Abstract
TMC207 is a new antituberculous drug belonging to the diarylquinoline class which very efficiently inhibits the ATP synthase of mycobacteria such as Mycobacterium tuberculosis, one of the most important pathogens in the world. In order to map the amino acid residues involved in the binding of the drug, we have selected in vitro TMC207-resistant mutants from M. tuberculosis and diverse atypical mycobacteria. Six distinct mutations, Asp28→Gly, Asp28→Ala, Leu59→Val, Glu61→Asp, Ala63→Pro, and Ile66→Met, have been identified in the subunit c forming a C ring in the ATP synthase. They were studied by evaluating the levels of resistance that they confer in the selected clones and by using an isogenic complementation system in Mycobacterium smegmatis. The rates of increase of TMC207 MIC values (8- to 133-fold) were interpreted by constructing by homology modeling a structure of the mycobacterial C ring which was used for docking simulations with TMC207. Our results suggest that the residues found to be mutated in the resistant clones, together with a tyrosine specifically conserved at position 64 in mycobacteria, define a cleft located between two adjacent c subunits in the C ring. This cleft, which encompasses the proton-binding site (Glu61), is well fitted to bind TMC207 at the level of the bromoquinoline moiety, with the drug being anchored by several ionic, hydrogen, and halogen bonds with residues Glu61, Tyr64, and Asp28, respectively. These data shed light on the molecular interactions allowing TMC207 to bind specifically and efficiently at the level of the proton-binding site of the mycobacterial C ring.
INTRODUCTION
Tuberculosis (TB) is a major cause of death from infectious disease, with more than 9 million incident cases and 1.3 million deaths in 2009, making TB a global public health issue. Because TB therapy is long, complex, and significantly hampered by the growing development of both multidrug-resistant TB (MDR) and extensively drug-resistant TB (XDR), the current antituberculosis arsenal is more and more compromised and new drugs acting against new targets are urgently required.
The diarylquinoline TMC207 (previously R207910; Fig. 1) is a very promising candidate drug for treatment of TB and mycobacterial infections (2) and is currently in phase IIb clinical trials for patients with multidrug-resistant tuberculosis (12). TMC207 targets ATP synthase with a range of activity specifically restricted to mycobacteria, with no significant activity against other bacteria (2) or against eukaryotic cells (18). The activity spectrum of TMC207 includes the mycobacterial species that are pathogenic to humans, such as Mycobacterium tuberculosis, but also atypical pathogenic species, such as Mycobacterium avium complex, Mycobacterium kansasii, and the fast growers Mycobacterium fortuitum and Mycobacterium abscessus (2).
Fig 1.
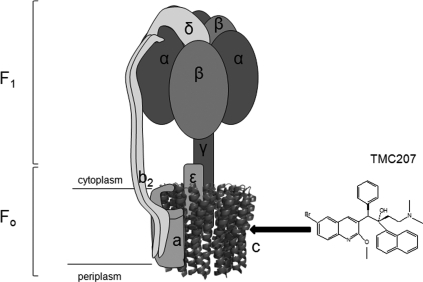
Structure of TMC207 (R,S geometry) and subunit arrangements of [α3β3γε-ab2-C ring] in the F-type ATP synthase. The region of interaction of the drug in the F-ATP synthase is shown by a black arrow pointing in subunit c to the location of the essential residue Asp or Glu allowing the ion transfer.
F-type ATP synthase, an ubiquitous enzyme in eukaryotic and prokaryotic cells, utilizes the energy stored in an H+ or Na+ electrochemical gradient to synthesize ATP (7, 13, 38, 42). The ATP synthase complex is composed of two sectors, i.e., a water-soluble sector (F1) that is bound to a membrane-embedded sector (FO) (Fig. 1). In bacteria, F1 is composed of five subunits with an α3β3γδε stoichiometry and contains three catalytic sites for ATP synthesis and/or hydrolysis in the α-β interface (Fig. 1). FO is formed by three subunits with an ab2c8-15 stoichiometry (Fig. 1) and functions as an ion-conducting pathway (9, 22, 26, 27, 32, 35, 40). Ion translocation through FO drives rotation of a cylindrical ring of subunits c that is coupled to rotation of the subunit γ within the α3β3 hexamer of F1, a rotation that drives synthesis of ATP (1, 4, 7, 13, 29, 38, 39, 42). Subunit c is a hairpin structure comprising two membrane-spanning α helices connected by a hydrophilic loop at the cytoplasmic side of the membrane and which carries the essential ion carrier (Asp or Glu) in the middle of the C-terminal α helix (Fig. 1) (26). This subunit is known to form an oligomeric ring, the stoichiometry of which varies between 8 and 15 depending on the species (39). Subunit a, composed of five transmembrane helices (36), interacts with subunit c at the periphery of the C ring, forming the a-c interface (3). The essential residue Arg210 (Escherichia coli numbering) of subunit a is postulated to facilitate the protonation/deprotonation cycle of the proton carrier Asp or Glu of subunit c and contributes to the rotation of the C ring during ion translocation through FO (13, 34, 38).
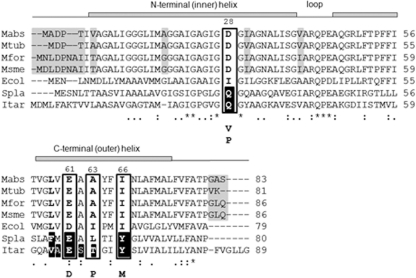
TMC207 targets the C ring of ATP synthase, as suggested by the identification of mutants of M. tuberculosis and Mycobacterium smegmatis resistant to the drug and having mutations in subunit c at positions 28 (Asp→Val/Pro), 61 (Glu→Asp), 63 (Ala→Pro), and 66 (Ile→Met) in the M. tuberculosis amino acid numbering system (Fig. 2) (2, 21, 30). The hypothesis is further supported by the fact that the mycobacterial species naturally resistant to TMC207, i.e., Mycobacterium xenopi, Mycobacterium novacastrense, and Mycobacterium shimoidei, display a Met at position 63 in subunit c in place of a conserved Ala in the species susceptible to the drug (20, 30).
Fig 2.
Multiple-sequence alignment of the subunits c from M. abscessus (Mabs), M. tuberculosis (Mtub), M. fortuitum (Mfor), M. smegmatis (Msme), E. coli (Ecol), Spirulina platensis (Spla), and I. tartaricus (Itar). Identical residues are marked with an asterisk. The gray shadow highlights variability between the amino acids from the mycobacterial species. The mutated positions previously reported in the drug-resistant mycobacterial strains (positions 28, 61, 63, and 66, M. tuberculosis numbering) are boxed. The mutated residues are indicated below the boxes. Amino acids shaded in black in the sequences of I. tartaricus and S. platensis correspond to the amino acids involved in the H+/Na+ binding in the subunits c of these species. Secondary-structure elements are indicated above the sequences.
