methods (14). All peptides were purified by high-pressure liquid chromatography (HPLC) to ∼95% homogeneity and satisfactorily characterized by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF-MS).
Bacterial viability assays.
Bacterial viability was assayed using the BacTiter-Glo microbial cell viability kit (Promega). Briefly, ECP or peptide [1-45] or [6-17]-Ahx-[23-36] was dissolved in 10 mM sodium phosphate buffer (pH 7.5), serially diluted from 10 to 0.1 μM, and tested against E. coli strains (optical density at 600 nm [OD600] = 0.2) for 4 h of incubation time. Fifty microliters of culture was mixed with 50 μl of BacTiter-Glo reagent in a microtiter plate according to the manufacturer's instructions and incubated at room temperature for 10 min. Luminescence was read on a Victor3 plate reader (Perkin-Elmer, Waltham, MA) with a 1-s integration time. Fifty percent inhibitory concentrations (IC50s) were calculated by fitting the data to a dose-response curve.
Kinetics of bacterial survival were determined using the LIVE/DEAD bacterial viability kit (Molecular Probes, Invitrogen) in accordance with the manufacturer's instructions. E. coli K-12 mutant strains were grown at 37°C to an OD600 of 0.2, centrifuged at 5,000 × g for 5 min, and stained in a 0.75% NaCl solution. Fluorescence intensity was continuously measured after protein or peptide addition (5 μM) using a Cary Eclipse spectrofluorimeter (Varian Inc., Palo Alto, CA) as described previously (30). To calculate bacterial viability, the signal in the range of 510 to 540 nm was integrated to obtain the Syto 9 signal (live bacteria) and that in the range of 620 to 650 nm was integrated to obtain the propidium iodide (PI) signal (dead bacteria). The percentage of live bacteria was represented as a function of time, and t50 values were calculated by fitting the data to a simple exponential decay function.
Minimal agglutination concentration (MAC).
E. coli cells were grown at 37°C to an OD600 of 0.2, centrifuged at 5,000 × g for 2 min, and resuspended in Tris-HCl buffer–0.1 M NaCl (pH 7.5) in order to give an absorbance at 600 nm of 10. An aliquot of 200 μl of the bacterial suspension was treated with increasing protein or peptide concentrations (from 0.01 to 10 μM) and incubated at room temperature for 1 h. The aggregation behavior was observed by visual inspection, and the agglutinating activity is expressed as the minimum agglutinating concentration of the sample tested, as previously described (35).
Bacterial cytoplasmic membrane depolarization assay.
Membrane depolarization was followed using a method described earlier (30). Briefly, E. coli K 12 strains were grown at 37°C to an OD600 of 0.2, centrifuged at 5,000 × g for 7 min, washed with 5 mM HEPES (pH 7.2) containing 20 mM glucose, and resuspended in 5 mM HEPES-KOH, 20 mM glucose, and 100 mM KCl at pH 7.2 to an OD600 of 0.05. DiSC3(5) was added to a final concentration of 0.4 μM, and changes in the fluorescence were continuously recorded after addition of protein or peptide (5 μM). The time required to achieve half of total membrane depolarization was estimated from nonlinear regression analysis. The same assays were performed in the presence of 2 mM EDTA to perturb the LPS organization by chelation of divalent cations.
SEM.
Briefly, E. coli K-12 strain cell cultures of 1 ml were grown at 37°C to mid-exponential phase (OD600 of 0.2) and incubated with 5 μM protein or peptides in phosphate-buffered saline (PBS) at room temperature. Sample aliquots were taken for up to 4 h of incubation and were prepared for analysis by scanning electron microscopy (SEM), as previously described (33).
Transmission electron microscopy (TEM).
E. coli K-12 strains were grown to an OD600 of 0.2 and incubated with 5 μM ECP or peptides for 4 h. After treatment, bacterial pellets were prefixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer at pH 7.4 for 2 h at 4°C and postfixed in 1% osmium tetroxide buffered in 0.1 M cacodylate at pH 7.4 for 2 h at 4°C. The samples were dehydrated in acetone (50, 70, 90, 95, and 100%). The cells were immersed in Epon resin, and ultrathin sections were examined in a Jeol JEM 2011 instrument (Jeol Ltd., Tokyo, Japan).
