Abstract
Alignment of DNA sequences found upstream of aphA6 and all blaNDM-1 genes displays 100% identity. This identity continues 19 bp into the blaNDM-1 gene such that the first 6 amino acids of aphA6 and blaNDM-1 are the same. Furthermore, the percent GC content (GC%) of aphA6 is considerably lower than that of blaNDM-1 and the GC% within the blaNDM-1 structural gene changes dramatically after the first 19 bp. This is unequivocal evidence that blaNDM-1 is a chimera.
TEXT
Carbapenems are potent broad-spectrum antibiotics that are reserved for life-threatening bacterial infections (7). However, their effectiveness is increasingly compromised by resistance due to carbapenem-hydrolyzing β-lactamases (11). One such enzyme, the New Delhi metallo-β-lactamase (NDM-1), was unknown before 2008, and yet media interest has enabled it to become a focus of world attention (18). We have previously shown that NDM-1 is widely disseminated in the United Kingdom and South Asia (5). We have also identified blaNDM-1 genes in a broad range of Gram-negative bacteria isolated from the New Delhi environment, including the serious pathogens Shigella boydii and Vibrio cholerae (19). Possession of blaNDM-1 confers resistance to all classes of β-lactam antibiotics (penicillins, cephalosporins, and carbapenems), with the sole exception of the monobactam aztreonam (20). In this, NDM-1 resembles previously characterized MBLs but differs from those in its ability to associate with the bacterial outer membrane and in its wide species distribution (4, 19). It is often associated with other antibiotic resistance genes on broad-host-range plasmids that provide transferable cross-class, and occasionally pan-antibiotic, resistance (5). In the continuing absence of new antibacterials effective against Gram-negative pathogens, the spread of multiply resistant NDM-1-producing strains is a cause for significant concern, as evidenced by the recent declaration by the World Health Organization of 7 April 2011 as World Antibiotic Resistance Day. However, the factors responsible for the unprecedented speed of dissemination of blaNDM-1 remain to be established. Here we use sequence comparisons to show that blaNDM-1 is a chimeric gene that has arisen by the in-frame fusion of a preexisting MBL gene with the aminoglycoside resistance gene aphA6. We propose that the resulting changes in NDM-1 expression, and the properties and processing of the expressed protein, partially explain the greater success with which NDM-1 has disseminated compared to other MBLs that confer a similar resistance phenotype.
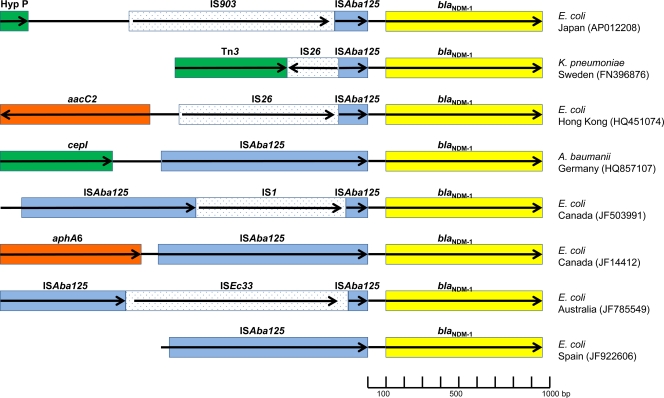
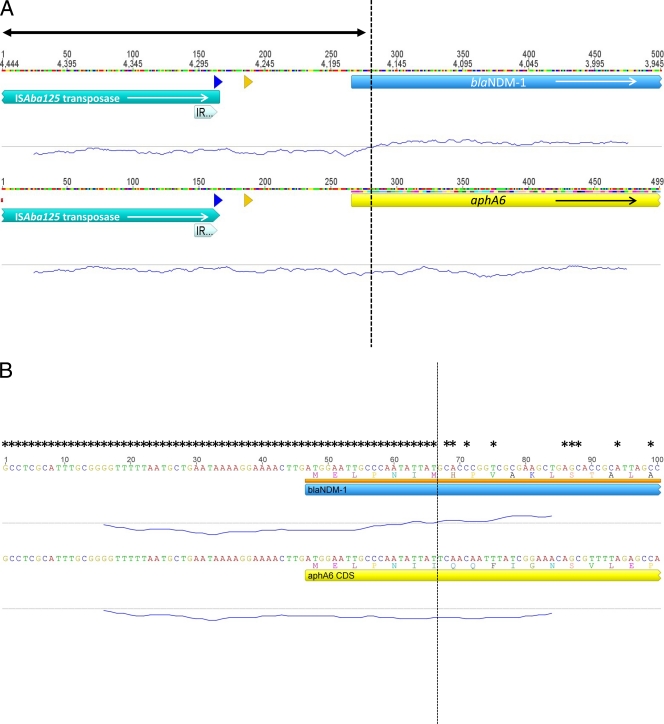
Alignment of several sequences surrounding the blaNDM-1 gene has recently shown that fragments of the insertion sequence ISAba125 are often found upstream of the start of the blaNDM-1 structural gene and provide part of its promoter (10, 13; L. Poirel, R. Bonnin, L. Villa, A. Carattoli, and P. Nordmann, presented at the 21st European Congress of Clinical Microbiology and Infectious Diseases [ECCMID] and 27th International Congress of Chemotherapy [ICC], Milan, Italy, 2011). We used Geneious Pro 5.5.3 software (Biomatters Ltd., New Zealand) to construct an alignment of all available structures found upstream of the blaNDM-1 gene and measured the percent GC content (GC%) at each position with a sliding window of 45 nucleotides. This revealed that ISAba125 sequences are indeed universally found 100 bp upstream of all blaNDM-1 genes for which sequence information is currently available. The sequences include three examples of the complete ISAba125, two of complete ISAba125 containing internal insertions of either IS1 or ISEc33, and several 5′ truncated versions that range in length from 157 to 179 nucleotides (Fig. 1). The alignment also indicates that the sequences found between the blaNDM-1 gene and ISAba125, or fragments thereof, are identical. The GC content of ISAba125, and of the 100-bp region that separates the 3′ end of ISAba125 from the 5′ end of blaNDM-1, is 37%. However, the blaNDM-1 gene has a considerably higher GC content of 61.5%. Of considerable interest is the fact that the GC% graph (Fig. 2A) changes dramatically within blaNDM-1 itself. Specifically, the GC% curve changes from below 50% to above 50% 19 to 20 bp inside the structural gene. Surprisingly, the first 6 amino acids of NDM-1 are identical to those of the aminoglycoside phosphotransferase AphA6, an occurrence that is highly unlikely to have happened by chance. To investigate this further, we searched the databases for information on the sequence found at the start of aphA6 and performed a second alignment of aphA6 and blaNDM-1 sequences Fig. 2). This included the first 240 nucleotides of the blaNDM-1 and aphA6 genes and 260 bp of sequence found upstream of both genes. That alignment indicated that ISAba125 is inserted at a point upstream of aphA6 identical to that of blaNDM-1 and that the intervening DNAs between the two genes are identical. The alignment breaks 19 bp within the blaNDM-1 gene, the exact point where the GC% graph changes (Fig. 2). This indicates that the first 19 bp of blaNDM-1, encoding the first 6 amino acids of the translated protein, are derived from aphA6 and that blaNDM-1 is a chimeric gene produced by an in-frame fusion of aphA6 with a preexisting MBL gene. Several facts strongly suggest that this fusion event happened in Acinetobacter baumanii. (i) The aphA6 gene is strongly associated with A. baumanii. aphA6 was first reported in 1988 in A. baumanii (8), and of the eight sequences in the genetic databases, four are from A. baumanii (1, 3) and a further two are from blaNDM-1-harboring plasmids. (ii) ISAba125 is also generally found in A. baumanii. Of 16 reported sequences, 12 are from A. baumanii, and a further 3 sequences are associated with blaNDM-1 plasmids. (iii) The first European isolate of A. baumanii containing blaNDM-1 (collected in Germany in 2007) contains both blaNDM-1 and an ISAba125 composite transposon with aphA6 (12). (iv) The sequence used for the alignment described above was taken from a composite transposon composed of direct repeats of ISAba125 found in A. baumanii (9). However, it is also possible that the fusion occurred in an environmental acinetobacter, since aphA6 is also found widespread in other Acinetobacter spp. (6).
Fig 1.