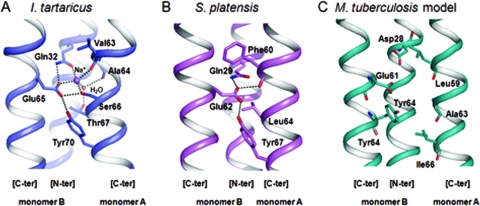
No crystal structure information on the mycobacterial C ring is available to date. The five crystal structures of C rings available in the Protein Data Bank (PDB) for F-type ATP synthases are the structures of Ilyobacter tartaricus (11 subunits c, PDB accession no. 2WGM) (25), spinach chloroplast (14 subunits c, PDB accession no. 2W5J) (37), the cyanobacterium Spirulina platensis (15 subunits c, PDB accession no. 2WIE) (32), Bacillus pseudofirmus OF4 (13 subunits c, accession no. PDB 2X2V) (33), and Saccharomyces cerevisiae yeast (10 subunits c, PDB accession no. 2WPD) (9). These C-ring structures are Na+-binding (I. tartaricus) or H+-binding (spinach chloroplast, S. platensis, B. pseudofirmus, yeast) ATP synthases. Despite the fact that the amino acid composition of the c subunits in ATP synthases is significantly variable (Fig. 2), these structures present common features related to the binding of the H+/Na+ ion. In the two structures representative of bacterial ATP synthases (S. platensis and I. tartaricus), each ion binding site lies in between two adjacent c subunits, and the conserved glutamate residue (Glu65 in I. tartaricus, Glu62 in S. platensis, corresponding to Glu61 in M. tuberculosis) (Fig. 3) provides the main stabilizing interaction for the H+/Na+ ion, which is also stabilized by another interaction with the side chain of a glutamine residue from the same subunit (Gln32 in I. tartaricus, Gln29 in S. platensis, corresponding to Asp28 in M. tuberculosis, carried by monomer B in Fig. 3). The backbone carbonyl oxygen atom of a residue held on by a neighboring subunit facing the conserved glutamate (Val63 in I. tartaricus, Phe60 in S. platensis, corresponding to Leu59 in M. tuberculosis, carried by monomer A in Fig. 3) also contributes to stabilize the ion (Fig. 3A and B). Furthermore, the conserved glutamate residue establishes a hydrogen bond with the hydroxyl group of a tyrosine residue (Tyr70 in I. tartaricus, Tyr67 in S. platensis, corresponding to Tyr64 in M. tuberculosis) (Fig. 3). These intramolecular interactions represent a common mode of ion binding in the C rings of several bacterial species, as well as of cyanobacteria and chloroplasts (24).
Fig 3.
Na+/H+ binding sites in the crystallographic structures of I. tartaricus (A) (35) and S. platensis (B) (34). The amino acids found in the ion coordination regions are shown in stick representation. Hydrogen bonds are represented by dashed lines. (C) Structural model of the same region in M. tuberculosis (this study). The amino acids shown in stick representation (apart from Tyr64) are those implicated in TMC207 resistance.
Here we report the isolation of TMC207-resistant mutants obtained (i) from M. tuberculosis H37Rv and three M. tuberculosis clinical strains, (ii) from M. smegmatis, and (iii) for the first time, from two other atypical mycobacteria which can cause diseases in humans: Mycobacterium abscessus and Mycobacterium fortuitum. Six different amino acid mutations implicated in TMC207 resistance were identified, three of them having never been observed previously. The level of resistance conferred by these mutations was evaluated by determining the MICs of the mutated strains as well as those of an M. smegmatis wild-type (wt) strain complemented with the mutated genes. Finally, by using the crystal structures of the homologous C ring of S. platensis (32), a model structure of the C ring of M. tuberculosis was built and used to predict the binding site of TMC207 on the C ring and the interactions that the drug could establish with its target.
MATERIALS AND METHODS
Bacterial strains, culture media, and antibiotics.
M. tuberculosis H37Rv and M. smegmatis mc2155 were used as reference strains. The other strains of M. tuberculosis, M. abscessus, and M. fortuitum were clinical isolates from our laboratory collection. M. smegmatis was cultured in brain heart infusion (BHI) medium (Difco) with 0.5% Tween 80 (Sigma) at 37°C with shaking. The other species were cultured in Middlebrook 7H9 medium (Difco) supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC Middlebrook) at 37°C for M. tuberculosis and 30°C for atypical mycobacteria. When required, the media were supplemented with 100 mg/liter of zeocin (Invitrogen). E. coli TOP10 was used as the host for plasmid transformation experiments. The cultures were incubated at 37°C in BHI medium supplemented with 25 mg/liter of zeocin.
The diarylquinoline TMC207 was provided by Johnson & Johnson Pharmaceutical Research and Development.
Determination of MICs of TMC207 against Mycobacterium tuberculosis and other mycobacterial species.
MICs were determined on 7H11 agar supplemented with 10% OADC containing serial 2-fold dilutions of TMC207 ranging from 0.0007 to 64 mg/liter. Plates were incubated at 30°C or 37°C for 3 to 42 days (depending on the mycobacterial species). The MIC values of the M. smegmatis strains complemented by the wild type and the mutated atpE genes were determined on BHI–Tween 80 agar supplemented with zeocin and containing increasing concentrations of TMC207 varying from 0.0007 to 32 mg/liter. Plates were incubated at 37°C for 5 days. The MIC was defined as the lowest concentration at which no growth was visible.
Isolation of resistant mutants and characterization of mutations.
For atypical mycobacteria, a 3-day culture of 4 ml of BHI–Tween 80 medium was inoculated in 300 ml of the same medium for M. smegmatis and of 7H9-OADC medium for M. fortuitum and M. abscessus. After 3 days, these cultures were used as inoculum either directly (containing approximately 109 CFU/ml) or after 10- to 100-fold concentration of the cells by centrifugation. Each inoculum (100 μl) was spread onto BHI–Tween 80 or 7H9-OADC agar containing different concentrations of TMC207 varying from 0.003 mg/liter to 1 mg/liter for M. smegmatis (MIC = 0.007 mg/liter), 0.015 mg/liter to 0.5 mg/liter for M. fortuitum (MIC = 0.01 mg/liter), and 0.25 mg/liter to 8 mg/liter for M. abscessus (MIC = 0.25 mg/liter). Cultures were incubated at 30°C for 3 days.
For M. tuberculosis, the strains were grown for 1 week without shaking at 37°C in 15 ml of Dubos-OADC medium. This culture was used to inoculate 300 ml of Dubos-OADC medium. After 7 days of growth, 100 μl of the culture (108 CFU/ml) was spread, directly or after concentration by a factor 10 or 100, onto 7H11-OADC agar containing six concentrations of TMC207 varying from 0.03 mg/liter to 0.5 mg/liter (MIC = 0.015 to 0.06 mg/liter). Cultures were incubated at 37°C for 6 weeks.
The DNA was extracted from a cell suspension of the mutants growing on the different concentrations of TMC207 by heat shock by five alternate cycles of 1 min incubation in boiling water and 1 min on ice. DNA amplification of the atpE gene coding for the ATP synthase subunit c was carried out using the degenerated primers atpBS and atpFAS (Table 1), as previously described (30). The atpB gene coding for the subunit a of ATP synthase of M. tuberculosis and M. abscessus was amplified with the primers atpBBKS and atpBBKAS and primers atpBABS_S and atpEABS_AS, respectively (Table 1). The entire ATP synthase operon was amplified with 8 primer pairs from three clones of M. tuberculosis: opeatpBKS1/atpBBKAS, atpEBKLYGS/opeatpBKAS2, opeatpBKS3/opeatpBKAS4, opeatpBKS5/opeatpBKAS6, opeatpBKS7/opeatpBKAS8, opeatpBKS9/opeatpBKAS10, opeatpBKS11/opeatpBKAS12, and opeatpBKS13/opeatpBKAS14 (Table 1). Each pair allows amplification of about 1,000 bp in the ATP synthase operon. All the PCR products were sequenced, and the sequences were compared with those of the wild-type genes to find mutations.