Docking.
Docking simulations were conducted with AutoDock 4.0 (Scripps Research Institute. La Jolla, CA). ECP structure 1DYT.pdb (22) was used as receptor molecule, and LPS ligand was obtained from 1FI1.pdb (13). Water molecules were removed from the structure; hydrogen atoms and atomic partial charges were added using Autodock Tools. In the first docking approach, receptor and ligand were kept rigid. The interaction of a probe group, corresponding to each type of atom found in the ligand, with the protein structure was computed at 0.5-Å grid positions in a 90-Å cubic box centered in the protein. The docking was accomplished using 150 Lamarckian genetic algorithm (LGA) runs, and the initial position of the ligand was random. The number of individuals in populations was set to 150. The maximum number of energy evaluations that the genetic algorithm should make was 2,500,000. The maximum number of generations was 27,000. The number of top individuals that are guaranteed to survive into the next generation was 1. Rates of gene mutation and crossover were 0.02 and 0.80, respectively. Following docking, all structures generated for the same compound were subjected to cluster analysis, with cluster families being based on a tolerance of 2 Å for an all-atom root mean square (RMS) deviation from a lower-energy structure.
For the second stage, the global minimum structure was subjected to redocking under the same conditions but allowing 8 rotatable bonds (see Fig. 7) and to cluster analysis using a box centered in the ligand with a 0.250-Å grid spacing.
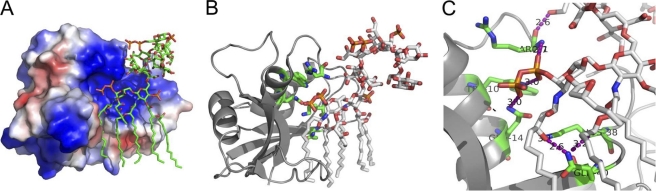
Fig 7.
Complex of ECP and LPS calculated by molecular docking. (A) Electrostatic potential map of ECP in complex with LPS. Cationic residues are colored in blue and anionic residues in red. (B and C) Detail of the complex between ECP and LPS. Interactions are highlighted in purple. Docking simulations were conducted with Autodock 4. Protein Data Bank (PDB) codes are 1DYT for ECP (22) and 1FI1 for LPS (13). The figure was constructed using PyMOL.
RESULTS
In order to study the role of LPS in the antimicrobial action of ECP, we have evaluated four LPS-defective strains derived from E. coli K-12 D21 lacking the O-antigen portion, the latter being used as a control (Fig. 1A and B). The Ra chemotype (strain D21) possesses the entire LPS core, the Rc chemotype (strains D21e7 and D21e19) does not display any further sugar moiety after the heptose portion (with only a Glu and/or Gal residue attached), the Rd chemotype (D21f1) contains the 3-deoxy-d-manno-octulosonic acid (Kdo) and heptose residues, while the D21f2 retains only the Kdo. The D21 strain is the only one that contains the phosphate residues associated with the LPS heptoses.
Bactericidal activity studies.
At a first approach, we examined bacterial viability at different protein concentrations using the luminescent BacTiter Pro reagent (Promega). Viable, metabolically active cells are measured by ATP quantification using a coupled luminescence detection assay. Thus, the luminescent signal is proportional to the amount of ATP required for the conversion of luciferin into oxyluciferin in the presence of luciferase. From 0.1 to 10 μM protein or peptide, IC50s, defined as the concentration at which cell viability is reduced to 50%, were determined. The results show that ECP antimicrobial activity is dependent on both LPS length and charge (Table 1). For strains D21 to D21e19, the antimicrobial activity is only slightly reduced, but a more severe decrease is observed for strain D21f2, having the fully truncated LPS. A comparison with the two representative ECP N-terminal peptides (Fig. 1C) provided complementary data on the bactericidal activity modulation by the LPS outer layer. For peptide [1-45], a modest increase in IC50s is detected when the LPS complexity is reduced. In turn, peptide [6-17]-Ahx-[23-36] displays an opposite trend compared with ECP, with strains D21f1 and D21f2 being the most sensitive to the peptide action (Table 1). Thus, LPS polysaccharide hinders peptide [6-17]-Ahx-[23-36] action rather than assisting it, contrary to what is observed for ECP and, to a lesser extent, for peptide [1-45].
