Abstract
Hypersusceptibility (HS) to inhibition by different antiretroviral drugs (ARVs) among diverse HIV-infected individuals may be a misnomer because clinical response to treatment is evaluated in relation to subtype B infections while drug susceptibility of the infecting virus, regardless of subtype, is compared to a subtype B HIV-1 laboratory strain (NL4-3 or IIIB). Mounting evidence suggests that HS to different ARVs may result in better treatment outcome just as drug resistance leads to treatment failure. We have identified key amino acid polymorphisms in the protease coding region of a non-B HIV-1 subtype linked to protease inhibitor HS, namely, 17E and 64M in CRF02_AG. These HS-linked polymorphisms were introduced in the BD6-15 CRF02_AG molecular clone and tested for inhibition using a panel of protease inhibitors. In general, suspected HS-linked polymorphisms did increase susceptibility to specific protease inhibitors such as amprenavir and atazanavir, but the combination of the 17E/64M polymorphisms showed greater HS. These two mutations were found at low frequencies but linked in a sequence database of over 700 protease sequences of CRF02_AG. In direct head-to-head virus competitions, CRF02_AG harboring the 17E/64M polymorphisms also had higher replicative fitness than did the 17E or the 64M polymorphism in the CFR02_AG clone. These findings suggest that subtype-specific, linked polymorphisms can result in hypersusceptibility to ARVs. Considering the potential benefit of HS to treatment outcome, screening for potential HS-linked polymorphisms as well as preexisting drug resistance mutations in treatment-naïve patients may guide the choice of ARVs for the best treatment outcome.
INTRODUCTION
Intense research over the last 2 decades of the HIV/AIDS pandemic has contributed to the development of several antiretroviral drugs (ARVs) which have significantly reduced HIV/AIDS morbidity and mortality. These drugs can be divided into six distinct classes, according to their viral target and mechanism of action. The nucleoside reverse transcriptase (RT) inhibitors (NRTIs), the nonnucleoside reverse transcriptase inhibitors (NNRTIs), and the protease (PR) inhibitors (PIs) were the first ARV classes approved by the FDA starting in the late 1980s and remain the mainstay for first-line highly active antiretroviral therapy (HAART), combining at least three different drugs from two ARV classes (9).
The vast genetic diversity within the HIV-1 epidemic can be categorized into four groups (M, N, O, and P), with group M HIV-1 responsible for the majority of worldwide infections. According to the current classification system (30), HIV-1 group M is divided into nine “pure” subtypes, at least 48 circulating recombinant forms (CRFs), and various unique mosaic strains. Subtype B is the most prevalent in developed countries (14), and consequently, it is the major target of drug design and resistance studies (19). Despite initial development to inhibit subtype B HIV-1, most FDA-approved protease (PR) and reverse transcriptase (RT) inhibitors are highly effective in blocking virus replication in treatment-naïve patients infected with HIV-1 non-B subtypes (1, 2, 44). ARV treatment imposes an immediate selective pressure on the infecting HIV-1 population within a patient and will favor outgrowth of drug-resistant variants with suboptimal drug levels (17). HIV-1 non-B subtypes generally acquire the same drug resistance mutations (DRMs) as those described in subtype B infections, yet quantitative and qualitative disparities have been described (11, 19, 35). Furthermore, the genetic diversity in the HIV-1 genes results in different baseline PR or RT amino acid sequence that can alter the absolute level of drug resistance conferred by identical drug resistance mutations in these drug-targeted genes (28, 31, 41). Infections with non-B subtype HIV-1 still represent a challenge for HAART, based on the relative paucity of treatment outcomes correlated with baseline HIV-1 sequence and relative levels of virus sensitivity to drug inhibitions. These factors could impact on the efficacy and durability of treatment during infection with these non-B HIV-1 variants. It is now well known that many secondary mutations selected under PI treatment in subtype B-infected patients are found as natural polymorphisms or even wild-type sequence in non-subtype B HIV-1 isolates (in the absence of treatment). In subtype B, these secondary mutations appear to enhance PI resistance levels and/or to compensate for fitness defects conferred by primary drug resistance mutations (16–18, 29).
Just as natural polymorphisms can enhance resistance or compensate for fitness loss, it is also possible that these genetic differences in non-subtype B HIV-1 strains may result in hypersusceptibility (HS) to ARV inhibition compared to subtype B viruses. Consistent with this hypothesis, Abecasis et al. (1) reported that some non-B subtypes demonstrate increased viral susceptibility to some PIs. For example, CRF02_AG strains presented higher sensitivity to indinavir and to ritonavir than did subtypes B, C, F, and G. In the present study, we evaluated the proportion of viral isolates with natural HS to PIs from treatment-naïve patients infected with five different genotypes of HIV-1. We also mapped the genetic polymorphisms in CRF02_AG that are linked to PI HS and tested them singly or paired in the context of a CRF02_AG infectious molecular clone. We show, for the first time, that specific PR natural polymorphisms in CRF02_AG confer HS on PIs as well as increased viral fitness.