Table 1.
Primers used for DNA amplification, sequencing, and site-directed mutagenesis of atp genes of Mycobacteria species
| Primer name | Sequence (5′ to 3′) |
|---|---|
| atpBS | TGTAYTTCAGCCARGCSATGG |
| atpFAS | CCGTTSGGDABGAGGAAGTTG |
| atpBBKS | ATGACTGAGACCATCCTGGCC |
| atpBBKAS | AATCGCGCTCACTTCACCCAT |
| atpBABS_S | TTCGGGGTTGCGTTGTTCCAGG |
| atpEABS_AS | AACAATTGTGGGGTCCGCCAT |
| opeatpBKS1 | CGGGTGGTTTCCGCTCCATCA |
| opeatpBKAS2 | TGATGCG CTCCGCCTCGACGTC |
| opeatpBKS3 | AGCGCACCGTGTTGTGGAAG |
| opeatpBKAS4 | TAGATACGGCTCAGCACTTCGG |
| opeatpBKS5 | TGCGGTGGCTCGCCGCGGCGAAA |
| opeatpBKAS6 | TTTGAAACCGGCGGA CTCCGACGC |
| opeatpBKS7 | TCGGGCAGAAGGGAACTACCATCG |
| opeatpBKAS8 | GCTATTTCTTCTTCTTCGGCGCCGG |
| opeatpBKS9 | GCTTCGCGGCCACCGGTGGCGGCT |
| opeatpBKAS10 | ACCTAGCTTGGGCCTAGCGGGCTTC |
| opeatpBKS11 | ACGCTGATGGCAAACCGCGAGCG |
| opeatpBKAS12 | GCCAGCGTGGGCTGGTATCCCAC |
| opeatpBKS13 | CAAGACGTATTGCTGTTC ATCGACAACA |
| opeatpBKAS14 | ACAACGACGAGCACGACCATGCCGAT |
| atpEsmeLYGE | GGAATTCTTACTGAAGGCCAGGAGTGGC |
| atpEsmeLYGS | TCCCCCGGGATCTCGATCCCAACGCCATCA |
| atpEBKLYGE | GGAATTCTTACTTGACGGGTGTAGCGAA |
| atpEBKLYGS | TCCCCCGGGACCCCACTATCGCTGCCGGCG |
| pLYG204 | GGTTTCATCCCCGATCCGGAG |
| Primers used to mutate the M. smegmatis atpE gene | |
| atpEsmeI66MS | GCCGCGTACTTCATGAACCTGGCCTTCATGGCG |
| atpEsmeI66MAS | CGCCATGAAGGCCAGGTTCATGAAGTACGCGGC |
| atpEsmeD28GS | GCCGGTATCGGCGGCGGTATCGCGGGTAACGCG |
| atpEsmeD28GAS | CGCGTTACCCGCGATACCGCCGCCGATACCGGC |
| atpEsmeD28VS | GCCGGTATCGGCGTCGGTATCGCGGGTAACGCG |
| atpEsmeD28VAS | CGCGTTACCCGCGATACCGACGCCGATACCGGC |
| atpEsmeD28AS | GCCGGTATCGGCGCCGGTATCGCGGGTAACGCG |
| atpEsmeD28AAS | CGCGTTACCCGCGATACCGGCGCCGATACCGGC |
| atpEsmeE61DS | GTCGGTCTGGTGGACGCCGCGTACTTCATC |
| atpEsmeE61DAS | GATGAAGTACGCGGCGTCCACCAGACCGAC |
| atpEsmeA63PS | GGTCTGGTGGAAGCCCCGTACTTCATCAACCTGGCC |
| atpEsmeA63PAS | GGCCAGGTTGATGAAGTACGGGGCTTCCACCAGACC |
| atpEsmeL59VS | CCGTTCTTCATCACCGTCGGTGTGGTGGAAGCCGCGTA |
| atpEsmeL59VAS | TACGCGGCTTCCACCACACCGACGGTGATGAAGAACGG |
| atpEsmeA63MS | GGTCTGGTGGAAGCCATGTACTTCATCAACCTGGCC |
| atpEsmeA63MAS | GGCCAGGTTGATGAAGTACATGGCTTCCACCAGACC |
| Primers used to mutate the M. tuberculosis atpE gene | |
| atpEBKA63PS | GGTTTGGTTGAGGCGCCATACTTCATCAACCTGGCG |
| atpEBKA63PAS | CGCCAGGTTGATGAAGTATGGCGCCTCAACCAAACC |
| atpEBKE61DS | GTCGGTTTGGTTGACGCGGCATACTTCATCAAC |
| atpEBKE61DAS | GTTGATGAAGTATGCCGCGTCAACCAAACCGAC |
| atpEBKL59VS | CGTTCTTCATCACCGTCGGTGTGGTTGAGGCGGCATAC |
| atpEBKL59VAS | GTATGCCGCCTCAACCACACCGACGGTGATGAAGAACG |
| atpEBKI66MS | GCGGCATACTTCATGAACCTGGCGTTTATGGCG |
| atpEKI66MAS | CGCCATAAACGCCAGGTTCATGAAGTATGCCGC |
| atpEBKD28GS | GCCGGTATCGGTGGCGGTGTCGCCGGTAACGCG |
| atpEBKD28GAS | CGCGTTACCGGCGACACCGCCACCGATACCGGC |
Complementation assay.
The wild-type atpE genes of M. smegmatis and M. tuberculosis were amplified by PCR with the primers atpEsmeLYGE and atpEsmeLYGS and primers atpEBKLYGE and atpEBKLYGS, respectively (Table 1). The amplified genes were directly inserted in the pMOSBlue vector using a pMOSBlue blunt-ended cloning kit (Amersham Biosciences). The recombinant vectors obtained were digested with EcoRI and SmaI to generate an insert compatible with the pLYG204.zeo plasmid (16) digested with enzymes EcoRI and MscI. After ligation, the recombinant pLYG204 vectors containing the atpE genes (pLYG-atpEsme for atpE from M. smegmatis and pLYG-atpEBK for atpE from M. tuberculosis) were introduced by transformation in E. coli TOP10. Electroporated cells were preincubated in LB medium for 1 h at 37°C, before being plated on BHI agar supplemented with zeocin (25 mg/ml) and grown for 1 day at 37°C. The presence of the recombinant vector in the colonies was checked by PCR and DNA sequencing using the primers pLYG204 and atpEsmeLYGE for M. smegmatis or atpEBKLYGE for M. tuberculosis (Table 1).
Point mutations were introduced in the pLYG204 recombinant vectors containing the atpE genes of M. smegmatis and M. tuberculosis by using a QuikChange site-directed mutagenesis kit (Stratagene). The mutagenic primers (Table 1) were designed from the sequence of the atpE wild-type gene. The mutated pLYGatpE plasmids were prepared from E. coli TOP10 transformants before being introduced by electroporation in M. smegmatis mc2155. After 2 h of culture at 37°C in BHI–Tween 80 medium, the M. smegmatis transformants were plated on BHI–Tween 80 agar supplemented with zeocin and grown at 37°C for 4 days.
Southern blot hybridization.