Table 1.
Antimicrobial activities of ECP and derived peptides with E. coli K-12 D21 and defective strains D21e7, D21e19, D21f1, and D21f2a
| Strain | IC50, μM (mean ± SD) |
||
|---|---|---|---|
| ECP | [1-45] | [6-17]-Ahx-[23-36] | |
| D21 | 0.4 ± 0.1 | 0.9 ± 0.4 | 2.5 ± 0.1 |
| D21e7 | 0.6 ± 0.1 | 0.9 ± 0.2 | 2.9 ± 0.5 |
| D21e19 | 0.8 ± 0.1 | 1.2 ± 0.3 | 3.5 ± 0.3 |
| D21f1 | 1.3 ± 0.4 | 1.4 ± 0.3 | 1.8 ± 0.6 |
| D21f2 | 2.9 ± 0.4 | 1.4 ± 0.2 | 1.8 ± 0.1 |
Antimicrobial activity was calculated by following the bacterial viability as described in Materials and Methods.
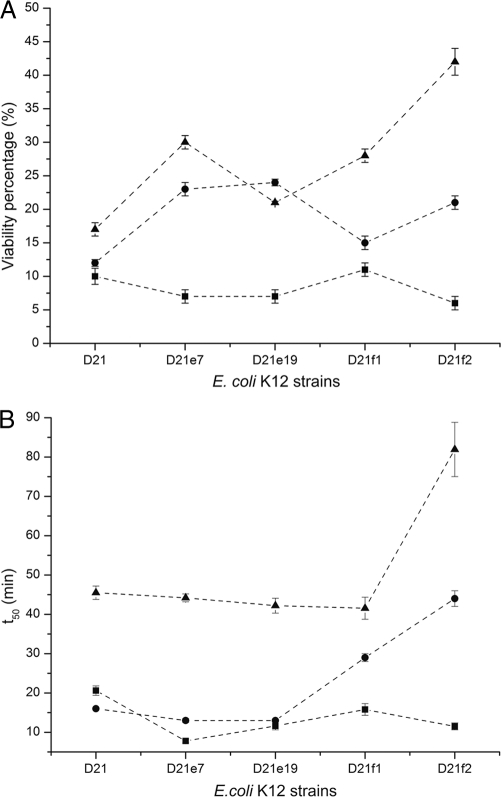
To further understand the role of LPS in the antimicrobial action of ECP, we followed the time course of the bactericidal process using the LIVE/DEAD bacterial viability kit, consisting of a 1:1 mixture of Syto 9 and propidium iodide (PI) nucleic acid dyes. Syto 9 can cross the cytoplasmic membrane and label all bacterial cells, while PI can access only the content of membrane-damaged cells, competing with and displacing bound Syto 9. The integration of Syto 9 and PI fluorescence provides an estimation of the viability percentage for monitoring the kinetics of the bactericidal process. On one hand, the values obtained for the cell viability percentage after 4 h of incubation (Fig. 2A) are consistent with the IC50s described above (Table 1). On the other hand, the time of half viability (t50), defined as the time needed to reduce cell viability to 50%, particularly increases for strain D21f2 with both ECP and peptide [1-45] (Fig. 2B). However, for [6-17]-Ahx-[23-36], t50 values are similar for all strains tested, suggesting that the bactericidal kinetics for the latter peptide are independent of LPS structure.
Fig 2.
Viability percentage and half time were determined with the LIVE/DEAD kit after 4 h of incubation of mid-log-phase-grown E. coli K-12 D21, D21e7, D21e19, D21f1, and D21f2 cultures with 5 μM protein and peptides. The percent viability (A) and half time of viability (B) after incubation with ECP (▴), [1-45] (●), or [6-17]-Ahx[23-36] (■) are depicted. The order of the strains on the x axis denotes increasing degrees of LPS truncation.