MATERIALS AND METHODS
Global data set of HIV-1 drug phenotypes from treatment-naïve patients.
We first analyzed the available phenotypic and genotypic drug resistance profiles of HIV-1 isolates from treatment-naïve subjects (1, 8, 42–45). The drug susceptibility assay employed the Antivirogram methodology (Virco, Belgium), which involves mammal-based recombination of a PCR-amplified DNA fragment (encompassing PR codons 1 to 99 and RT codons 1 to 400) into a proviral clone of HIV-1 subtype B, ΔPR-TR400 (15). The susceptibility of these chimeric viruses was then measured in MT-2 cells with increasing concentrations of amprenavir (APV), indinavir (IDV), nelfinavir (NFV), lopinavir (LPV), saquinavir (SQV), and tipranavir (TPV), all PIs. A wild-type (susceptible) virus of HIV-1 subtype B (IIIb) was used as a control. Phenotypic results were expressed in fold change (FC), defined as the ratio between the 50% effective concentration (EC50) value for the recombinant HIV-1 chimeric virus harboring the patient PR-RT and the EC50 values for the control IIIb. The EC50 value represents the drug concentration needed to inhibit 50% of viral replication. Of the 165 viral isolates with phenotyping results, 72 were subtype B, 23 were subtype C, 26 were subsubtype F1, 29 were subtype G, and 34 were CRF02_AG.
Proportion of HS to PIs in HIV-1 subtypes and HS mapping.
A virus was defined as hypersusceptible (HS) to a drug (PI) when the FC value was less than 0.4, i.e., the EC50 value for the query virus was at least 2.5× lower than that of the control IIIb virus. HS was determined for each drug and for each HIV-1 strain/mutant compared to IIIb. Based on FC values for each drug and with the HIV-1 isolates of each subtype/CRF, an HS group (FC, ≤0.4) and a non-HS group (FC, >0.4) were compared by aligning the nucleotide sequences and predicted amino acids of the PR coding region using BioEdit v.7.0 (39). Potential differences in amino acid polymorphism between the two groups were then confirmed by two-tailed Fisher's exact test, and P values of ≤0.05 were considered significant.
Generation of mutant CRF02_AG molecular clones.
The HIV-1 CRF02_AG infectious molecular clone BD6-15, derived from an X4-tropic isolate, was used to phenotypically test the CRF02_AG-specific polymorphisms linked to HS characterized in this study (38). The mutations G16E, G17E, I64M, K70R, and I72V were introduced in the PR coding region of pBD6-15 by site-directed mutagenesis using the QuikChange II XL site-directed mutagenesis kit (Stratagene) and the set of primers listed in Table S1 in the supplemental material. Mutants were confirmed by PCR amplification and DNA sequencing of the entire PR region of pBD6-15 (38).
Transfections, virus propagation, and titration.
The original and mutated BD6-15 plasmids were transfected into 293T cells using the Effectene transfection reagent (Qiagen), with 0.4 μg of plasmid DNA, according to the manufacturer's instructions. After 48 h, aliquots of the transfection supernatant were tested for RT activity using a radioisotopic RT assay (24). RT-positive transfection-derived viruses were used to infect five different human cell lines, namely, TZM-bl, U87-CD4-CXCR4, C8166, MT-2, and MT-4. Propagation of BD6-15 and its derivatives was best on MT-2 cells based on RT assays conducted on days 3, 6, 8, and 10 postinfection. After viral propagation, infected MT-2 cells were harvested and genomic DNA was extracted using the QIAamp DNA blood kit (Qiagen). The entire viral PR region was again PCR amplified and sequenced to rule out any reversion or other changes during virus production. All viruses generated in this study were titrated on MT-2 cells as previously described (34).
Protease inhibitor phenotyping assay.
In vitro drug sensitivity assays were performed to determine the EC50 values for six PIs (APV, atazanavir [ATV], IDV, LPV, NFV, and SQV) and with all viruses generated for this study. As a control, the phenotypic profiles of the parental wild-type virus (BD6) and all the derived mutants as well as NL4-3 were tested against zidovudine (AZT), an NRTI (see Fig. S1 in the supplemental material). Drug susceptibility was measured by exposing each virus to 5-fold serial dilutions of each drug, in quadruplicate, in MT-2 cell cultures. On day 5 postinfection, supernatant aliquots were collected and the virus titer was measured in RT assays. The HIV-1 subtype B infectious molecular clone pNL4-3 was used as an additional control. The EC50 value of each virus for each drug was calculated in a logarithmic curve where the y axis represented the percentage of infectivity, the x axis represented the drug concentration (as previously described), and the fold change (FC) represented the ratio between the EC50 value of the mutant virus and the EC50 value of the parental BD6-15 (to a particular drug). Hypersusceptibility (HS) was characterized when the FC of the mutant virus was ≤0.4. The difference between the mean EC50 value of each mutant CRF02_AG and that of the original clone was evaluated using two-tailed Student t tests, and P values of ≤0.05 were considered significant.
