Abstract
One hundred sixty-nine nonreplicate imipenem-resistant Pseudomonas aeruginosa strains isolated in a large hospital on the coastal region of Croatia were studied. The most active antibiotics were colistin and amikacin. Most of the isolates were multiresistant. The most prevalent serotype was O12, followed by O11. Six strains carried the blaVIM-2 gene located in a novel class 1 integron composed in its variable part of the blaVIM-2-blaoxa-10-ΔqacF-aacA4 genes. Metallo-β-lactamase-producing strains belonged to sequence types ST235 and ST111.
TEXT
Acquired metallo-β-lactamases (MBLs) are emerging worldwide as powerful resistance determinants in Gram-negative bacteria (3). So far, six MBL enzyme types have been described in clinical isolates of Pseudomonas aeruginosa—VIM type, IMP type, SPM type, GIM type, AIM type, and, most recently, the NDM type found among clinical isolates from Serbia, a Balkan country (3, 14).
VIM-type enzymes appeared in the Mediterranean area with the first report of VIM-1 in Italy from a P. aeruginosa strain isolated in 1997, and then the observation of an allelic variant, VIM-2, in France from a P. aeruginosa strain isolated in 1996 (3). To date, all Mediterranean countries on the European side reported the presence of MBL in P. aeruginosa, mainly of the VIM type (Spain, France, Italy, Croatia, Greece, and Turkey) but also of the IMP type (Italy and France). Some of the VIM-producing P. aeruginosa strains, particularly in Italy and Greece, were involved in nosocomial outbreaks (3, 29). Most acquired MBL genes are carried on mobile gene cassettes inserted in integrons, associated with mobile DNA elements (transposons and plasmids) (20). Those integrons often harbored additional gene cassettes carrying resistance determinants for other antibiotic classes (e.g., aminoglycosides), disinfectants, or other β-lactamase genes (7). Therefore, multidrug-resistant (MDR) strains with colistin-only-susceptible (COS) phenotypes are becoming highly prevalent (3, 8).
In Croatia, a Mediterranean country situated on the eastern coast of the Adriatic Sea, and in the west Balkans, two studies so far have reported the presence of VIM-2 MBL in P. aeruginosa (1, 28) without sufficient characterization or comparison with isolates from other countries. Here, a larger-scale analysis of the prevalence, type, and genetic support of MBL genes in a collection of imipenem-resistant P. aeruginosa isolates from Croatia is presented.
(Part of this study was presented as a poster [P773] at the 20th European Congress of Clinical Microbiology and Infectious Diseases 2010, Vienna, Austria.)
A total of 169 consecutive nonredundant P. aeruginosa isolates resistant to imipenem (MIC, >8 μg/ml) were collected from various clinical specimens in Split University Hospital, the second largest tertiary-care hospital (1,400 beds) in Croatia, during the period 2001 to 2007. Isolates from 2001 and 2002 were periodically collected because of exceptional resistance, and from 2003 to 2007, most consecutively isolated imipenem-resistant strains were collected and conserved for further analyses. Testing of susceptibility to common antipseudomonal antibiotics was done using broth microdilution, and the results were interpreted according to CLSI standards (2). P. aeruginosa ATCC 27853 was used as a quality control strain (2). Multiresistance was defined as resistance to imipenem and to at least one antibiotic representative of two more classes (i.e., ureidopenicillins, cephalosporins, aminoglycosides, or fluoroquinolones), according to the last proposal for standard definitions (18).