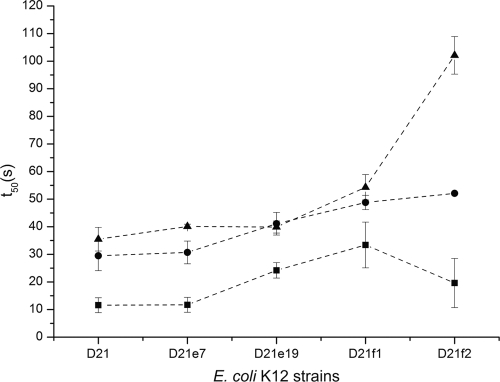
To examine the influence of LPS structure on ECP action at the bacterial cell membrane, we used the DiSC3(5) fluorescent probe, which is sensitive to membrane potential. The DiSC3(5) fluorescence is quenched upon interaction with intact cell membranes. When the membrane potential is lost, the probe is released to the medium, resulting in an increase of fluorescence that can be recorded as a function of time. Thus, we have measured the half-depolarization time (t50), defined as the time needed to achieve a half-depolarization effect. For ECP, the depolarization is delayed as a function of the LPS truncation grade (Fig. 3). However, peptide [6-17]-Ahx-[23-36] shows similar t50 values for all the strains tested, suggesting that depolarization is not affected by LPS composition. Furthermore, the presence or absence of EDTA in the incubation medium has no effect on membrane depolarization (data not shown). EDTA is a chelating agent that disrupts LPS structure by sequestrating divalent cations that tightly pack the polysaccharide moiety of LPS. These results suggest that both ECP and peptides by themselves may efficiently displace the divalent cations from the outer layer.
Fig 3.
Depolarization activity on E. coli K-12 D21, D21e7, D21e19, D21f1, and D21f2 cells was followed using the DiSC3(5) dye depolarization assay with a 5 μM protein concentration. The time needed to achieve a half-depolarizing effect is depicted for ECP (▴), [1-45] (●), and [6-17]-Ahx[23-36] (■). The order of the strains on the x axis denotes increasing degrees of LPS truncation.
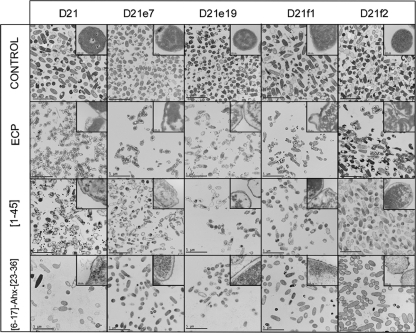
Finally, E. coli K-12 mutants were visualized by transmission electron microscopy (TEM) after incubation (Fig. 4). For ECP a complete disruption of cell integrity was observed. Outer bacterial layers were detached, and the cell shape was considerably altered, suggesting that ECP induces a mechanical disruption of the cell. Material condensation appears as electron-dense granular areas inside the cell, even in the D21f2 mutant strain. For peptide [1-45], a similar behavior is observed, though the outer layers seem to suffer less from mechanical disruption. Mutant strains D21f1 and D21f2 appear to be less condensed, and, particularly in strain D21f2, the outer layers seem to be well preserved. In contrast, for peptide [6-17]-Ahx-[23-36], the outer layer morphology is not significantly altered compared with that of control cells, and neither condensation nor material spillage outside the cell is observed. It is important to highlight that no bacterial agglutination can be observed by TEM due to sample preparation.
Fig 4.
Transmission electron micrographs of E. coli K-12 strains (D21, D21e7, D21e19, D21f1, and D21f2) incubated in the presence of 5 μM ECP, [1-45], or [6-17]-Ahx[23-36] for 4 h. The magnification scale is indicated at the bottom of each micrograph.
These results together suggest that ECP and peptide [1-45] display similar mechanisms of action, needing the entire LPS structure to ensure bacterial killing. In contrast, the action of peptide [6-17]-Ahx-[23-36] action is hindered by complex LPS structures, suggesting a different mechanism of action.
Studies of agglutinating activity.