Growth competitions assays and determination of replicative fitness.
Viral fitness of CRF02_AG-derivative mutant viruses was determined by full pairwise dual-infection/competitions between all combinations of viruses. MT-2 cells were infected with equal virus titers (multiplicity of infection [MOI] of 0.0005) from each of the competing viruses. Monoinfections were also performed using a similar MOI. All mono- and dual competitions were conducted in triplicate. On day 5 postinfection, which was the peak of virus growth, RT activity was monitored by an RT assay, and the cells were harvested and genomic DNA was extracted as described previously (37). The entire viral PR region was PCR amplified, and the fragments were analyzed by a Luminex-based oligonucleotide ligation assay (OLA) as previously described and validated (20, 37). Briefly, three tagged-fluorescence primers were used: two upstream primers that discriminate the original and mutant sequences and a universal downstream primer. A ligation reaction was performed, and the emission of a double fluorescence (upstream- and downstream-tagged probes; see Table S2 in the supplemental material) was screened in a Bioplex 200 apparatus (Bio-Rad), using Luminex xMAP technology. The relative proportion of each double fluorescence signal was used to determine the proportion of each virus in the competing culture. The final ratio of the two viruses produced from each dual infection was determined by comparing the virus production in the competition to the virus production in the monoinfection. The relative fitness of each virus was determined as described previously (37).
RESULTS
Hypersusceptibility of HIV-1 isolates from treatment-naïve patients.
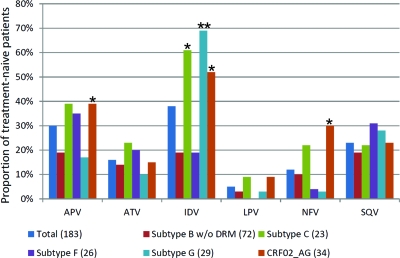
We analyzed the PR sequence compared to drug susceptibility of 165 HIV-1 isolates from treatment-naïve patients infected with five distinct HIV-1 subtypes/CRFs (subtypes B, C, F, and G and CRF02_AG) to verify if PI susceptibilities were different between HIV-1 subtypes. Hypersusceptibility was most evident among non-B subtypes with the PI IDV (38%) and the least frequent among all HIV-1 isolates with LPV (6%) (Fig. 1). The CRF02_AG cluster had the highest proportion of isolates hypersusceptible to PIs, namely, APV, IDV, and NFV, compared to subtype B isolates (P = 0.046, 0.001, and 0.019, respectively). Other subtypes displayed hypersusceptibilities to PIs that were more drug dependent, e.g., more subtype C and G isolates than subtype B isolates were hypersusceptible to IDV (P < 0.001), whereas subtypes B, C, and G had the same low proportions of isolates that were HS to ATV, LPV, and SQV. Subtype F1 was similar to subtype B in the proportion of HIV-1 isolates hypersusceptible to all PIs. As described below, the genetic distance in the PR coding region between subtypes F1 and B was similar to that between subtypes B, C, and G and CRF02_AG. Thus, we suspected that specific polymorphisms rather than general genetic divergence between these subtypes were responsible for the observed hypersusceptibility to the PIs.
Fig 1.
Proportions of five HIV-1 group M subtypes or CRFs from drug-naïve subjects with hypersusceptibility to several protease inhibitors. Single asterisks represent a P value of <0.05, while double asterisks depict P values of <0.001, for a Fisher exact test comparing the proportions between a non-B subtype and subtype B. Numbers in parentheses represent the total number of strains analyzed for each HIV-1 subtype/CRF variant.
Mapping polymorphisms that affect HS phenotypes.
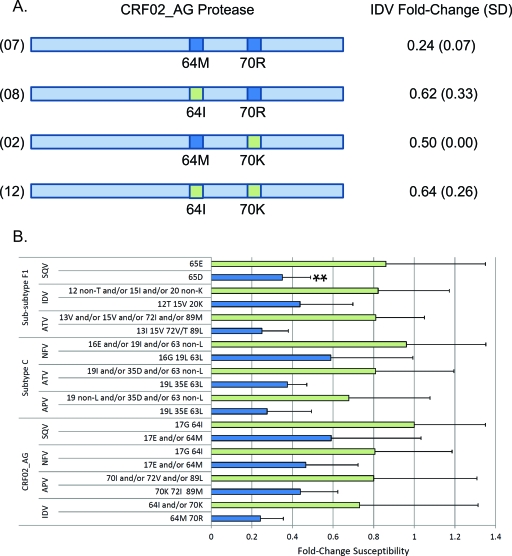
Hypersusceptibility may be linked to natural polymorphisms present in the PR of different HIV-1 subtypes and recombinant forms. In silico comparisons of translated PR regions from these virus isolates revealed that HS was associated not with a single polymorphism but rather with two or more linked amino acids within the consensus of a particular subtype. We compared the fold change in EC50 values conferred by different compositions of mapped PR polymorphisms for each subtype. Figure 2 shows the FC values of definite amino acid compositions displaying significant (P ≤ 0.05) comparisons between HS and non-HS phenotypes. Interestingly, in many cases, the same polymorphisms were linked to HS for two or more PIs in a given HIV-1 subtype or CRF. Examples included 19L/63L conferring HS to APV and NFV on subtype C isolates and 17E/64M conferring HS to IDV, NFV, and SQV on CRF02_AG isolates. We observed that single changes in HS-linked polymorphisms did not result in hypersusceptibility, which again emphasizes the linkage of two or more mutations that may have coevolved within a subtype (Fig. 2B). Moreover, in some cases the amino acid positions linked to HS with specific drugs were located in similar PR regions such as amino acid positions 19 and 63 in subtype C, 17 and 64 in CRF02_AG, and 15 and 72 in subtype F1.