Genomic DNA was extracted from M. smegmatis and M. tuberculosis as previously described (31), except that M. tuberculosis DNA was extracted with chloroform-isoamyl alcohol and N-cetyl-N,N,N-trimethylammonium bromide (Sigma). Enzyme digestions were performed with PstI and BstEII for M. smegmatis DNA and with MscI and NcoI for M. tuberculosis DNA. The restriction fragments were separated by electrophoresis on a 1% agarose gel at 100 V for 15 min and then at 40 V for 15 h. Southern blot hybridizations were performed as previously described (31) using as probes the amplified atpE genes of M. smegmatis and M. tuberculosis.
Construction of model structures of C rings of M. tuberculosis and M. smegmatis and docking simulations.
The models of the C rings of the ATP synthases of M. tuberculosis and M. smegmatis were built by homology modeling from the AtpE amino acid sequences of strains H37Rv and mc2155, respectively, using the software MODELLER (15) and the crystal structure of the c subunit of S. platensis as the template. The entire structure of the C ring was obtained using the Secondary Structure Matching algorithm from the Coot tools (14). The structures obtained were energy minimized in 100 steps using the program CNS (5).
The TMC207 structure was constructed using the module Modeler in the Insight II program (Accelrys). Hydrogen atoms were added, and the molecule was energy minimized using the CHARMM program and the force field PARAM22. The ligand was docked into the C ring structure using the AUTODOCK program (version 4) (28). The grid of the docking simulation was defined by a cube 40 by 40 by 40 Å centered at a position located in the vicinity of the Glu61 residue within the binding site. The docking simulations were performed using a Lamarckian genetic algorithm search routine whose parameters were set using the Autodock tools (ADT): in total, 150 runs were made, with a population size of 250 million and 25 million energy evaluations. The best protein-ligand docked complexes were selected on the basis of various parameters: the Autodock clustering scenario (root mean square based), the Autodock intermolecular energy, and, most importantly, the reasonableness of the interactions between the ligand and the protein.
RESULTS
Selection of mutants.
Mutants of M. tuberculosis, M. fortuitum, M. abscessus, and M. smegmatis resistant to TMC207 were selected in vitro on various concentrations of the drug. The presence of mutations in subunit c was searched for by amplifying and determining the nucleotide sequence of the atpE gene and the 78-bp upstream and 121-bp downstream regions, using the degenerated primers atpBS and atpFAS (Table 1) (30). As shown in Table 2, the mutation frequencies varied from 10−8 to 5.10−8, whatever was the species considered. Among the mutants obtained, the number of TMC207-resistant colonies showing a mutation in atpE varied according to the strains used for selection. In M. tuberculosis and M. abscessus, the ratio of strains showing a mutation relative to the total number of clones tested ranged from 16 to 83%. In the two atypical species M. fortuitum and M. smegmatis, 100% of the mutants had a mutation in AtpE (Table 2).
Table 2.
Phenotypic and genotypic characteristics of mutants resistant to TMC207 obtained from two reference strains (M. tuberculosis H37Rv and M. smegmatis mc2155) and five clinical isolates (M. tuberculosis P, D, and LQ, M. fortuitum B, and M. abscessus V)
| Strain | Mutation frequency | No. of atpE mutants/no. of colonies tested | Mutation found in subunit C (no. of mutants) | MIC (mg/liter) |
Rate of MIC increase | |
|---|---|---|---|---|---|---|
| Mutant | Wild type | |||||
| M. tuberculosis | 0.03 | |||||
| H37Rv | 10−8 | 4/25 | A63P (4) | 4 | 133 | |
| P | 5 × 10−8 | 5/7 | D28G (4) | 0.5 | 16 | |
| E61D (1) | NDa | |||||
| D | 10−8 | 5/10 | A63P (2) | 4 | 133 | |
| L59V (1) | 0.25 | 8 | ||||
| E61D (2) | 0.5 | 16 | ||||
| LQ | 10−8 | 5/6 | D28G (1) | 0.5 | 16 | |
| E61D (2) | 1 | 33 | ||||
| A63P (1) | 4 | 133 | ||||
| I66M (1) | 1 | 33 | ||||
| M. fortuitum B | 1.5 × 10−8 | 9/9 | D28A (9) | 4 | 0.010 | 400 |
| M. abscessus V | 1.5 × 10−8 | 8/17 | A63P (1) | 16 | 0.25 | 64 |
| D28A (7) | 8 | 32 | ||||
| M. smegmatis mc2155 | ND | 8/8 | I66M (8) | 16 | 0.12 | 133 |
ND, not determinable.
Four of 25 mutants obtained for M. tuberculosis H37Rv which had an amino acid substitution in AtpE exhibited the same mutation, Ala63→Pro. In contrast, a higher diversity of mutations was observed in the 23 mutants obtained from the M. tuberculosis clinical isolates, with 5 different amino acid substitutions detected: Asp28→Gly, Leu59→Val, Glu61→Asp, Ala63→Pro, and Ile66→Met (Table 2). Regarding the mutants selected from atypical mycobacteria, a single type of mutation was detected in the nine mutants of M. fortuitum (Asp28→Ala) and the eight mutants of M. smegmatis (Ile66→Met), and two types were detected in the 8 mutants of M. abscessus (Asp28→Ala in seven clones and Ala63→Pro in one clone).
The MIC values of TMC207 for the mutant strains and for the corresponding wild-type mycobacterial species are given in Table 2. Whatever was the concentration of TMC207 used in the selection, the mutants harboring the same mutation in a given species displayed identical MIC values. The highest MIC values were observed in M. tuberculosis mutants having the Ala63→Pro mutation (MIC = 4 mg/liter, 133-fold higher than the MIC of the wild-type strain H37Rv). The M. tuberculosis mutants carrying the mutations Glu61→Asp, Ile66→Met, and Asp28→Gly had MIC values varying from 0.5 to 1 mg/liter, i.e., 16- to 33-fold higher than the MIC for H37Rv. The lowest MIC value was measured for the M. tuberculosis mutant having the mutation Leu59→Val (MIC = 0.25 mg/liter, 8-fold increase). Regarding the mutants selected from atypical mycobacteria, the MIC values ranged from 4 to 16 mg/liter, with a noticeable 400-fold increase for the M. fortuitum mutants carrying Asp28→Ala compared to the M. abscessus mutants carrying the same mutation (32-fold increase). The two other mutations, Ala63→Pro in M. abscessus and Ile66→Met in M. smegmatis, raised the MIC values by 64- and 133-fold, respectively.
Concerning the 29 M. tuberculosis colonies selected on TMC207 which had no mutation in atpE, Southern hybridization and DNA amplification and sequencing were used to seek out other possible resistance mechanisms. We first checked by Southern hybridization analysis that only one copy of atpE was present on the chromosome. Using MscI- and NcoI-digested chromosomal DNA fragments of M. tuberculosis, an atpE probe was found to hybridize to a single MscI fragment of approximately 1,200 bp and to a single NcoI fragment of approximately 3,670 bp, respectively (data not shown), indicating that there is only one copy of this gene in the M. tuberculosis genome. Considering the previously proposed hypothesis suggesting that the drug could bind at the interface formed between the a and c subunits (11), the presence of mutations in the subunit a encoded by atpB in the 29 M. tuberculosis and the 9 M. abscessus mutants displaying resistance to TMC207 without a mutation in atpE was searched for by determining the nucleotide sequence of atpB in all the mutants. We also assessed if mutations could be present in the other ATP synthase subunits by amplifying and sequencing the entire ATP synthase operon in three M. tuberculosis strains: the reference strain H37Rv, a TMC207-resistant mutant carrying a mutation in atpE, and a resistant mutant without a mutation in atpE, selected on 0.5 mg/liter of TMC207. In all cases, the comparison of the nucleotide sequences obtained showed that no mutation was present in the atpB gene or in any of the other genes of the ATP synthase operon (data not shown).