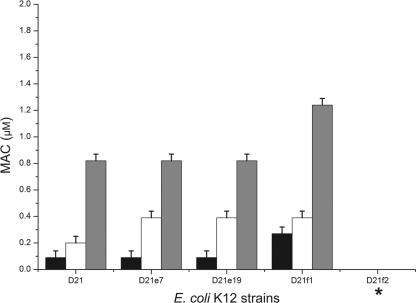
It has been described that ECP shows high LPS affinity and can agglutinate E. coli cells in the micromolar range (30). To quantify the agglutinating activity, we determined the minimal agglutination concentration (MAC), defined as the minimal peptide concentration able to induce agglutination in bacteria (35). Again, a correlation can be observed between LPS core complexity and agglutinating activity (Fig. 5). Strain D21f2, with the shortest LPS, shows no agglutination even after 12 h of incubation with 5 μM ECP. This behavior was observed for both ECP and peptides. However, peptide [6-17]-Ahx-[23-36] needed a much higher concentration to promote bacterial cell agglutination, perhaps due to its lower affinity for LPS (35).
Fig 5.
The agglutination activity was evaluated by the calculation of the minimal agglutination concentration (MAC) of the sample tested, corresponding to the first condition where bacterial aggregates are visible to the naked eye. The results for ECP (black bars), [1-45] (white bars), and [6-17]-Ahx[23-36] (gray bars) are shown. *, strain D21f2 does not show agglutination up to a concentration of 10 μM. The order of the strains on the x axis denotes increasing degrees of LPS truncation.
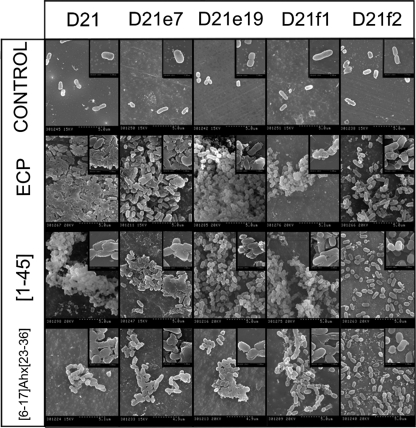
With the aim of studying cell surface morphology, we examined bacterial cultures by scanning electron microscopy (SEM). For all E. coli K-12 mutants, consistent cell damage was observed after incubation with both ECP and peptides (Fig. 6). For ECP, a high degree of cell agglutination, for strains D21 to D21f1, is observed. While the characteristic baton-shaped cell morphology is maintained, the bacterial cell wall appears to be considerably damaged and only strain D21f2 displays a low degree of agglutination, in agreement with the MAC experiments described above. For peptide [1-45], similar agglutination levels are observed, also showing a high level damage at the cell surface, except for strain D21f2. However, for peptide [6-17]-Ahx-[23-36], only small clusters of bacteria were observed, with the cell wall being only locally damaged. These observations agree with the TEM images, where neither material spillage nor material condensation was found.
Fig 6.
Scanning electron micrographs of E. coli K-12 strains (D21, D21e7, D21e19, D21f1, and D21f2) incubated in the presence of 5 μM ECP, [1-45], or [6-17]-Ahx[23-36] for 4 h. The magnification scale is shown at the bottom of each micrograph.
DISCUSSION
LPS is thought to act as a permeability barrier for antibiotics, detergents, and host proteins (25). It has been described that Gram-positive bacteria are more susceptible to specific antibiotics than Gram-negative, less permeable bacteria (29). Several hypotheses can explain how LPS modulates Gram-negative outer envelope permeability. Nikaido and coworkers have suggested that the carbohydrate moiety would act as a barrier against hydrophobic molecules (25), thus explaining why the permeativity of some antibiotics (e.g., cephalosporins) is proportional to their hydrophobicity (19). In fact, X-ray diffraction studies suggest that LPS thickness (i.e., its polysaccharide content) is inversely proportional to outer membrane permeability (29), supporting the aforementioned hypothesis (25).
On the other hand, some AMPs (e.g., lactoferrin or lysozyme) seem to decrease their antimicrobial activity with a progressively truncated LPS core (26), similarly to what was observed for ECP in the present study (Table 1). Additionally, ECP can induce the agglutination of Gram-negative bacteria, a behavior that is not shared with many other AMPs. Indeed, the agglutinating activity of ECP seems to be crucial for its antimicrobial action, suggesting that agglutination is required for bacterial killing (35).