Fig 2.
Schematic representation of the main hypersusceptible CRF02_AG genotypes at positions 64 and 70, showing their respective average FC values with standard deviation (SD) to a given PI. All FC and SD values were derived from triplicate drug susceptibility analyses with each virus and with each drug. Only the FC of IDV used as an example in this figure is shown. All FC values were derived by comparing the EC50 values to the control virus (HxB2). Numbers in parentheses at the left of genotypes represent the number of isolates for which the average was calculated. (B) FC relative to the wild-type HIV-1 CRF02_AG, subtype C, and subtype F1 carrying definite protease polymorphisms or combinations thereof. Bars represent the mean FC of each polymorphism composition displayed on the y axis with associated standard deviation. Only polymorphism compositions significantly associated with decreased susceptibility to definite PIs for each subtype are shown. The double asterisk denotes a significant P value of >0.001 in the Student t test. In both panels A and B, dark blue indicates polymorphisms or combinations associated with increased susceptibility to a PI, while pale green shows nonassociated combinations.
Phenotypic effects conferred by polymorphisms in a CRF02_AG infectious clone.
To determine the effect of various PR mutations on PI HS in CRF02_AG, we produced eight mutant clones from a CRF02_AG infectious molecular clone (BD6-15), five of them with a single mutation (16E, 17E, 64M, 70R, and 72V) and three of them double mutants (16E/64M, 17E/64M, and 64M/70R). Only 16E had a weak association with HS in CRF02_AG. Control experiments with all the mutants, their parental wild-type virus, and the NL4-3 control virus showed similar sensitivities to AZT inhibition. This CRF was selected for these mutagenesis analyses and HS studies because of its high prevalence in the worldwide epidemic (8%) and the availability of an infectious molecular clone. Moreover, CRF02_AG is the most frequent non-B HIV-1 subtype in the developed world, particularly in Europe. CRF02_AG is also the fastest-expanding HIV-1 genetic form in the worldwide epidemic according to the most recent HIV molecular epidemiology survey (14). As described in Fig. 2, it appears that HS to PIs was specific to a set of polymorphisms and drug. The 17E polymorphism in HIV-1 BD6-15 showed a modest increase in susceptibility to APV, ATV, and NFV compared to the parental BD6-15 virus, while the 64M mutant virus displayed greater HS to APV, NFV, and LPV (Fig. 3). The 70R mutation conferred HS to APV and IDV, while 72V slightly increased susceptibility to SQV and LPV. Double mutant viruses provided the most interesting results. In general, the double mutations showed the greatest HS but, again, only to specific drugs. For example, increased HS to ATV, NFV, and SQV was observed with polymorphisms 17E and 64M (Fig. 3). However, there was no additive effect of 17E and 64M on HS to APV, IDV, or LPV (Fig. 3).
Fig 3.
Phenotyping FC values of distinct CRF02_AG BD6 mutant viruses harboring distinct mutations for HS to six different PIs in comparison with the CRF02_AG wild-type molecular clone BD6. Mutants with distinct PR polymorphism combinations are depicted on the x axis, while boxed polymorphisms represent those introduced by site-directed mutagenesis. Asterisks above bars represent mutants with statistically significantly reduced susceptibility compared to the wild-type BD6 clone (double asterisks represent a P value of >0.001). The horizontal bar represents the cutoff used to define hypersusceptibility (FC, ≤0.4). The green bars rightmost in each graph represent the FC of a prototypic subtype B clone (NL4-3) for comparison. All FC and SD values were derived from quadruplicate determinations.
Replicative fitness and HS.