Complementation experiments.
To further investigate the role played by the different mutations found in the mutants resistant to TMC207, a method of gene complementation was developed to assess the level of TMC207 resistance that they are able to confer in an isogenic context. This was achieved by introducing in M. smegmatis a plasmid carrying the atpE gene of M. smegmatis harboring the mutations found in all the species included in this study or, alternatively, the atpE gene of M. tuberculosis harboring the mutations found in the M. tuberculosis mutants. The trans-dominant effect exerted by the plasmid carrying the atpE mutant gene, when coexpressed with the chromosomal atpE wild-type gene, allowed us to estimate the level of resistance conferred by this mutant.
As shown in Table 3, the four control strains, i.e., M. smegmatis mc2155, the strain complemented with pLYG204 without the insert, and the strains of M. smegmatis complemented with pLYG-atpEsmewt and pLYG-atpEBKwt, all displayed the same MIC value (0.12 mg/liter). The M. smegmatis strains complemented with the atpE gene expressing the M. smegmatis subunit c containing the mutation Asp28→Val, Asp28→Ala, or Asp28→Gly showed a high level of resistance, with MIC values of 16 mg/liter (a 133-fold increase compared to the control strains). The M. smegmatis strains expressing the subunit c with the mutations Glu61→Asp and Ile66→Met also showed significant increases in the level of resistance to the drug, with MICs of 4 and 8 ml/liter, respectively (33- to 66-fold higher than the MICs of the control strains). Finally, the complementation of M. smegmatis with the M. smegmatis atpE gene expressing a subunit c containing the mutation Leu59→Val, Ala63→Met, or Ala63→Pro resulted in a 4-fold increase of the MIC values (MIC = 0.5 mg/liter, compared to 0.12 mg/liter for the control strains).
Table 3.
MIC of TMC207 for M. smegmatis complemented strainsa
| Gene and source | Mutation | Recombinant plasmid | MIC (mg/liter) |
|---|---|---|---|
| None | None | None | 0.12 |
| None | None | pLYG204 | 0.12 |
| atpE of M. smegmatis | None | pLYG-atpEsmewt | 0.12 |
| atpE of M. tuberculosis | None | pLYG-atpEBKwt | 0.12 |
| atpE of M. smegmatis | D28V | pLYG-atpEsmeD28V | 16 |
| atpE of M. smegmatis | D28A | pLYG-atpEsmeD28A | 16 |
| atpE of M. smegmatis | D28G | pLYG-atpEsmeD28G | 16 |
| atpE of M. smegmatis | I66M | pLYG-atpEsmeI66 M | 8 |
| atpE of M. smegmatis | E61D | pLYG-atpEsmeE61D | 4 |
| atpE of M. smegmatis | L59V | pLYG-atpEsmeL59V | 0.5 |
| atpE of M. smegmatis | A63P | pLYG-atpEsmeA63P | 0.5 |
| atpE of M. smegmatis | A63M | pLYG-atpEsmeA63M | 0.5 |
The host strain was M. smegmatis mc2155.
Unexpectedly, when the atpE gene of M. tuberculosis harboring the mutations identified in this species (Asp28→Gly, Leu59→Val, Glu61→Asp, Ala63→Pro, and Ile66→Met) was introduced in M. smegmatis, the complemented bacteria showed a susceptible phenotype with MIC values equal to the MIC value of the control strains (MIC = 0.12 mg/liter; data not shown).
C-ring models of M. tuberculosis and M. smegmatis.
The C-ring models of M. tuberculosis and M. smegmatis were built by homology modeling using the C-ring structure coordinates of the H+-ATP synthase of S. platensis, which shares 30% sequence identity with the amino acid sequences of the mycobacterial c subunit. As expected from the very high sequence identity found between M. tuberculosis and M. smegmatis, the models of the C rings from the two mycobacterial species displayed very similar shapes (data not shown), with the only significant structural difference being located in the N-terminal region, where 4 additional amino acids are found in M. smegmatis compared to M. tuberculosis, leading to an additional long tail extending toward the periplasm in M. smegmatis (visible in Fig. 2). Another significant difference was observed at the level of the C-terminal residue, where a positively charged Lys is found in M. tuberculosis instead of the Gln in M. smegmatis (Fig. 2). The four other amino acid differences found between M. tuberculosis and M. smegmatis (A/T6, A/G18, V/I30, V/I39; Fig. 2) are scattered throughout the sequence of the c subunit and do not lead to significant structural differences in the C-ring models of M. tuberculosis and M. smegmatis (data not shown).
A detailed view of the region of the conserved residue Glu61 in the C-ring model of the mycobacterial ATP synthase is given in Fig. 3C. As expected, the side chain of Glu61, which is carried by the C-terminal helix of a given monomer in the C ring (monomer B [C-ter] in Fig. 3C), protrudes above the N-terminal helix of the same monomer (monomer B [N-ter]), with the carboxylate group pointing toward the residues of the C-terminal helix of a neighboring monomer (monomer A [C-ter] in Fig. 3C). Strikingly, compared to the structures of the C ring from I. tartaricus and S. platensis, which both have a tyrosine residue pointing toward the carboxylate moiety of the conserved glutamate (Tyr70 and Tyr67 in Fig. 3A and B, respectively), the mycobacterial C ring possesses a tyrosine residue in position 64 that would be the residue equivalent to Tyr70 and Tyr67 in I. tartaricus and S. platensis (Fig. 3A to C). Therefore, for a given proton-binding site, the side chains of Glu61 and Tyr64 in mycobacteria are held by the same polypeptide chain (monomer B [C-ter] in Fig. 3A) and are both oriented in the same direction, while in I. tartaricus and S. platensis, the corresponding side chains (Glu65/62 and Tyr70/67) are carried by two different monomers (monomer B [C-ter] for Glu and monomer A [C-ter] for Tyr) with opposite orientations, with the oxygen atom of the tyrosinate being located close to the oxygen atom of the carboxylate moiety (Fig. 3A and B). Despite these differences, the spatial location of the tyrosinate moiety in the three cavity models is globally similar (Fig. 3A to C).
It must be pointed out here that most of the residues located around Glu61 in M. tuberculosis and M. smegmatis are those that are involved in resistance to TMC207: Asp28, which is carried by the monomer holding the side chain of Glu61 in a given proton-binding site (monomer B in Fig. 3C), and Leu59, Ala63, and Ile66, which come from a neighboring monomer (monomer A in Fig. 3C). Compared to the S. platensis three-dimensional (3D) structure (Fig. 3B), the specific presence of these residues in the c-subunit model of M. tuberculosis significantly increases the room around the glutamate residue at position 61 (Fig. 3C), which could allow the formation of a cleft delimited by residues Leu59, Glu61, Ala63, Tyr64, and Ile66 at its entrance and Asp28 at its bottom. Among these residues, Ala63 and Asp28, which are specifically found in the mycobacterial c subunit instead of Leu64 and Gln29 in S. platensis, have the greatest contribution to the formation of a cleft at the level of Glu61 in the mycobacterial C ring (Fig. 3B and C).
Docking of TMC207.