The agglutinating activity is evidenced by SEM micrographs showing that rough mutants, but not the D21f2 deep-rough mutant, become clumped after treatment with ECP (Fig. 6). We thus suggest that the LPS polysaccharide moiety is essential to trigger Gram-negative bacterial cell agglutination, which is directly correlated with antimicrobial activity (Table 1; Fig. 5). For the D21f2 deep-rough mutant, agglutination is almost insignificant, which is translated to high MIC values, increased times to reduce cell culture viability, and slow depolarization kinetics (Table 1 and Fig. 2 and 3). We conclude from these data that ECP needs to first interact with the polysaccharide moiety of LPS, triggering cell agglutination. This process will mechanically disrupt the bacterial outer layer (Fig. 4), thus disturbing the membrane potential and bacterial homeostasis. Binding affinity to LPS was previously confirmed for the whole protein (33) and for the [1-45] peptide (32).
Simulations of docking of ECP to LPS show a high affinity for the complex (−16.53 kcal/mol). LPS is docked on the N-terminal cationic patch of ECP (Fig. 7A), with the protein interacting in the interface between lipid A and the polysaccharide moiety of LPS (Fig. 7B). Specific hydrogen-bonding contacts are observed for Arg1, Trp10, Gln14, Lys38, and Gln40 (Fig. 7C). These results may explain the severe decrease in the antimicrobial and agglutinating activities observed for the D21f2 mutant, which lacks the sugar molecules shown to bind to ECP.
The results were then compared with those for the ECP [1-45] peptide, which corresponds to the ECP antimicrobial domain located at the N terminus (32). We have observed here that the agglutinating activity is maintained, though it is slightly reduced (Fig. 5). The antimicrobial activity and the mechanical disruption of bacterial cells are also conserved (Table 1; Fig. 4). In fact, peptide [1-45] conserves the residues described to be crucial to bind sulfate anions (E. Boix et al., unpublished results), as well as phosphorylated and sulfated polysaccharides (16, 34), which also correspond to the domain identified by molecular docking (Fig. 7).
In the search for the minimum template that retains the antimicrobial action of ECP, peptide [6-17]-Ahx-[23-36] was designed and shown to conserve the antimicrobial activity against a wide battery of bacterial strains (35). Nonetheless, we find here that the mechanism of action is not conserved despite its equal activity. Indeed, the shorter peptide construct has limited LPS affinity (35). Peptide [6-17]-Ahx-[23-36] displays a significant reduction of bacterial agglutination and does not cause notable mechanical cell disruption (Fig. 4 and 5). Peptide [6-17]-Ahx-[23-36] is more efficient in membrane depolarization (Fig. 3), which is not affected by LPS composition, a behavior that seems to correlate with cell viability kinetics (Fig. 2). In this case, LPS length is inversely correlated with MIC values (Table 1) suggesting that the polysaccharide LPS moiety would act as a permeation barrier. Indeed, peptide [6-17]-Ahx-[23-36] is the most hydrophobic compound, with a hydrophobic-to-hydrophilic ratio of 0.9 (ratios of 0.6 and 0.8 were calculated for ECP and peptide [1-45], respectively), in agreement with the hypothesis that LPS would act as a barrier for more-hydrophobic compounds. The mechanism of action displayed by peptide [6-17]-Ahx-[23-36] is also more lytic, though it turns out to be less selective for bacterial membranes, increasing the hemolytic activity compared with that of ECP (35). Thus, ECP and its N-terminal domain show a higher selectivity for bacterial structures, favoring agglutination and cell wall damage instead of having a mere cell membrane-lytic mechanism.
In summary, we show that both ECP and its N terminus display antimicrobial and agglutinating activities that are dependent on LPS structure. However, the shorter peptide [6-17]-Ahx-[23-36] displays lower agglutinating activity though retaining the antimicrobial activity, suggesting a different mechanism of action. We conclude that the antimicrobial domain of ECP exerts a global action on bacteria by mechanically disrupting the outer layer, whereas shorter derived peptides have a more unspecific action directly at the inner membrane level.