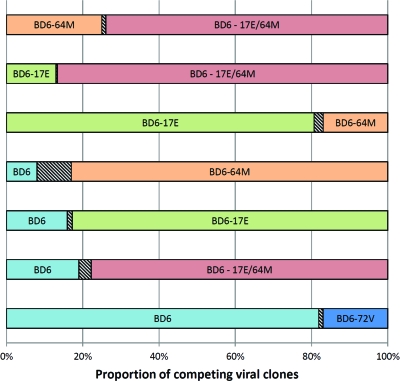
We measured the impact of HS-linked polymorphisms on the ex vivo fitness in the BD6-15 backbone by performing head-to-head competitions in MT-2 cells. The relative proportion of each competitor after 5 days in culture was measured by an oligonucleotide ligation assay as described in Materials and Methods and as previously published (34, 37). The BD6 variant with the highest replicative fitness was BD6-17E/64M, while the least fit was the parental BD6-15 virus (Fig. 4). With the exception of BD6-72V, all of the BD clones carrying the HS polymorphisms were more fit than the parental BD6 virus with a final fitness ranking of 17E/64M > 17E > 64M > BD6-15. As described below, increased fitness appears to be directly related to the introduction of polymorphisms that increase susceptibility to PIs. In contrast, acquired mutations conferring resistance to drugs generally encode reduced HIV-1 replicative fitness. However, it is important to note that drug HS is obviously a consequence of divergent HIV-1 evolution and not related to an exogenous selective force, considering that these polymorphisms existed long before the introduction of any drug treatment in West and Central Africa, i.e., the epicenter of CRF02_AG.
Fig 4.
Proportions of BD6 viruses derived from head-to-head competitions. Proportions were measured on day 5 after infection with equal amounts of two viruses. Results were obtained from 3 independent experiments. The hatched boxes represent standard errors.
DISCUSSION
Despite the introduction of HAART in many developing countries, its medium- and long-term impact on control of viremia in non-B subtype HIV-1 infections remains largely unknown. According to the work of Abecasis et al., subtype C and G and CRF02_AG isolates are more susceptible to IDV than are subtype B isolates (1). In the present study, we also observed a higher sensitivity of CRF02_AG viruses than subtype B viruses to APV and NFV. In addition, polymorphisms 17E, 64M, and 70R, found at frequencies of 8, 10, and 13% in CRF02_AG, respectively, can result in HS to APV, NFV, and IDV within this CRF. CRF02_AG is the dominant HIV-1 subtype in West Africa and accounts for 8% of infections worldwide.
Several studies have shown that drug resistance mutations specific to certain drugs can confer HS to others. A classical example is M184V in HIV-1 reverse transcriptase, which confers major resistance to lamivudine (3TC) but increases viral susceptibility to thymidine analogs (zidovudine [AZT] and stavudine [d4T]) and delays the emergence of thymidine analog-associated resistance mutations (TAMs) (3, 5, 22, 27). In the PR region, the major resistance mutation to APV, I50L, confers HS to all remaining PIs (46), while N88S confers resistance to atazanavir (ATV) and NFV and HS to APV (48). Nonetheless, mapping of natural HS-linked polymorphisms in HIV-1 isolates from treatment-naïve patients is rare in the literature. Leigh Brown et al. determined that polymorphisms at PR positions 10, 13, 37, and 61 were linked to HS in subtype B isolates (25), but a subsequent study did not confirm these results (26). Finally, Abecasis et al. showed a correlation between PR polymorphisms at positions 35, 37, 57, 70, and 89 and HS in non-B subtypes (1). Our findings are in partial agreement with this study, as the polymorphism 35E was linked to HS to APV and NFV in subtype C isolates and the polymorphism 70R was linked to HS to IDV in CRF02_AG isolates. Our analysis further showed, in the majority of cases, that a set of two or more PR polymorphisms were linked to HS for a given PI. In other instances, we were unable to determine the genetic components behind the HS phenotype, indicating that a complex pattern of HS-linked polymorphisms is likely required for this drug hypersusceptibility.
It is important to note that mutations in Gag, specifically at the cleavage site, can also confer reduced susceptibility to PI and have been described in subtype B (4, 6, 23) but not in CRF02_AG or other non-subtype B isolates (representing >90% of the infections worldwide). Based on subtype B analyses, our infectious clone BD6-15 may have a compensatory mutation for PI resistance in the Gag region (436R). Upon analysis of the Los Alamos National Laboratory (LANL) database, we found that approximately 17% of CRF01_AG isolates carry that polymorphism. Ghosn et al. (10) tested a large number of Gag polymorphisms typical of non-B subtypes (including CRF02_AG) with respect to association with virological failure under PI treatment but failed to associate this mutation with treatment failure outcome.