The most effective antipseudomonal antibiotics were colistin (85% susceptible strains) and amikacin (83% susceptible strains), followed by ceftazidime, piperacillin-tazobactam, and ciprofloxacin (with 31%, 28%, and 27% of susceptible strains). One hundred thirty-eight P. aeruginosa isolates (82%) were multiresistant. Eight strains (5%) exhibited a colistin-only susceptible (COS) phenotype. To assess the possible role of derepressed AmpC or upregulated efflux in resistance, MICs of ceftazidime of COS isolates were determined in the presence of 300 μg/ml of phenylboronic acid (Sigma) (32), while MICs of meropenem were determined in the presence of 12.5 μM carbonyl cyanide-m-chlorophenylhydrazone (CCCP) (Sigma) (24). The presence of OprD in COS isolates was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 4% stacking and 11% separating gels, as previously described (4). The carbapenemase activity of crude cell extracts of COS strains was assessed spectrophotometrically, as previously described (20). In seven out of eight COS strains, phenylboronic acid significantly (4 times or more) decreased MICs of ceftazidime, while in six out of eight COS isolates, CCCP significantly decreased MICs of meropenem, as shown in Table 1. The majority of COS isolates were found to lack an OprD outer membrane protein (data not shown). No imipenem hydrolysis was detected in the spectrophotometric assay. The multiresistance of COS P. aeruginosa strains appeared to be due to an interplay of hyperexpressed AmpC, upregulation of efflux pumps, and the lack of the outer membrane D1 pathway, as previously described (6). MICs of imipenem were also determined in the presence of EDTA and phenanthroline using the EPI (EDTA-phenanthroline-imipenem) test, with P. aeruginosa VA-182 as a control strain (19, 20). MBL genes were screened by multiplex PCR using pairs of VIM and IMP primers amplifying specifically blaVIM and blaIMP genes, using blaVIM- and blaIMP-harboring strains (VA-182, and AC-54/97, respectively) as positive controls (20, 25). Additional PCRs, with primers specific for blaSPM, blaGIM, and blaNDM genes were performed with COS isolates (14, 33). Class 1 integrons were amplified by PCR using INT primers which targeted conserved regions, while variable arrays of integrons were characterized by a “primer-walking” strategy with specific primers (20).
Table 1.
MICs of antipseudomonal antibiotics and effects of phenylboronic acid and CCCP, OprD presence, and serotypes of colistin-only-susceptible Pseudomonas aeruginosa isolates
| Strain no. | MIC (μg/ml) ofa: |
OprD presence | Serotype | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | CAZ + BA | TZP | IPM | MEM | MEM + CCCP | AMK | CIP | |||
| 31 | 32 | 4 | 128 | 16 | 32 | 8 | 32 | 64 | No | O12 |
| 146 | 32 | 4 | >128 | 16 | >128 | 64 | 32 | 32 | No | O11 |
| 149 | 128 | 16 | >128 | 16 | 64 | 16 | 32 | 32 | No | O11 |
| 150 | 64 | 8 | >128 | 16 | 32 | 8 | 64 | 32 | No | O3 |
| 153 | 64 | 16 | >128 | 16 | 64 | 8 | 32 | 32 | No | O11 |
| 154 | 64 | 16 | >128 | 16 | 32 | 32 | 32 | 32 | No | O11 |
| 156 | 64 | 32 | >128 | 16 | 128 | 32 | 32 | 32 | No | O11 |
| 175 | 32 | 32 | >128 | 16 | 64 | 16 | 32 | 32 | Yes | O12 |
CAZ, ceftazidime; BA, phenylboronic acid; TZP, piperacillin-tazobactam; IPM, imipenem; MEM, meropenem; CCCP, carbonyl cyanide-m-chlorophenylhydrazone; AMK, amikacin; CIP, ciprofloxacin.
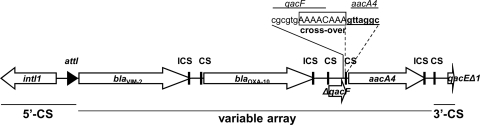
Six isolates (3%) showed a significant decrease of imipenem MICs in the presence of EDTA and phenanthroline ranging from 16 to >32-fold and carried the blaVIM-2 MBL gene. MBL-producing strains were susceptible to colistin, and all except one (susceptible to aztreonam) were susceptible to piperacillin-tazobactam. None of the COS strains revealed the presence of an MBL gene, indicating other mechanisms of resistance besides carbapenemase were involved, as discussed previously. According to PCR mapping and sequence analysis, the blaVIM-2 gene was located within the first gene cassette of a class 1 integron cassette array, which also included a blaOXA-10, a truncated qacF, and an aacA4 cassette, followed by qacEΔ1 typical of 3′-conserved segments (Fig. 1). All VIM-producing isolates carried the same integron. According to the sequences, the Pc promoter of this integron was a variant designated PcWTGM-10, providing strong expression of the gene cassette array, but associated with weak integrase IntI activity (15). This could possibly explain the stable integron content observed during a 6-year period with a likely repetitive independent acquisition of the integron from environmental sources (7, 21). The cassette array of this class 1 integron was unusual in two aspects: first, although the coexistence of different β-lactamases is frequent in MBL-carrying integrons, in this one, two neighboring gene cassettes encoded two different β-lactamases (VIM-2 and OXA-10), a situation similar to the one described for P. aeruginosa isolates harboring VIM-6 and OXA-10 in Asia (16); second, a remnant of the qacF gene, seldomly reported in integrons harboring MBL genes, could have a residual function in promoting transcription of adjacent genes, as already described for other qac genes (qacEΔ1 and qacH) (7, 9, 13). The truncated qacF cassette possibly originated through an anomalous recombination event at its 3′ end (Fig. 1). The fourth gene cassette, aacA4, encoding a 6′-N-aminoglycoside acetyltransferase, is frequently found in integrons containing MBL-encoding genes (20, 29).