Sequence information found upstream of blaNDM-1 in various bacterial species isolated in different geographical regions. Colored blocks indicate regions of identity to various open reading frames or IS elements. The arrows indicate the direction of transcription and length of coding sections. Sections of ISAba125 are colored blue.
Fig 2.
(A) Alignment of 500-bp sections of DNA, including the 5′ ends of blaNDM-1 and aphA6 genes and 260 bp of upstream sequence. Open reading frames (ORFs) are depicted as colored blocks with arrows indicating the direction of transcription. The double-ended arrow indicates the region of sequences that are 100% identical. The right inverted repeat of ISAba125 is depicted as a thick arrow at the end of the transposase ORFs. Promoter sequences are depicted as a blue arrow at −35 and a yellow arrow at −10. The trace underneath each sequence represents the GC%, which was calculated using a sliding window of 45 bp, and the dashed vertical line indicates the point at which the GC% changes from below 50% to above 50% within the blaNDM-1 sequence. (B) Amplification of 100-bp section of alignment A, indicating the shared first six amino acids of NDM-1 and AphA6 and the exact nucleotide sequence of each region. Asterisks indicate nucleotides that are identical in the two loci; the dashed vertical line indicates the exact site of the gene fusion for all other annotations as described above.
The in-frame fusion of aphA6 with an MBL gene to form blaNDM-1 is likely to have several consequences for the expression and function of the NDM-1 enzyme. First, the success of NDM-1, in terms of its expression, and the ability to confer resistance in diverse bacterial species are likely linked to possession of a new promoter arising from the natural genetic engineering described here. It has recently been shown that control by the endogenous blaNDM-1 promoter gives carbapenem resistance in Escherichia coli higher than that seen when blaNDM-1 is controlled by either the plac or pBAD promoter (2). Second, the gene fusion event may exert functional changes linked to the altered N terminus. The fusion adds six new amino acids to the previous MBL, likely changing the N-terminal sequence from MHPVAKL to MELPNIM. A putative type I lipidation signal peptide has been identified that appears to be involved in directing the mature protein to the bacterial outer membrane (3). If the methionine at position M7 is in fact the initiation methionine codon of the previous MBL, then the previous MBL has a more impressive lipidation signal score of 9.87 (http://www.cbs.dtu.dk/services/LipoP/) and the change in lipidation signal may have consequences for functionality.
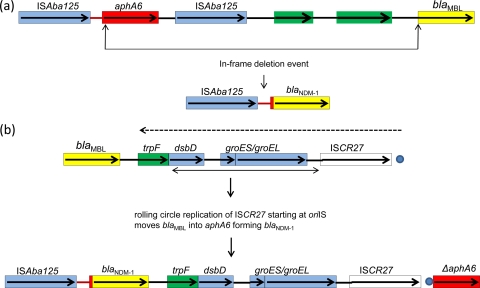
The fusion of a blaNDM-1 to the aminoglycoside gene, aphA6, forming a chimera can be postulated to have arisen by one of two routes. A deletion event occurring between an upstream aphA6 composite transposon and the blaNDM-1 gene would remove the intervening ISAba125 gene and the majority of aphA6 together with all sequence found upstream of the ancestral MBL gene (Fig. 3a). Alternatively, an ISCR element could have accumulated these genes by a rolling-circle mechanism (Fig. 3b) (14–17). When found in a favorable position adjacent to blaNDM-1, a movement event would move the ancestral gene without its upstream sequence to a new location within the aphA6 gene (Fig. 3b). Comparison of the genetic environments of blaNDM-1 in different bacterial species, including A. baumanii, has identified a common downstream structure consisting of an ISCR16-like ISCR element here named ISCR27(Fig. 3) adjacent to the groEL and groES genes (12). We suggest that this element is responsible for both the original acquisition of the preexisting MBL gene and the construction of the aphA6-blaNDM-1 chimera.
Fig 3.