ACKNOWLEDGMENTS
The work was supported by the Ministerio de Educación y Cultura (grant number BFU2009-09371), cofinanced by FEDER funds and by the Generalitat de Catalunya (2009 SGR 795). D.P is a recipient of a UAB predoctoral fellowship, and M.T. is a recipient of an Alianza Cuatro Universidades postdoctoral fellowship.
Spectrofluorescence measurements were performed at the Laboratori d'Anàlisi i Fotodocumentació, UAB. We thank Alejandro Sánchez-Chardi for his assistance with the electron microscopy samples at the Servei de Microscopia, UAB.
Footnotes
Published ahead of print 13 February 2012
REFERENCES
- 1. Boix E, et al. 1999. Kinetic and product distribution analysis of human eosinophil cationic protein indicates a subsite arrangement that favors exonuclease-type activity. J. Biol. Chem. 274:15605–15614 [DOI] [PubMed] [Google Scholar]
- 2. Boix E, Nogues MV. 2007. Mammalian antimicrobial proteins and peptides: overview on the RNase A superfamily members involved in innate host defence. Mol. Biosyst. 3:317–335 [DOI] [PubMed] [Google Scholar]
- 3. Boix E, Torrent M, Sanchez D, Nogues MV. 2008. The antipathogen activities of eosinophil cationic protein. Curr. Pharm. Biotechnol. 9:141–152 [DOI] [PubMed] [Google Scholar]
- 4. Brandenburg K, Andra J, Garidel P, Gutsmann T. 2011. Peptide-based treatment of sepsis. Appl. Microbiol. Biotechnol. 90:799–808 [DOI] [PubMed] [Google Scholar]
- 5. Brandenburg K, et al. 2010. Molecular basis for endotoxin neutralization by amphipathic peptides derived from the alpha-helical cationic core-region of NK-lysin. Biophys. Chem. 150:80–87 [DOI] [PubMed] [Google Scholar]
- 6. Brandenburg K, et al. 1993. Influence of the supramolecular structure of free lipid A on its biological activity. Eur. J. Biochem. 218:555–563 [DOI] [PubMed] [Google Scholar]
- 7. Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238–250 [DOI] [PubMed] [Google Scholar]
- 8. Brown KL, Hancock RE. 2006. Cationic host defense (antimicrobial) peptides. Curr. Opin. Immunol. 18:24–30 [DOI] [PubMed] [Google Scholar]
- 9. Caroff M, Karibian D. 2003. Structure of bacterial lipopolysaccharides. Carbohydr Res. 338:2431–2447 [DOI] [PubMed] [Google Scholar]
- 10. Carreras E, Boix E, Rosenberg HF, Cuchillo CM, Nogues MV. 2003. Both aromatic and cationic residues contribute to the membrane-lytic and bactericidal activity of eosinophil cationic protein. Biochemistry 42:6636–6644 [DOI] [PubMed] [Google Scholar]
- 11. Cohen J. 2002. The immunopathogenesis of sepsis. Nature 420:885–891 [DOI] [PubMed] [Google Scholar]
- 12. Delcour AH. 2009. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 1794:808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferguson AD, et al. 2001. Active transport of an antibiotic rifamycin derivative by the outer-membrane protein FhuA. Structure 9:707–716 [DOI] [PubMed] [Google Scholar]
- 14. Fields GB, Noble RL. 1990. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept Protein Res. 35:161–214 [DOI] [PubMed] [Google Scholar]
- 15. Frecer V, Ho B, Ding JL. 2004. De novo design of potent antimicrobial peptides. Antimicrob. Agents Chemother. 48:3349–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. García-Mayoral MF, et al. 2010. NMR structural determinants of eosinophil cationic protein binding to membrane and heparin mimetics. Biophys. J. 98:2702–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gutsmann T, et al. 2010. New antiseptic peptides to protect against endotoxin-mediated shock. Antimicrob. Agents Chemother. 54:3817–3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hancock RE. 1997. Peptide antibiotics. Lancet 349:418–422 [DOI] [PubMed] [Google Scholar]
