Abstract
As a consequence of multidrug resistance, clinicians are highly dependent on fluoroquinolones for treating the serious systemic infection typhoid fever. While reduced susceptibility to fluoroquinolones, which lessens clinical efficacy, is becoming ubiquitous, comprehensive resistance is exceptional. Here we report ofloxacin treatment failure in typhoidal patient infected with a novel, highly fluoroquinolone-resistant isolate of Salmonella enterica serovar Typhi. The isolation of this organism has serious implications for the long-term efficacy of ciprofloxacin and ofloxacin for typhoid treatment.
TEXT
Antimicrobial therapy is critical for typhoid treatment, but the circulation of antimicrobial-resistant organisms has become ubiquitous in many regions of endemicity, and the presence of multidrug-resistant Salmonella enterica serovars Typhi and Paratyphi A (showing resistance to chloramphenicol, trimethoprim-sulfamethoxazole, and ampicillin) precludes treatment with these antimicrobials (6). Clinicians are now highly dependent on the fluoroquinolones for typhoid therapy. Yet, predictably, widespread fluoroquinolone usage has been followed by the emergence of isolates with elevated MICs (1). These isolates are characterized by point mutations within the gyrA (DNA gyrase) gene and occasionally an additional nucleotide substitution in the parC gene (7). The propagation of these organisms is particularly concerning when one considers a lack of feasible alternatives and an explicit correlation between increasing MICs to fluoroquinolones and treatment failure (8). Yet, despite the widespread dissemination of such organisms, the isolation of organisms that exhibit MICs of >1.0 μg/ml to ofloxacin has been, until now, negligible.
In June 2011, a 13-year-old male presented to the outpatient department at Patan Hospital in Kathmandu, Nepal, with a fever that had started 10 days previously, peaking at around 39°C daily. The patient also developed a headache and mild abdominal discomfort with nausea. On the eighth day of fever, the patient was taken to a local medical store, where it was recommended that he be treated with 200 mg of ofloxacin to be taken twice daily. After 2 days of ofloxacin treatment, the patient's symptoms became more pronounced, and he showed increased restlessness and a temperature in excess of 40°C. He presented to the outpatient department on the tenth day of illness but had no signs of icterus, anemia, lymphadenopathy, cyanosis, edema, or dehydration and no rash on the trunk. His temperature was recorded at 38.8°C, with a pulse of 104/min and a respiratory rate of 24/min. A clinical diagnosis of typhoid was made, and a complete blood count and culture and sensitivity were requested. The treating clinician increased the ofloxacin dosage to 300 mg (20 mg/kg of body weight/day) twice daily for 7 days. However, the patient returned to the outpatient department after the additional 7 days of ofloxacin treatment without improvement. In the intervening period, his blood culture had yielded Salmonella Typhi, which was highly resistant to ofloxacin (zone size, 11 mm), ciprofloxacin (zone size, 11 mm), and nalidixic acid (zone size, 0 mm); a secondary blood culture was not taken. On the basis of these findings, he was prescribed oral azithromycin (20 mg/kg/day) once daily for 7 days. The fever declined on the third day of the azithromycin treatment (the twentieth day of fever), and ultimately, the patient made a fortuitous and uneventful recovery.
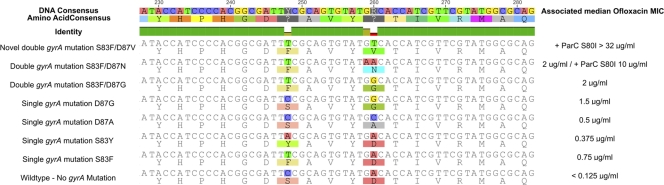
After the patient had recovered from the infection, and with written consent, we investigated the Salmonella Typhi isolate. The resulting fluoroquinolone MICs were exceptional; in excess of 256 μg/ml against nalidixic acid, greater than 32 μg/ml against ofloxacin and ciprofloxacin, 6 μg/ml against levofloxacin, and 2 μg/ml against gatifloxacin. The Salmonella Typhi isolate, was not, however, multidrug resistant. We purified DNA from the Salmonella Typhi isolate, aiming to define the molecular basis of the fluoroquinolone resistance. We PCR amplified the gyrA, gyrB, and parC genes and sequenced the resultant PCR amplicons (1). We also attempted to amplify the common Gram-negative plasmid-mediated quinolone resistance (PMQR) determinants: qnrA, qnrB, qnrS, aac(6)lb-cr, and qepA (5). We were unable to detect any of the five common PMQR genes. However, DNA sequencing of the three fluoroquinolone target loci identified a single previously described mutation in the parC gene, changing serine to isoleucine at codon 80 (S80I), and a double mutation in the gyrA gene (Fig. 1). The two gyrA mutations were both within the quinolone resistance-determining region (QRDR). The primary gyrA substitution was common, inducing a replacement of serine with phenylalanine at codon 83. However, the second mutation was novel, the substitution of cytosine for thymine at nucleotide 248 had the effect of changing aspartic acid to valine at codon 87 (Fig. 1). The resulting DNA sequence was submitted to EMBL.