This study provides the first evidence for effects of individual or dual HS-linked polymorphisms on viral susceptibility to ARVs and on viral fitness. We demonstrated that CRF02_AG viruses carrying HS-linked polymorphisms 17E and 64M had higher replicative fitness than did viruses harboring either 17E or 64M and that these mutant viruses showed a higher fitness than did the parental virus. These results may indicate that CRF02_AG PR carrying HS-linked polymorphisms 17E and/or 64M may have increased PR cleavage activity and/or higher affinity for the CRF02_AG Gag and Gag-Pol substrates, which may in turn result in higher binding affinity for PR mimetic inhibitors. Additional investigation is required to confirm this hypothesis. It is also important to note that HS to PIs is an unintended by-product of divergent evolution within these CRF or subtype lineages. In this case, divergent evolution would likely maintain HIV-1 fitness but survival or expansion of the virus in the human population would be dependent on a combination of different factors such as transmission efficiency and virulence, of which replicative fitness may play only a minor role (36). When analyzing 1,414 viral sequences of CRF02_AG from treatment-naïve subjects retrieved from the Los Alamos National Laboratory HIV Database (http://hiv-web.lanl.gov, accessed in July 2011), we observed low frequencies of these polymorphisms. The 17E polymorphism was found in only 112 sequences (7.9%), while another 145 (10.3%) harbored 64M. Both polymorphisms were found together in higher proportion (31 of 1,414, or 2.2%) than in random distribution (0.6%), suggesting a linkage and/or coevolution. These findings are supported by the higher fitness of virus harboring 17E/64M than of virus harboring either 17E or 64M alone (based on head-to-head competitions). Our fitness results based on direct competition of replication-competent CRF02_AG viruses are in contrast with previous studies using a chimeric virus (subtype B backbone with non-subtype B PR and/or RT coding regions) derived from treatment-naïve patients. These analyses suggest that HS to multiple PIs was related to a lower replicative capacity (RC). These studies measured RC from cells transfected with defective proviral DNA constructs, which are then used in single-cycle, monoinfection assays (25, 26). Furthermore, these RC values of the chimeric virus are compared to an NL4-3 laboratory strain. In our study, we related replicative fitness of the HS-linked polymorphisms in an HIV-1 clone, derived from the same subtype, and then directly competed the wild type and the mutated HIV-1 clones to best approximate the differences in replicative fitness of the CRF02_AG HIV-1 viruses found in the epidemic.
The clinical benefit of HS has been only recently evaluated in terms of treatment outcomes. Patients with HIV-1 subtype B infection, exposed to NRTIs and presenting HS to efavirenz, had a better virologic response during therapeutic salvage that included NNRTIs (33). Such a benefit was corroborated by other clinical trials of salvage therapy (7, 12, 13, 40). The PR drug resistance mutation N88S, which causes resistance to ATV and NFV, also confers HS to APV. N88S in NFV-experienced patients was correlated with a better response to APV-containing salvage therapy, and the presence of N88S appeared to counteract the resistance conferred by I50V, the major mutation linked to drug resistance to APV (21, 47). Another potential benefit of HS-linked polymorphisms could be a delay in the emergence of drug resistance mutations. Recent studies by our group have shown differential correlation between the times to appearance of drug resistance mutations within different HIV-1 subtypes (8, 32, 35), a phenomenon likely influenced by the preexistence of HS-linked polymorphisms. However, the mechanism by which HS-linked polymorphisms counteract, attenuate, and/or delay the emergence of major drug resistance mutations is poorly understood. One obvious scenario may relate to the lower levels of drug required for virus inhibition, and thus, any reductions in trough drug levels between dosing times (magnified with missed dosing/poor adherence) may be less consequential for HIV-1 isolates with HS-linked polymorphisms. Based on analyses of CRF02_AG isolates exposed to NFV, 44% (8/18) of viral isolates without NFV-associated mutations carried 17E and/or 64M, while only 8% (1/12) of those presenting NFV resistance harbored any of these polymorphisms (P = 0.049). These findings suggest that maintenance of drug susceptibility and delay of resistance development are associated with HS-linked polymorphisms.
According to the UNAIDS, 5.2 million people living in developing countries by the end of 2009 were receiving treatment (http://www.unaids.org) The emergence of drug resistance could have devastating consequences due in part to poor treatment monitoring and drug resistance screening. In these countries, where HIV-1 non-B subtypes are prevalent, the beneficial impact of HS-linked polymorphisms could better direct initial and/or salvage therapy. However, additional studies are needed to evaluate the clinical impact of these natural HIV polymorphisms.