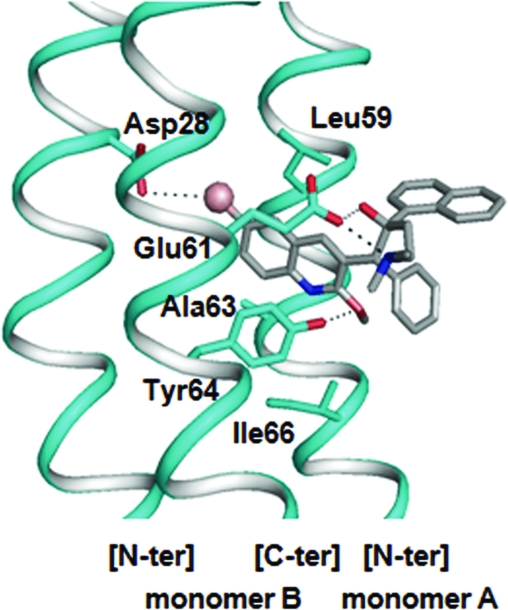
The coordinates of the wild-type M. tuberculosis C-ring model were used for docking simulations with TMC207 (R,S stereoisomer; Fig. 1). The binding site for docking was centered on the amino acid Glu61, and the solutions obtained were clustered according to the root-mean-square criterion. The best docking solution, which is detailed in Fig. 4, was extracted from the cluster containing the highest number of equivalent solutions displaying the lowest value of estimated free energy binding (−8.89 kcal/mol). According to this mode of docking, the TMC207 molecule would be anchored at the level of the quinoline ring into the cleft delimited by Asp28, Glu61, and Tyr64 on one side (carried by monomer B in Fig. 4) and Leu59, Ala63, and Ile66 on the other side (carried by monomer A in Fig. 4), with the drug being stabilized by three distinct types of interactions: (i) a hydrogen bond and a salt bridge linking Glu61 to TMC207 at the level of the hydroxyl and the dimethylamino groups of the drug, respectively, (ii) a hydrogen bond between the hydroxyl group of Tyr64 and the methyloxy moiety of the drug, and (iii) a halogen bond between the electronegative bromine atom of TMC207 and the residue Asp28 buried at the bottom of the pocket (Fig. 4). In this conformation, the naphthyl and phenyl rings of TMC207 protrude at the external surface of the C ring (Fig. 4) and would be, in this way, stabilized by hydrophobic interactions with the lipid bilayer surrounding the α helices of the C ring.
Fig 4.
Optimal docking result obtained for the R,S stereoisomer of TMC207 in the C ring of M. tuberculosis. The polypeptide chains forming the binding cleft are shown in ribbon representation. The amino acid residues contributing to the binding region are shown in stick representation. Oxygen and nitrogen atoms are indicated in red and dark blue, respectively. The TMC207 molecule is represented by using CPK colors, and the bromine atom is shown as a sphere. Hydrogen and halogen bonds are represented by dotted lines.
DISCUSSION
In this study, we selected in vitro independent and spontaneous TMC207-resistant mutants from various mycobacterial species, including M. tuberculosis, M. abscessus, and M. fortuitum, and interpreted the effect of the mutations by generating a structural model of the TMC207-binding site in the C ring of the mycobacterial ATP synthase.
Whatever was the mycobacterial species used, TMC207-resistant mutants were obtained at an apparent rate of mutation varying from 10−8 to 5.10−8, a value comparable to the mutation rates previously reported for isoniazid and rifampin (10, 23, 41). Strikingly, 38 of the 82 mutants (46%) obtained from M. tuberculosis and M. abscessus displayed no mutation in the atpE gene. These mutants were selected on low concentrations of TMC207 (i.e., 2- to 8-fold the wild-type MIC value). Considering the hypothesis made by de Jonge et al. (11), who previously suggested that the compound is embedded at the level of the a-c subunit interface, we sequenced atpB in the 82 mutants but found no mutations in the gene encoding the a subunit. Finally, for 1 of the 38 resistant clones without a mutation in atpE and atpB, selected on 0.5 mg/liter of TMC207, the sequences of the genes encoding the complete ATP synthase operon (FO and F1 domains) were all found to be wild type compared to the reference sequence from H37Rv, highlighting that mechanisms not related to mutations in the ATP synthase operon can also contribute to resistance to TMC207 in M. tuberculosis and M. abscessus. Alternative hypotheses can be formulated to explain the resistance observed in the nonmutated clones, which include inactivation of the drug (a mechanism that cannot be excluded) or, alternatively, a change in pleiotropic transcriptional regulators affecting the intrinsic resistance of mycobacteria, such as WhiB7, affecting cellular metabolism (6). Further investigations about the possible other mechanisms of resistance to TMC207 are under way to elucidate this point.
The effect of the mutations selected in the mutants obtained in vitro was assessed (i) by determining the MICs of TMC207 for the various mutants, (ii) by evaluating the effect of the mutations in an isogenic complementation assay, and (iii) by analyzing the structural modifications induced by the mutations in a 3D model of the mycobacterial C ring. With regard to the TMC207-resistant mutants showing amino acid substitutions in the c subunit, the diversity of the mutations selected from the three clinical strains of M. tuberculosis (strains P, D, and LQ; Table 2) and, to a lesser extent, from the clinical strain V of M. abscessus was markedly higher than that observed in the clinical strain B of M. fortuitum and in the two laboratory strains M. tuberculosis H37Rv and M. smegmatis mc2155 (Table 2), suggesting the presence in the strains of clinical origin and particularly in M. tuberculosis of mechanisms resulting in an increased mutation diversity (8). Regarding the results obtained in the complementation assay, a striking observation is the absence of complementation observed when the mutated c subunits of M. tuberculosis were introduced in M. smegmatis, whatever was the mutation harbored by the c subunit. Considering the very high similarity between the amino acid sequences of the c subunits of M. tuberculosis and M. smegmatis (85%), it is very likely that the absence of variation of MIC values observed when the M. tuberculosis c subunits are expressed in M. smegmatis is linked to the specific N- and C-terminal sequences characterizing the former species (MDPT and VK, respectively) compared to the latter (MDLDPNAI and GLQ, respectively). Assuming that such sequences are essential to stabilize the c subunits at the level of the external part of the lipid bilayer, it can be hypothesized that the insertion of the M. tuberculosis c subunit into the M. smegmatis membrane is unfavorable because of the absence of an acidic N-terminal tail characterizing the M. smegmatis c subunit, as well as the presence of a positively charged lysine residue at the C-terminal part in the M. tuberculosis c subunit.