Two possible routes of blaNDM-1 construction. (a) blaNDM-1 could have been constructed by a deletion event that occurred between an upstream aphA6-ISAba125 composite transposon and an MBL blaMBL gene. This would remove all sequence between the arrows, including most of aphA6, and all sequence upstream of blaMBL. (b) A rolling-circle replication event involving the ISCR27 element starting at the oriIS site could mobilize the upstream DNA that includes blaMBL into the aphA6 composite transposon, causing an insertion into the aphA6 gene and a fusion producing blaNDM-1. Genes are depicted as colored boxes with arrows indicating the direction of their transcription and intervening DNA as horizontal lines. The intervening DNA between the aphA6 gene and the ISAba125 element is colored red. The dotted horizontal arrow indicates both the direction of rolling-circle replication of ISCR27 and the common sequence found in several blaNDM-1 plasmids and on the chromosome in A. baumanii. The filled dot indicates the oriIS, and the double-ended arrow indicates the DNA that has been mobilized from a Xanthomonas-type organism (14, 16) by ISCR27.
Finally, the 100% sequence identity found between the aphA6 transposon and the blaNDM-1 upstream sequence indicates that the fusion event described here was a very recent event in line with what is known about the emergence of blaNDM-1.
ACKNOWLEDGMENT
This work was funded by the European FP7 “Predicting antibiotic resistance” initiative.
Footnotes
Published ahead of print 6 February 2012
REFERENCES
- 1. Ghazawi A, et al. 2012. NDM-2 carbapenemase-producing Acinetobacter baumannii in the United Arab Emirates. Clin. Microbiol. Infect. 18:E34–E36 [DOI] [PubMed] [Google Scholar]
- 2. Hornsey M, Phee L, Wareham DW. 2011. A novel variant, NDM-5, of the New Delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55:5952–5954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kaase M, et al. 2011. NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J. Antimicrob. Chemother. 66:1260–1262 [DOI] [PubMed] [Google Scholar]
- 4. King D, Strynadka N. 2011. Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance. Protein Sci. 20:1484–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kumarasamy KK, et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lambert T, Gerbaud G, Bouvet P, Vieu JF, Courvalin P. 1990. Dissemination of amikacin resistance gene aphA6 in Acinetobacter spp. Antimicrob. Agents Chemother. 34:1244–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Livermore DM. 2009. Has the era of untreatable infections arrived? J. Antimicrob. Chemother. 64(Suppl. 1):i29–i36 [DOI] [PubMed] [Google Scholar]
- 8. Martin P, Jullien E, Courvalin P. 1988. Nucleotide sequence of Acinetobacter baumannii aphA-6 gene: evolutionary and functional implications of sequence homologies with nucleotide-binding proteins, kinases and other aminoglycoside-modifying enzymes. Mol. Microbiol. 2:615–625 [DOI] [PubMed] [Google Scholar]
- 9. Nigro SJ, Post V, Hall RM. 2011. Aminoglycoside resistance in multiply antibiotic-resistant Acinetobacter baumannii belonging to global clone 2 from Australian hospitals. J. Antimicrob. Chemother. 66:1504–1509 [DOI] [PubMed] [Google Scholar]
- 10. Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol. 19:588–595 [DOI] [PubMed] [Google Scholar]
- 11. Patel G, Bonomo RA. 2011. Status report on carbapenemases: challenges and prospects. Expert Rev. Anti Infect. Ther. 9:555–570 [DOI] [PubMed] [Google Scholar]
- 12. Pfeifer Y, et al. 2011. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J. Antimicrob. Chemother. 66:1998–2001 [DOI] [PubMed] [Google Scholar]
- 13. Solé M, et al. 2011. First description of an Escherichia coli strain producing NDM-1 carbapenemase in Spain. Antimicrob. Agents Chemother. 55:4402–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Toleman MA, Walsh TR. 2011. Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol. Rev. 35:912–935 [DOI] [PubMed] [Google Scholar]
- 16. Toleman MA, Walsh TR. 2008. Evolution of the ISCR3 group of ISCR elements. Antimicrob. Agents Chemother. 52:3789–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Toleman MA, Walsh TR. 2010. ISCR elements are key players in IncA/C plasmid evolution. Antimicrob. Agents Chemother. 54:3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walsh TR, Toleman MA. 2011. The new medical challenge: why NDM-1? Why Indian? Expert Rev. Anti Infect. Ther. 9:137–141 [DOI] [PubMed] [Google Scholar]
- 19. Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11:355–362 [DOI] [PubMed] [Google Scholar]
- 20. Yong D, et al. 2009. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]