Fig 1.
Schematic presentation of the class 1 integron carrying blaVIM-2 gene cassette (In493; no. AM392427). CS, core site; ICS, inverse core site; 5′- and 3′-CS, 5′- and 3′-conserved segments.
Southern blot hybridization was performed on whole genomic DNA of two representatives of MBL-carrying strains using a PCR-generated fragment of the entire blaVIM-2 gene (P. aeruginosa strain VA-182) labeled with 32P by the random priming technique (Rediprime II DNA labeling system; Amersham Biosciences), as previously described (20). For both isolates, Southern blot hybridization yielded a positive signal indicative of the presence of the blaVIM-2 determinant on the chromosomal DNA (data not shown).
O serotypes were determined by the slide agglutination method using commercial antisera O1 to O16 (Bio-Rad, France). The most common serotype was O12 (99 isolates; 59% of all isolates), followed by O11 (29 isolates; 17%) and O1 (7 isolates; 4%), respectively. Eighteen strains (11%) were nontypeable. Most of the O12 (85%) and O11 (69%) isolates were multiresistant and also represented the majority of COS isolates, which is in accordance with previous studies suggesting a possible role of these serotypes in dissemination of multiresistance (11, 12, 23).
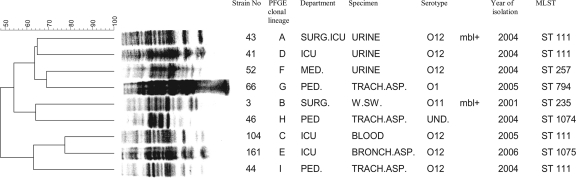
Clonal diversity was assessed by pulsed-field gel electrophoresis (PFGE) using XbaI restriction enzyme and a CHEF-DRIII apparatus (Bio-Rad) as previously described (27). Clonal relatedness was determined using GELCompar software (Applied Maths, Kortrijk, Belgium) with 2% position tolerance and Dice coefficient. A dendrogram representing the relatedness between the isolates was built by the unweighted-pair group method using arithmetic averages (UPGMA) based on the similarity values calculated with Dice coefficient. A cutoff of ≥80%, equivalent to the criterion of “possibly related” by Tenover et al., was applied (31). Among the 169 isolates, PFGE analysis allowed us to define nine major clonal lineages. Seventy-seven isolates (45%) belonged to lineage A, which was found to be the most prevalent, while 61 isolates (36%) belonged to clonal lineage B. The dominant clone A consisted mainly of O12 isolates (86%), whereas clone B was more heterogeneous, consisting mostly of O11 (40%) and O12 (21%) isolates and suggesting that serotyping can lead to a misleading classification of the isolates. Variable serotypes among a clonal lineage could involve recent genetic changes and recombination events (12, 22). All COS strains belonged to the same clone (clone B). Five MBL-positive strains were of the O11 serotype and belonged to clone B, while the remaining MBL-producing strain was of the O12 serotype and belonged to clone A. Further analysis of P. aeruginosa infraspecific diversity was performed by multilocus sequence typing (MLST) according to the published protocols (5, 10). MLST was applied to two VIM-carrying P. aeruginosa isolates on representative strains of the remaining clonal lineages C to I (Fig. 2). Nucleotide sequences were determined on both strands and searched against the MLST database (http://pubmlst.org/paeruginosa/) for the assignment of allelic profiles and sequence types (STs). MLST analysis identified six STs among nine strains with distinct PFGE profiles and two newly described STs—ST1074 (closest to ST212) and 1075 (closest to ST111). The MBL-producing O12 strain belonging to PFGE clone A was typed as ST111, while the MBL-producing O11 strain of clone B belonged to ST235. Representatives of clones A, C, D, and I were part of ST111, while other clones' representatives displayed distinct STs, as shown in Fig. 2. The MLST findings were in support of previous analysis proving the presence of two international clonal complexes—CC235, represented by the predicted founder, ST235, and CC111, represented by ST111—in dissemination of blaVIM-harboring P. aeruginosa (34). VIM-producing P. aeruginosa strains belonging to these two international clones had already been reported from countries bordering Croatia, like Serbia (ST235), Hungary (ST235 and ST230), and Italy (ST111, ST235, and ST227) (10, 17, 34). The prevalence of MBL-producing P. aeruginosa isolates in this part of Europe ranged from 0.1% in Spain and 0.8% in Greece to 1.3% in Italy (3, 26, 29). Taking into account the 10% of imipenem-resistant isolates reported during the last decade in Croatia (30), the estimated prevalence of MBL-producing P. aeruginosa strains would be 0.3% in Croatia. In spite of these numbers, P. aeruginosa can evolve rapidly and can select a beneficial genetic configuration that can lead to dramatic hospital outbreaks, as already reported in the Mediterranean basin and throughout the world (3, 4, 8, 34).