Before addressing in detail the structural effects of the mutations, it is worth mentioning here that the 3D organization of the proton-binding site at the level of residue Glu61 in M. tuberculosis and M. smegmatis compared to the structural model of S. platensis presents a striking difference in the positioning of the essential tyrosine residue, which is located at position 64 in the mycobacterial C ring, while it is found at position 67 and forms an H-bond interaction with Glu62 in S. platensis (Fig. 3B and C). In the mycobacterial ATP synthase, the absence of Tyr at position 66 (equivalent to position 67 of S. platensis), combined with the specific presence of Asp28 and Ala63 instead of Gln and Leu in S. platensis, respectively, significantly enlarges the groove found at the level of Glu61 between two adjacent c subunits in the mycobacterial C ring (Fig. 3B and C), thus generating a pocket the size of which may be large enough to accommodate the diarylquinolines (Fig. 4). Accordingly, simulations of TMC207 docking to a minimized 3D model of the mycobacterial C ring indicate that the putative pocket could accommodate the bromoquinoline moiety of the drug, which would be stabilized by three different types of interactions: (i) two H bonds with the side chains of Glu61 and Tyr64, respectively, (ii) a salt bridge with the carboxylate moiety of Glu61, and (iii) a halogen bond between the Br atom of the drug and the side chain of Asp28 (Fig. 4). Previous studies have suggested that TMC207 most likely interferes with proton transfer by interacting with the mycobacterial c subunit at the level of a binding niche, including Glu61 (11, 19). Our results corroborate this model by demonstrating that there is a nearly perfect correspondence between the amino acid residues defining the proton-binding cavity in the ATP synthase C ring, on one hand, and those found to be mutated in TMC207-resistant mutants, on the other, which define the binding pocket around the drug (i.e., Leu59, Glu61, Ala63, and Ile66 on the edge of the pocket and Asp28 at the level of the ground of the pocket). It must be highlighted here that de Jonge et al. previously proposed a structural model of the binding pocket of TMC207 in the mycobacterial ATP synthase in which the drug binds at the interface between subunits a and c (11). Our results, combined with those previously obtained by Huitric et al., who showed the absence of mutation in subunit a of TMC207-resistant clones (21), strongly suggest that the binding pocket of TMC207 is, rather, located between two c subunits at the level of the proton-binding cavity.
The mutation Ala63→Pro in the mycobacterial c subunit is the substitution displaying the greatest effect among the mutants selected in vitro on TMC207 (the MIC value increased by 133-fold in M. tuberculosis and by 64-fold in M. abscessus). This result is readily explained by the fact that proline residues act as structural disruptors when they are introduced in the middle of an alpha helix, which is the case in this study, since the mutation Ala63→Pro occurs in the middle of the C-terminal helix of subunit c (monomer A [C-ter] in Fig. 3C). Such modifications may also account for the fact that Ala63→Pro in the complementation assay caused only a moderate level of resistance (4-fold increase of the TMC207 MIC) compared to the 133-fold increase observed in the M. tuberculosis mutants. This could be related to the fact that the plasmid-encoded c subunit containing the mutation would be incorporated in the C ring less efficiently that the wild-type c subunit determined by the chromosomal copy of the atpE gene. One has to note that Ala63, which is highly conserved in the mycobacterial species susceptible to TMC207, is naturally replaced by a Met residue in the three mycobacteria known to display intrinsic resistance to the drug, i.e., M. xenopi, M. novacastrense, and M. shimoidei (20, 30), reinforcing the idea that Ala63 plays a key role in the development of resistance to TMC207.
Asp28 is the residue showing the highest degree of diversity in the TMC207-resistant mutants. Two new mutations (Asp28→Gly and Asp28→Ala) beyond those previously reported in other studies (Asp28→Val or Pro) (2, 21) were identified in our study. These mutations conferred a marked increase in the MIC values of TMC207, whatever was the species in which they occurred (16- to 400-fold increase; Table 2). In the in vitro complementation assay, the three tested mutations (Asp28Val, -Ala, and -Gly) were those displaying the most marked effects (133-fold). This indicates that the increase of the TMC207 resistance level in the Asp28 mutants is more related to the loss of the carboxylic moiety of Asp28 than to the variations of steric hindrance linked to the substitution. Our docking results strongly support the hypothesis of a binding site accepting the quinoline ring, with the anchoring of the drug being ensured by a halogen bond between the bromine atom and the carboxylate of Asp28 (Fig. 4). Interestingly, a recent report of Guillemont and colleagues on the quantitative structure-activity relationship of diarylquinolines series has highlighted that omitting the bromine atom in the 6 portion of the quinoline ring was highly deleterious to the antimycobacterial activity of the drug (17). It is also of note that none of the species identified by Andries et al. (2) to be naturally resistant to TMC207 have an aspartate residue at the equivalent position 28 in the c subunit, such as Escherichia coli (Ile28, MIC > 32 mg/liter) or Corynebacterium jeikeium (Val28, MIC = 4 mg/liter).
The only amino acid replacement tolerated at position 61 corresponds to the mutation Glu61→Asp, which preserves the carboxylate group of the proton carrier in the H+-ATP synthases. In our study, this mutation occurred in five TMC207-resistant colonies for which the level of resistance was increased by 16- to 33-fold in the in vitro mutants, with a 4-fold increase being observed in the complementation assay. These effects are very likely related to the shortening of the side chain carrying the carboxylate moiety, which is crucial for the anchoring of the drug by providing two bonding interactions with the hydroxyl (H bond) and the dimethylamino (ionic bond) groups of TMC207 (Fig. 4). This hypothesis is supported by previous reports mentioning (i) that the replacement of the dimethylamino group with nonbasic functionality or, alternatively, the shortening or the lengthening of the lateral chain carrying this group significantly alters the activity of the drug (17) and (ii) that electrostatic forces are an important factor for binding of TMC207 to the subunit c of the mycobacterial ATP synthase (19).
The replacement of Ile by Met at position 66 introduces a bulky side chain carrying a sulfur atom at the level of the TMC207-binding site, resulting in a significant increase of the MIC values of TMC207 in the in vitro mutants and in our complementation assay as well. The presence of Met66 likely induces significant steric clashes between the sulfur-containing group of the residue and the tyrosinate moiety of Tyr64 (Fig. 4B), with two possible consequences: an altered fit of the quinoline moiety between two C-terminal helices and a disruption of the H bond linking the hydroxyl group of Try64 to the 2-methoxy oxygen atom of the drug. A recent QSAR study reported that the methoxy group in the 2 position of the quinoline ring plays a crucial role in TMC207 activity, with its replacement by other substituents being highly deleterious to the inhibiting potency of the drug (17).
Finally, the mutation Leu59→Val, which is identified for the first time in this study, is associated with a moderate increase of resistance to TMC207 in the in vitro mutants and in the complemented M. smegmatis strains (4-fold increase). The main structural effect of this replacement is to shift the isopropyl group toward the bottom of the TMC207-binding site, with consequences for steric adjustment of the diarylquinoline in the groove delimited by two C-terminal helices in the C ring.
In conclusion, our data support the hypothesis that TMC207 blocks the mycobacterial ATP synthase by interacting with the C ring at the level of a cleft centered on the essential residue Glu61. This binding site would be located in the groove delimited by two C-terminal helices from two adjacent c subunits, with its topological features mostly being modeled by residues located on both sides (Leu59, Glu61, Ala63, and Ile66) and at the bottom (Asp28) of the groove that correspond to those involved in the development of resistance to the drug.
ACKNOWLEDGMENT
This work was supported by an ESCMID research grant (2009).