- 19. Hiruma R, Yamaguchi A, Sawai T. 1984. The effect of lipopolysaccharide on lipid bilayer permeability of beta-lactam antibiotics. FEBS Lett. 170:268–272 [DOI] [PubMed] [Google Scholar]
- 20. Jeong H, et al. 2009. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J. Mol. Biol. 394:644–652 [DOI] [PubMed] [Google Scholar]
- 21. Laurents DV, et al. 2009. The (1)H, (13)C, (15)N resonance assignment, solution structure, and residue level stability of eosinophil cationic protein/RNase 3 determined by NMR spectroscopy. Biopolymers 91:1018–1028 [DOI] [PubMed] [Google Scholar]
- 22. Mallorqui-Fernandez G, et al. 2000. Three-dimensional crystal structure of human eosinophil cationic protein (RNase 3) at 1.75 A resolution. J. Mol. Biol. 300:1297–1307 [DOI] [PubMed] [Google Scholar]
- 23. Mangoni ML. 2011. Host-defense peptides: from biology to therapeutic strategies. Cell. Mol. Life Sci. 68:2157–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matsuzaki K, Sugishita K, Miyajima K. 1999. Interactions of an antimicrobial peptide, magainin 2, with lipopolysaccharide-containing liposomes as a model for outer membranes of gram-negative bacteria. FEBS Lett. 449:221–224 [DOI] [PubMed] [Google Scholar]
- 25. Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Prokhorenko IR, Zubova SV, Ivanov AY, Grachev SV. 2009. Interaction of Gram-negative bacteria with cationic proteins: dependence on the surface characteristics of the bacterial cell. Int. J. Gen. Med. 2:33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Raetz CR. 1990. Biochemistry of endotoxins. Annu. Rev. Biochem. 59:129–170 [DOI] [PubMed] [Google Scholar]
- 28. Sanchez D, et al. 2011. Mapping the eosinophil cationic protein antimicrobial activity by chemical and enzymatic cleavage. Biochimie 93:331–338 [DOI] [PubMed] [Google Scholar]
- 29. Snyder DS, McIntosh TJ. 2000. The lipopolysaccharide barrier: correlation of antibiotic susceptibility with antibiotic permeability and fluorescent probe binding kinetics. Biochemistry 39:11777–11787 [DOI] [PubMed] [Google Scholar]
- 30. Torrent M, et al. 2010. Comparison of human RNase 3 and RNase 7 bactericidal action at the Gram-negative and Gram-positive bacterial cell wall. FEBS J. 277:1713–1725 [DOI] [PubMed] [Google Scholar]
- 31. Torrent M, et al. 2007. Topography studies on the membrane interaction mechanism of the eosinophil cationic protein. Biochemistry 46:720–733 [DOI] [PubMed] [Google Scholar]
- 32. Torrent M, de la Torre BG, Nogues VM, Andreu D, Boix E. 2009. Bactericidal and membrane disruption activities of the eosinophil cationic protein are largely retained in an N-terminal fragment. Biochem. J. 421:425–434 [DOI] [PubMed] [Google Scholar]
- 33. Torrent M, Navarro S, Moussaoui M, Nogues MV, Boix E. 2008. Eosinophil cationic protein high-affinity binding to bacteria-wall lipopolysaccharides and peptidoglycans. Biochemistry 47:3544–3555 [DOI] [PubMed] [Google Scholar]
- 34. Torrent M, Nogues MV, Boix E. 2011. Eosinophil cationic protein (ECP) can bind heparin and other glycosaminoglycans through its RNase active site. J. Mol. Recognit. 24:90–100 [DOI] [PubMed] [Google Scholar]
- 35. Torrent M, et al. 2011. Refining the eosinophil cationic protein antibacterial pharmacophore by rational structure minimization. J. Med. Chem. 54:5237–5244 [DOI] [PubMed] [Google Scholar]
- 36. Torrent M, et al. 2009. Comparison of the membrane interaction mechanism of two antimicrobial RNases: RNase 3/ECP and RNase 7. Biochim. Biophys. Acta 1788:1116–1125 [DOI] [PubMed] [Google Scholar]
- 37. Venge P. 2010. The eosinophil and airway remodelling in asthma. Clin. Respir. J. 4(Suppl 1):15–19 [DOI] [PubMed] [Google Scholar]