Fig 2.
Dendrogram showing PFGE profiles and corresponding STs of Pseudomonas aeruginosa strains representative of the clonal lineages. SURG.ICU, Surgical Intensive Care Unit; MED., Department of Medicine; PED., Department of Pediatrics; TRACH.ASP., tracheal aspirate; W.SW., wound swab; BRONCH.ASP., bronchial aspirate; UND., undetermined.
In conclusion, a low prevalence of metallo-β-lactamase-producing P. aeruginosa isolates in Croatia was observed, but the reported strains belonged to epidemic clones that spread over the whole of Mediterranean Europe.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper was assigned GenBank accession no. AM392427. The integron sequence reported in this paper was submitted to the INTEGRALL database (http://integrall.bio.ua.pt/) and was assigned no. In493.
ACKNOWLEDGMENTS
This work was supported by the Croatian Ministry of Science, Education and Sport (grant no. 108-1080-0015).
We thank Dubravko Sijak and Stjepan Katic for technical assistance.
Footnotes
Published ahead of print 27 February 2012
REFERENCES
- 1. Bosnjak Z, et al. 2010. VIM-2 β-lactamase in Pseudomonas aeruginosa isolates from Zagreb, Croatia. Scand. J. Infect. Dis. 42:193–197 [DOI] [PubMed] [Google Scholar]
- 2. Clinical and Laboratory Standards Institute 2009. Performance standards for antimicrobial susceptibility testing: 19th informational supplement M100-S19. CLSI, Wayne, PA [Google Scholar]
- 3. Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect. Dis. 11:381–393 [DOI] [PubMed] [Google Scholar]
- 4. Cornaglia G, Mazzariol A, Lauretti L, Rossolini GM, Fontana R. 2000. Hospital outbreak of carbapenem resistant Pseudomonas aeruginosa producing VIM-1, a novel transferable metallo-β-lactamase. Clin. Infect. Dis. 31:1119–1125 [DOI] [PubMed] [Google Scholar]
- 5. Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42:5644–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. El Amin N, et al. 2005. Carbapenem resistance mechanisms in Pseudomonas aeruginosa: alterations of porin OprD and efflux proteins do not fully explain resistance patterns observed in clinical isolates. APMIS 113:187–196 [DOI] [PubMed] [Google Scholar]
- 7. Fluit AC, Schmitz F-J. 2004. Resistance integrons and super-integrons. Clin. Microbiol. Infect. 10:272–288 [DOI] [PubMed] [Google Scholar]
- 8. Fonseca EL, Freitas dos Santos F, Vicente AC. 2010. The colistin-only-sensitive Brazilian Pseudomonas aeruginosa clone SP (sequence type 277) is spread worldwide. Antimicrob. Agents Chemother. 54:2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gillings MR, Xuejun D, Hardwick SS, Holley MP, Stokes HW. 2009. Gene cassettes encoding resistance to quaternary ammonium compounds: a role in the origin of clinical class 1 integrons? ISME J. 3:209–215 [DOI] [PubMed] [Google Scholar]
- 10. Giske CG, et al. 2006. Establishing clonal relationships between VIM-1-like metallo-β-lactamase-producing Pseudomonas aeruginosa strains from four European countries by multilocus sequence typing. J. Clin. Microbiol. 44:4309–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Higgins PG, Fluit AC, Milatovic D, Verhoef J, Schmitz FJ. 2002. Antimicrobial susceptibility of imipenem-resistant Pseudomonas aeruginosa. J. Antimicrob. Chemother. 50:299–301 [DOI] [PubMed] [Google Scholar]
- 12. Jamasbi RJ, Proudfoot EM. 2008. Phenotypic and genotypic characteristics of clinical isolates of Pseudomonas aeruginosa: rate of occurrence and distribution of different serotypes, antimicrobial susceptibility profiles. Lab. Med. 39:155–161 [Google Scholar]
- 13. Jeong JH, Shin KS, Lee JW, Park EJ, Son SY. 2009. Analysis of a novel class 1 integron containing metallo-β-lactamase gene VIM-2 in Pseudomonas aeruginosa. J. Microbiol. 47:753–759 [DOI] [PubMed] [Google Scholar]