Footnotes
Published ahead of print 21 February 2012
REFERENCES
- 1. Abrahams JP, Leslie AG, Lutter R, Walker JE. 1994. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370:621–628 [DOI] [PubMed] [Google Scholar]
- 2. Andries K, et al. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223–227 [DOI] [PubMed] [Google Scholar]
- 3. Angevine CM, Herold KA, Vincent OD, Fillingame RH. 2007. Aqueous access pathways in ATP synthase subunit a. Reactivity of cysteine substituted into transmembrane helices 1, 3, and 5. J. Biol. Chem. 282:9001–9007 [DOI] [PubMed] [Google Scholar]
- 4. Boyer PD. 1989. A perspective of the binding change mechanism for ATP synthesis. FASEB J. 3:2164–2178 [DOI] [PubMed] [Google Scholar]
- 5. Brunger AT, et al. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921 [DOI] [PubMed] [Google Scholar]
- 6. Burian J, et al. 2012. The mycobacterial transcriptional regulator whiB7 gene links redox homeostasis and intrinsic antibiotic resistance. J. Biol. Chem. 287:299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Capaldi RA, Aggeler R. 2002. Mechanism of the F(1)F(0)-type ATP synthase, a biological rotary motor. Trends Biochem. Sci. 27:154–160 [DOI] [PubMed] [Google Scholar]
- 8. Chang JR, et al. 2011. Genotypic analysis of genes associated with transmission and drug resistance in the Beijing lineage of Mycobacterium tuberculosis. Clin. Microbiol. Infect. 17:1391–1396 [DOI] [PubMed] [Google Scholar]
- 9. Dautant A, Velours J, Giraud MF. 2010. Crystal structure of the Mg · ADP-inhibited state of the yeast F1c10-ATP synthase. J. Biol. Chem. 285:29502–29510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. David HL. 1970. Probability distribution of drug-resistant mutants in unselected populations of Mycobacterium tuberculosis. Appl. Microbiol. 20:810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Jonge MR, Koymans LH, Guillemont JE, Koul A, Andries K. 2007. A computational model of the inhibition of Mycobacterium tuberculosis ATPase by a new drug candidate R207910. Proteins 67:971–980 [DOI] [PubMed] [Google Scholar]
- 12. Diacon AH, et al. 2009. The diarylquinoline TMC207 for multidrug-resistant tuberculosis. N. Engl. J. Med. 360:2397–2405 [DOI] [PubMed] [Google Scholar]
- 13. Dimroth P, von Ballmoos C, Meier T. 2006. Catalytic and mechanical cycles in F-ATP synthases. Fourth in the Cycles Review Series. EMBO Rep. 7:276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 15. Eswar N, et al. 2006. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics Chapter 5:Unit 5.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao LY, et al. 2003. Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival. Infect. Immun. 71:922–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guillemont J, Meyer C, Poncelet A, Bourdrez X, Andries K. 2011. Diarylquinolines, synthesis pathways and quantitative structure-activity relationship studies leading to the discovery of TMC207. Future Med. Chem. 3:1345–1360 [DOI] [PubMed] [Google Scholar]
- 18. Haagsma AC, et al. 2009. Selectivity of TMC207 towards mycobacterial ATP synthase compared with that towards the eukaryotic homologue. Antimicrob. Agents Chemother. 53:1290–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haagsma AC, et al. 2011. Probing the interaction of the diarylquinoline TMC207 with its target mycobacterial ATP synthase. PLoS One 6:e23575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huitric E, Verhasselt P, Andries K, Hoffner SE. 2007. In vitro antimycobacterial spectrum of a diarylquinoline ATP synthase inhibitor. Antimicrob. Agents Chemother. 51:4202–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huitric E, et al. 2010. Rates and mechanisms of resistance development in Mycobacterium tuberculosis to a novel diarylquinoline ATP synthase inhibitor. Antimicrob. Agents Chemother. 54:1022–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang W, Hermolin J, Fillingame RH. 2001. The preferred stoichiometry of c subunits in the rotary motor sector of Escherichia coli ATP synthase is 10. Proc. Natl. Acad. Sci. U. S. A. 98:4966–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson R, et al. 2006. Drug resistance in Mycobacterium tuberculosis. Curr. Issues Mol. Biol. 8:97–111 [PubMed] [Google Scholar]
- 24. Krah A, Pogoryelov D, Meier T, Faraldo-Gomez JD. 2010. On the structure of the proton-binding site in the F(o) rotor of chloroplast ATP synthases. J. Mol. Biol. 395:20–27 [DOI] [PubMed] [Google Scholar]
- 25. Meier T, et al. 2009. Complete ion-coordination structure in the rotor ring of Na+-dependent F-ATP synthases. J. Mol. Biol. 391:498–507 [DOI] [PubMed] [Google Scholar]
- 26. Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. 2005. Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus. Science 308:659–662 [DOI] [PubMed] [Google Scholar]
- 27. Mitome N, Suzuki T, Hayashi S, Yoshida M. 2004. Thermophilic ATP synthase has a decamer c-ring: indication of noninteger 10:3 H+/ATP ratio and permissive elastic coupling. Proc. Natl. Acad. Sci. U. S. A. 101:12159–12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morris G, et al. 1998. Automated docking using a Lamarckian genetic algorithm and empirical free energy function. J. Comput. Chem. 19:1639–1662 [Google Scholar]
- 29. Noji H, Yasuda R, Yoshida M, Kinosita K., Jr 1997. Direct observation of the rotation of F1-ATPase. Nature 386:299–302 [DOI] [PubMed] [Google Scholar]
- 30. Petrella S, et al. 2006. Genetic basis for natural and acquired resistance to the diarylquinoline R207910 in mycobacteria. Antimicrob. Agents Chemother. 50:2853–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Petrella S, Clermont D, Casin I, Jarlier V, Sougakoff W. 2001. Novel class A beta-lactamase Sed-1 from Citrobacter sedlakii: genetic diversity of beta-lactamases within the Citrobacter genus. Antimicrob. Agents Chemother. 45:2287–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pogoryelov D, Yildiz O, Faraldo-Gomez JD, Meier T. 2009. High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nat. Struct. Mol. Biol. 16:1068–1073 [DOI] [PubMed] [Google Scholar]
- 33. Preiss L, Yildiz O, Hicks DB, Krulwich TA, Meier T. 2010. A new type of proton coordination in an F(1)F(o)-ATP synthase rotor ring. PLoS Biol. 8:e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Steed PR, Fillingame RH. 2009. Aqueous accessibility to the transmembrane regions of subunit c of the Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 284:23243–23250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stock D, Leslie AG, Walker JE. 1999. Molecular architecture of the rotary motor in ATP synthase. Science 286:1700–1705 [DOI] [PubMed] [Google Scholar]
- 36. Vik SB, Ishmukhametov RR. 2005. Structure and function of subunit a of the ATP synthase of Escherichia coli. J. Bioenerg. Biomembr. 37:445–449 [DOI] [PubMed] [Google Scholar]
- 37. Vollmar M, Schlieper D, Winn M, Buchner C, Groth G. 2009. Structure of the c14 rotor ring of the proton translocating chloroplast ATP synthase. J. Biol. Chem. 284:18228–18235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. von Ballmoos C, Cook GM, Dimroth P. 2008. Unique rotary ATP synthase and its biological diversity. Annu. Rev. Biophys. 37:43–64 [DOI] [PubMed] [Google Scholar]
- 39. von Ballmoos C, Wiedenmann A, Dimroth P. 2009. Essentials for ATP synthesis by F1F0 ATP synthases. Annu. Rev. Biochem. 78:649–672 [DOI] [PubMed] [Google Scholar]
- 40. Watt IN, Montgomery MG, Runswick MJ, Leslie AG, Walker JE. 2010. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc. Natl. Acad. Sci. U. S. A. 107:16823–16827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Werngren J, Hoffner SE. 2003. Drug-susceptible Mycobacterium tuberculosis Beijing genotype does not develop mutation-conferred resistance to rifampin at an elevated rate. J. Clin. Microbiol. 41:1520–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yoshida M, Muneyuki E, Hisabori T. 2001. ATP synthase—a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2:669–677 [DOI] [PubMed] [Google Scholar]