Supplementary Material
Footnotes
Published ahead of print 13 February 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Abecasis AB, et al. 2006. Investigation of baseline susceptibility to protease inhibitors in HIV-1 subtypes C, F, G and CRF02_AG. Antivir. Ther. 11:581–589 [PubMed] [Google Scholar]
- 2. Agwale SM, et al. 2006. Genotypic and phenotypic analyses of human immunodeficiency virus type 1 in antiretroviral drug-naive Nigerian patients. AIDS Res. Hum. Retroviruses 22:22–26 [DOI] [PubMed] [Google Scholar]
- 3. Ait-Khaled M, et al. 2002. M184V is associated with a low incidence of thymidine analogue mutations and low phenotypic resistance to zidovudine and stavudine. AIDS 16:1686–1689 [DOI] [PubMed] [Google Scholar]
- 4. Banke S, Lillemark MR, Gerstoft J, Obel N, Jorgensen LB. 2009. Positive selection pressure introduces secondary mutations at Gag cleavage sites in human immunodeficiency virus type 1 harboring major protease resistance mutations. J. Virol. 83:8916–8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boyer PL, Sarafianos SG, Arnold E, Hughes SH. 2002. The M184V mutation reduces the selective excision of zidovudine 5′-monophosphate (AZTMP) by the reverse transcriptase of human immunodeficiency virus type 1. J. Virol. 76:3248–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dam E, et al. 2009. Gag mutations strongly contribute to HIV-1 resistance to protease inhibitors in highly drug-experienced patients besides compensating for fitness loss. PLoS Pathog. 5:e1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Demeter LM, et al. 2008. Association of efavirenz hypersusceptibility with virologic response in ACTG 368, a randomized trial of abacavir (ABC) in combination with efavirenz (EFV) and indinavir (IDV) in HIV-infected subjects with prior nucleoside analog experience. HIV Clin. Trials 9:11–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dumans AT, et al. 2002. Prevalence of protease and reverse transcriptase drug resistance mutations over time in drug-naive human immunodeficiency virus type 1-positive individuals in Rio de Janeiro, Brazil. Antimicrob. Agents Chemother. 46:3075–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dunning J, Nelson M. 2009. Novel strategies to treat antiretroviral-naive HIV-infected patients. J. Antimicrob. Chemother. 64:674–679 [DOI] [PubMed] [Google Scholar]
- 10. Ghosn J, et al. 2011. Polymorphism in Gag gene cleavage sites of HIV-1 non-B subtype and virological outcome of a first-line lopinavir/ritonavir single drug regimen. PLoS One 6:e24798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grossman Z, et al. 2004. Mutation D30N is not preferentially selected by human immunodeficiency virus type 1 subtype C in the development of resistance to nelfinavir. Antimicrob. Agents Chemother. 48:2159–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hammer SM, et al. 2002. Dual vs single protease inhibitor therapy following antiretroviral treatment failure: a randomized trial. JAMA 288:169–180 [DOI] [PubMed] [Google Scholar]
- 13. Haubrich RH, et al. 2002. The clinical relevance of non-nucleoside reverse transcriptase inhibitor hypersusceptibility: a prospective cohort analysis. AIDS 16:F33–F40 [DOI] [PubMed] [Google Scholar]
- 14. Hemelaar J, Gouws E, Ghys PD, Osmanov S. 2011. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 25:679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hertogs K, et al. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holguin A, Alvarez A, Soriano V. 2002. High prevalence of HIV-1 subtype G and natural polymorphisms at the protease gene among HIV-infected immigrants in Madrid. AIDS 16:1163–1170 [DOI] [PubMed] [Google Scholar]
- 17. Johnson VA, et al. 2010. Update of the drug resistance mutations in HIV-1: December 2010. Top. HIV Med. 18:156–163 [PubMed] [Google Scholar]
- 18. Kantor R, Katzenstein D. 2003. Polymorphism in HIV-1 non-subtype B protease and reverse transcriptase and its potential impact on drug susceptibility and drug resistance evolution. AIDS Rev. 5:25–35 [PubMed] [Google Scholar]
- 19. Kantor R, et al. 2005. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med. 2:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lalonde MS, et al. 2007. Sensitive oligonucleotide ligation assay for low-level detection of nevirapine resistance mutations in human immunodeficiency virus type 1 quasispecies. J. Clin. Microbiol. 45:2604–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lam E, Parkin NT. 2003. Amprenavir resistance imparted by the I50V mutation in HIV-1 protease can be suppressed by the N88S mutation. Clin. Infect. Dis. 37:1273–1274 [DOI] [PubMed] [Google Scholar]
- 22. Larder BA, Kemp SD, Harrigan PR. 1995. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 269:696–699 [DOI] [PubMed] [Google Scholar]
- 23. Larrouy L, et al. 2010. Gag mutations can impact virological response to dual-boosted protease inhibitor combinations in antiretroviral-naive HIV-infected patients. Antimicrob. Agents Chemother. 54:2910–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee MH, Sano K, Morales FE, Imagawa DT. 1987. Sensitive reverse transcriptase assay to detect and quantitate human immunodeficiency virus. J. Clin. Microbiol. 25:1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leigh Brown AJ, et al. 2004. Genetic basis of hypersusceptibility to protease inhibitors and low replicative capacity of human immunodeficiency virus type 1 strains in primary infection. J. Virol. 78:2242–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martinez-Picado J, Savara AV, Shi L, Sutton L, D'Aquila RT. 2000. Fitness of human immunodeficiency virus type 1 protease inhibitor-selected single mutants. Virology 275:318–322 [DOI] [PubMed] [Google Scholar]