Fig 1.
DNA and predicted amino acid alignments of the DNA gyrase gene (gyrA) from Salmonella Typhi isolates with reduced susceptibility to fluoroquinolones. Shown is an outline of seven identified conformations of the QRDR of the DNA gyrase gyrA gene in clinical Salmonella Typhi isolates based on the 2010 study by Parry et al. (7) and the novel S83F D87V mutation isolated here. The DNA and the corresponding amino acid consensus are shown in the first and second rows of the figure, respectively, with the DNA identity between sequences shown beneath. For each conformation, the DNA sequence is shown at the top and the predicted amino acid sequence is shown beneath. The median MIC of ofloxacin for each of the mutations (with and without a tertiary S80I mutation in the ParC topoisomerase) is shown to the right, based on data from this report and reference 3.
Antimicrobials are crucial for treating typhoid, and resistance is the main constraint that compels antimicrobial therapy preferences to change with time. The current WHO guidelines suggest that the fluoroquinolones are the optimal group of antimicrobials for uncomplicated typhoid treatment in adults (10). However, Salmonella Typhi and Salmonella Paratyphi A isolates with reduced susceptibility to fluoroquinolones are now common in Asia and are becoming increasingly common in Africa (2, 9), yet complete resistance is rare (4). Here we report an isolate exhibiting extensive resistance against fluoroquinolones, marked by a novel gyrA mutation and no additional PMQR sequences. Clearly, additional characterization of this isolate is required to precisely understand the basis of its phenotype, yet its isolation should ring alarm bells within the typhoid community. Indeed, one may speculate that other such strains will emerge rapidly, should the organism have acceptable biological fitness and favorable dissemination conditions. Anecdotally, we find that chloramphenicol is making a comeback in the community in Nepal as a consequence of reducing resistance levels, yet, azithromycin will likely become the preferred drug as ofloxacin and ciprofloxacin become ineffective (3).
Nucleotide sequence accession number.
The DNA sequence resulting from the cytosine for thymine substitution at nucleotide 248 has been submitted to EMBL (accession no. HE588040).
ACKNOWLEDGMENTS
We thank the microbiology laboratories at Patan Hospital in Nepal and the Oxford University Clinical Research Unit in Vietnam.
This work was supported by The Wellcome Trust of Great Britain, S.B. is supported by a fellowship from the OAK Foundation through Oxford University.
Footnotes
Published ahead of print 27 February 2012
REFERENCES
- 1. Chau TT, et al. 2007. Antimicrobial drug resistance of Salmonella enterica serovar Typhi in Asia and molecular mechanism of reduced susceptibility to the fluoroquinolones. Antimicrob. Agents Chemother. 51:4315–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chuang CH, et al. 2009. Surveillance of antimicrobial resistance of Salmonella enterica serotype Typhi in seven Asian countries. Epidemiol. Infect. 137:266–26918474127 [Google Scholar]
- 3. Dolecek C, et al. 2008. A multi-center randomised controlled trial of gatifloxacin versus azithromycin for the treatment of uncomplicated typhoid fever in children and adults in Vietnam. PLoS One 3:e2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dutta S, et al. 2008. Emergence of highly fluoroquinolone-resistant Salmonella enterica serovar Typhi in a community-based fever surveillance from Kolkata, India. Int. J. Antimicrob. Agents 31:387–389 [DOI] [PubMed] [Google Scholar]
- 5. Le TM, et al. 2009. High prevalence of plasmid-mediated quinolone resistance determinants in commensal members of the Enterobacteriaceae in Ho Chi Minh City, Vietnam. J. Med. Microbiol. 58:1585–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parry CM, Threlfall EJ. 2008. Antimicrobial resistance in typhoidal and nontyphoidal salmonellae. Curr. Opin. Infect. Dis. 21:531–538 [DOI] [PubMed] [Google Scholar]
- 7. Parry CM, et al. 2010. Suitable disk antimicrobial susceptibility breakpoints defining Salmonella enterica serovar Typhi isolates with reduced susceptibility to fluoroquinolones. Antimicrob. Agents Chemother. 54:5201–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parry CM, et al. 2011. The influence of reduced susceptibility to fluoroquinolones in Salmonella enterica serovar Typhi on the clinical response to ofloxacin therapy. PLoS Negl. Trop. Dis. 5:e1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith AM, Govender N, Keddy KH. 2010. Quinolone-resistant Salmonella Typhi in South Africa, 2003–2007. Epidemiol. Infect. 138:86–90 [DOI] [PubMed] [Google Scholar]
- 10. World Health Organization 2003. Background document: the diagnosis, treatment and prevention of typhoid fever. WHO/V&B/03.07. World Health Organization, Geneva, Switzerland [Google Scholar]