- 14. Jovcic B, et al. 2011. Emergence of NDM-1 metallo-β-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob. Agents Chemother. 55:3929–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jove T, Da Re S, Denis F, Mazel D, Ploy M-C. 2010. Inverse correlation between promoter strength and excision activity in class 1 integron. PLoS Genet. 6:e1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koh TH, Yamaguchi K, Ishii Y. 2008. Characterisation of the metallo-β-lactamase VIM-6 and its genetic support. Int. J. Antimicrob. Agents 32:446–449 [DOI] [PubMed] [Google Scholar]
- 17. Maatallah M, et al. 2011. Population structure of Pseudomonas aeruginosa from five Mediterranean countries: evidence for frequent recombination and epidemic occurrence of CC235. PLoS One 6:e25617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Magiorakos AP, et al. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18:268–281 [DOI] [PubMed] [Google Scholar]
- 19. Migliavacca R, et al. 2002. Simple microdilution test for detection of metallo-β-lactamase production in Pseudomonas aeruginosa. J. Clin. Microbiol. 40:4388–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pallecchi L, Riccio ML, Docquier JD, Fontana R, Rossolini GM. 2001. Molecular heterogeneity of blaVIM-2-containing integrons from Pseudomonas aeruginosa plasmids encoding the VIM-2 metallo-β-lactamase. FEMS Microbiol. Lett. 195:145–150 [DOI] [PubMed] [Google Scholar]
- 21. Pellegrini C, et al. 2009. Identification of bla(IMP-22) in Pseudomonas spp. in urban wastewater and nosocomial environments: biochemical characterization of a new IMP metallo-enzyme variant and its genetic location. J. Antimicrob. Chemother. 63:901–908 [DOI] [PubMed] [Google Scholar]
- 22. Pirnay JP, et al. 2009. Pseudomonas aeruginosa population structure revisited. PLoS One 13:e7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pitt TL, Livermore DM, Miller G, Vatopoulos A, Legakis NJ. 1990. Resistance mechanism of multiresistant serotype O12 Pseudomonas aeruginosa isolated in Europe. J. Antimicrob. Chemother. 26:319–328 [DOI] [PubMed] [Google Scholar]
- 24. Pournaras S, et al. 2005. Spread of efflux pump-overexpressing, non-metallo-β-lactamase-producing, meropenem-resistant but ceftazidime-susceptible Pseudomonas aeruginosa in a region with blaVIM endemicity. J. Antimicrob. Chemother. 56:761–764 [DOI] [PubMed] [Google Scholar]
- 25. Riccio ML, et al. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of bla(IMP) allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rossolini GM, et al. 2008. First countrywide survey of acquired metallo-β-lactamases in gram-negative pathogens in Italy. Antimicrob. Agents Chemother. 52:4023–4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sambrook J, Fritsch EF, Maniatis T. (ed). 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28. Sardelic S, Pallecchi L, Punda Polic V, Rossolini GM. 2003. Carbapenem-resistant Pseudomonas aeruginosa carrying VIM-2 metallo-β-lactamase determinants, Croatia. Emerg. Infect. Dis. 9:1022–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Strateva T, Yordanov D. 2009. Pseudomonas aeruginosa—a phenomenon of bacterial resistance. J. Med. Microbiol. 58:1133–1148 [DOI] [PubMed] [Google Scholar]
- 30. Tambic Andrasevic A, Tambic T. 2009. Antibiotic resistance in 2008, p 9–98 In Tambic T, Tambic Andrasevic A. (ed), Antibiotic resistance in Croatia, 2008. The Croatian Academy of Medical Sciences, Zagreb, Croatia [Google Scholar]
- 31. Tenover FC, et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Upadhyay S, Sen MR, Bhattacharjee A. 2011. Diagnostic utility of boronic acid inhibition with different cephalosporins against Escherichia coli producing AmpC β-lactamases. J. Med. Microbiol. 60:691–693 [DOI] [PubMed] [Google Scholar]
- 33. Woodford N. 2010. Rapid characterization of β-lactamases by multiplex PCR. Methods Mol. Biol. 642:181–192 [DOI] [PubMed] [Google Scholar]
- 34. Woodford N, Turton F, Livermore D. 2011. Multiresistant Gram-negative bacteria: the role of high risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35:736–755 [DOI] [PubMed] [Google Scholar]