- 27. Mouroux M, et al. 2001. Low-rate emergence of thymidine analogue mutations and multi-drug resistance mutations in the HIV-1 reverse transcriptase gene in therapy-naive patients receiving stavudine plus lamivudine combination therapy. Antivir. Ther. 6:179–183 [PubMed] [Google Scholar]
- 28. Ode H, et al. 2007. Mechanism of drug resistance due to N88S in CRF01_AE HIV-1 protease, analyzed by molecular dynamics simulations. J. Med. Chem. 50:1768–1777 [DOI] [PubMed] [Google Scholar]
- 29. Pieniazek D, et al. 2000. Protease sequences from HIV-1 group M subtypes A-H reveal distinct amino acid mutation patterns associated with protease resistance in protease inhibitor-naive individuals worldwide. HIV Variant Working Group. AIDS 14:1489–1495 [DOI] [PubMed] [Google Scholar]
- 30. Robertson DL, et al. 2000. HIV-1 nomenclature proposal. Science 288:55–56 [DOI] [PubMed] [Google Scholar]
- 31. Sanches M, et al. 2007. Structural characterization of B and non-B subtypes of HIV-protease: insights into the natural susceptibility to drug resistance development. J. Mol. Biol. 369:1029–1040 [DOI] [PubMed] [Google Scholar]
- 32. Santos AF, Abecasis AB, Vandamme AM, Camacho RJ, Soares MA. 2009. Discordant genotypic interpretation and phenotypic role of protease mutations in HIV-1 subtypes B and G. J. Antimicrob. Chemother. 63:593–599 [DOI] [PubMed] [Google Scholar]
- 33. Shulman N, et al. 2001. Phenotypic hypersusceptibility to non-nucleoside reverse transcriptase inhibitors in treatment-experienced HIV-infected patients: impact on virological response to efavirenz-based therapy. AIDS 15:1125–1132 [DOI] [PubMed] [Google Scholar]
- 34. Soares EA, et al. 2009. Mutation T74S in HIV-1 subtype B and C proteases resensitizes them to ritonavir and indinavir and confers fitness advantage. J. Antimicrob. Chemother. 64:938–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soares EA, et al. 2007. Differential drug resistance acquisition in HIV-1 of subtypes B and C. PLoS One 2:e730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tebit DM, Arts EJ. 2011. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect. Dis. 11:45–56 [DOI] [PubMed] [Google Scholar]
- 37. Tebit DM, et al. 2010. Divergent evolution in reverse transcriptase (RT) of HIV-1 group O and M lineages: impact on structure, fitness, and sensitivity to nonnucleoside RT inhibitors. J. Virol. 84:9817–9830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tebit DM, Zekeng L, Kaptue L, Krausslich HG, Herchenroder O. 2003. Construction and characterisation of a full-length infectious molecular clone from a fast replicating, X4-tropic HIV-1 CRF02.AG primary isolate. Virology 313:645–652 [DOI] [PubMed] [Google Scholar]
- 39. Tippmann HF. 2004. Analysis for free: comparing programs for sequence analysis. Brief. Bioinform. 5:82–87 [DOI] [PubMed] [Google Scholar]
- 40. Tozzi V, et al. 2004. Mutations in HIV-1 reverse transcriptase potentially associated with hypersusceptibility to nonnucleoside reverse-transcriptase inhibitors: effect on response to efavirenz-based therapy in an urban observational cohort. J. Infect. Dis. 189:1688–1695 [DOI] [PubMed] [Google Scholar]
- 41. Velazquez-Campoy A, Todd MJ, Vega S, Freire E. 2001. Catalytic efficiency and vitality of HIV-1 proteases from African viral subtypes. Proc. Natl. Acad. Sci. U. S. A. 98:6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vergne L, et al. 2003. Low rate of genotypic HIV-1 drug-resistant strains in the Senegalese government initiative of access to antiretroviral therapy. AIDS 17(Suppl. 3):S31–S38 [DOI] [PubMed] [Google Scholar]
- 43. Vergne L, et al. 2000. Genetic diversity of protease and reverse transcriptase sequences in non-subtype-B human immunodeficiency virus type 1 strains: evidence of many minor drug resistance mutations in treatment-naive patients. J. Clin. Microbiol. 38:3919–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vergne L, et al. 2006. Natural polymorphism in protease and reverse transcriptase genes and in vitro antiretroviral drug susceptibilities of non-B HIV-1 strains from treatment-naive patients. J. Clin. Virol. 36:43–49 [DOI] [PubMed] [Google Scholar]
- 45. Vidal N, et al. 2006. HIV type 1 pol gene diversity and antiretroviral drug resistance mutations in the Democratic Republic of Congo (DRC). AIDS Res. Hum. Retroviruses 22:202–206 [DOI] [PubMed] [Google Scholar]
- 46. Weinheimer S, Discotto L, Friborg J, Yang H, Colonno R. 2005. Atazanavir signature I50L resistance substitution accounts for unique phenotype of increased susceptibility to other protease inhibitors in a variety of human immunodeficiency virus type 1 genetic backbones. Antimicrob. Agents Chemother. 49:3816–3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zachary KC, Hanna GJ, D'Aquila RT. 2001. Human immunodeficiency virus type 1 hypersusceptibility to amprenavir in vitro can be associated with virus load response to treatment in vivo. Clin. Infect. Dis. 33:2075–2077 [DOI] [PubMed] [Google Scholar]
- 48. Ziermann R, et al. 2000. A mutation in human immunodeficiency virus type 1 protease, N88S, that causes in vitro hypersensitivity to amprenavir. J. Virol. 74:4414–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.