Abstract
Nonnucleoside reverse transcriptase (RT) inhibitors (NNRTIs) are important components of current combination therapies for human immunodeficiency virus type 1 (HIV-1) infection. In screening of chemical libraries, we found 6-azido-1-benzyl-3-(3,5-dimethylbenzyl)uracil (AzBBU) and 6-amino-1-benzyl-3-(3,5-dimethylbenzyl)uracil (AmBBU) to be highly active and selective inhibitors of HIV-1 replication in vitro. To determine the resistance profiles of these compounds, we conducted a long-term culture of HIV-1-infected MT-4 cells with escalating concentrations of each compound. After serial passages of the infected cells, escape viruses were obtained, and they were more than 500-fold resistant to the uracil derivatives compared to the wild type. Sequence analysis was conducted for RT of the escape viruses at passages 12 and 24. The amino acid mutation Y181C in the polymerase domain of RT was detected for all escape viruses. Docking studies using the crystal structure of RT showed that AmBBU requires the amino acid residues Leu100, Val106, Tyr181, and Trp229 for exerting its inhibitory effect on HIV-1. Four additional amino acid changes (K451R, R461K, T468P, and D471N) were identified in the RNase H domain of RT; however, their precise role in the acquisition of resistance is still unclear. In conclusion, the initial mutation Y181C seems sufficient for the acquisition of resistance to the uracil derivatives AzBBU and AmBBU. Further studies are required to determine the precise role of each mutation in the acquisition of HIV-1 resistance.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) is responsible for synthesizing a double-stranded integrative cDNA from the single-stranded viral genomic RNA in the early virus life cycle. HIV-1 RT is a heterodimer composed of two subunits, p66 and p51, and p51 is generated by the proteolytic processing and removal of C terminus (amino acids 441 to 560) from p66 (32). The p66 subunit is composed of two spatially distinct domains: polymerase (residues 1 to 426) and RNase H (residues 427 to 560). The polymerase domain is composed of four subdomains: fingers (residues 1 to 85 and 118 to 155), palm (residues 86 to 117 and 156 to 236), thumb (237 to 318), and connection (319 to 426) (46). The polymerase domain creates a copy of the viral genome, while the RNase H domain promotes RNA degradation from the DNA/RNA duplex during reverse transcription. Other RNase H functions include the removal of tRNA3Lys, and the polypurine tract, which are primers for minus- and plus-strand DNA synthesis (11, 47, 58).
The polymerase domain is currently targeted by two classes of antiretroviral drugs, nucleoside RT inhibitors (NRTIs) and non-nucleoside RT inhibitors (NNRTIs). NNRTIs are important components of current antiretroviral therapies for HIV-1. To date, more than 50 structurally diverse classes of compounds have been identified as genuine NNRTIs (60). Earlier NNRTIs include nevirapine (NVP), delavirdine (DLV), and efavirenz (EFV), and recent NNRTIs include etravirine (ETR). Several other NNRTIs, such as emivirine (MKC-442), underwent clinical trials, and yet they were not approved due to unfavorable pharmacokinetics, insufficient efficacy, and/or safety concerns (50). Recently, rilpivirine (RPV; formerly TMC278) has been formally licensed for clinical use in treatment-naive adult patients (56), while other NNRTIs, including IDX899, RDEA-428, and lersivirine, are currently under clinical development (1, 12, 30).
Although NNRTIs are generally well tolerated, a major limitation for all currently available NNRTIs is the low genetic barrier to resistance, which allows rapid emergence of drug resistance caused by a small number of amino acid mutations in the target region. HIV-1 drug resistance mutations in RT are extensively characterized for NRTIs and NNRTIs (29). NNRTIs inhibit HIV-1 by allosteric binding to a hydrophobic pocket in the RT about 10 Å behind the catalytic site (48). The positions associated with NNRTI resistance that make up the central NNRTI binding pocket are L100, K101, K103, V106, V108, V179, Y181, Y188, G190, F227, and W229. Additional positions that make up the pocket include E138, which is contributed by the p51 subunit, and M230, L234, P236, K238, and Y318, which form part of an extended pocket. Additional accessory NNRTI resistance abutting positions that form the NNRTI binding pocket include A98 and P225.
Mutations that are selected after failure of treatment with NNRTIs are located in the enzyme hydrophobic pocket, and they reduce the binding affinity of the inhibitors to the enzyme (7). A single mutation in the NNRTI-binding pocket may confer high-level resistance to one or more NNRTIs. Since NVP, DLV, and EFV have similar binding modes to RT, viruses resistant to one of these compounds develop cross-resistance to the others. ETR maintains its activity against NVP-, DLV-, or EFV-resistant mutants due to its ability of multiple binding modes to RT.
In order to improve the design of novel NNRTIs for future clinical development, in vitro isolation and analyses of drug-resistant viruses are necessary to obtain valuable information on the resistance patterns of novel compounds. We previously synthesized and evaluated nine novel uracil analogues as NNRTIs, including four 1-substituted 3-(3,5-dimethylbenzyl)-5-fluorouracils and five 6-substituted 1-benzyl-3-(3,5-dimethylbenzyl)uracils (28). Two of these compounds—6-azido-1-benzyl-3-(3,5-dimethylbenzyl)uracil (AzBBU) and 6-amino-1-benzyl-3-(3,5-dimethylbenzyl) uracil (AmBBU)—were found to be highly active and selective inhibitors of HIV-1 replication in vitro.
In the present study, we conducted a long-term culture experiment with HIV-1-infected MT-4 cells with escalating concentrations of AzBBU and AmBBU. After serial passages of the infected cells, escape viruses were obtained, which displayed complete resistance to these compounds. Sequence analysis of RT from escape viruses was performed to determine the mutations related to the acquisition of resistance.
MATERIALS AND METHODS
Compounds.
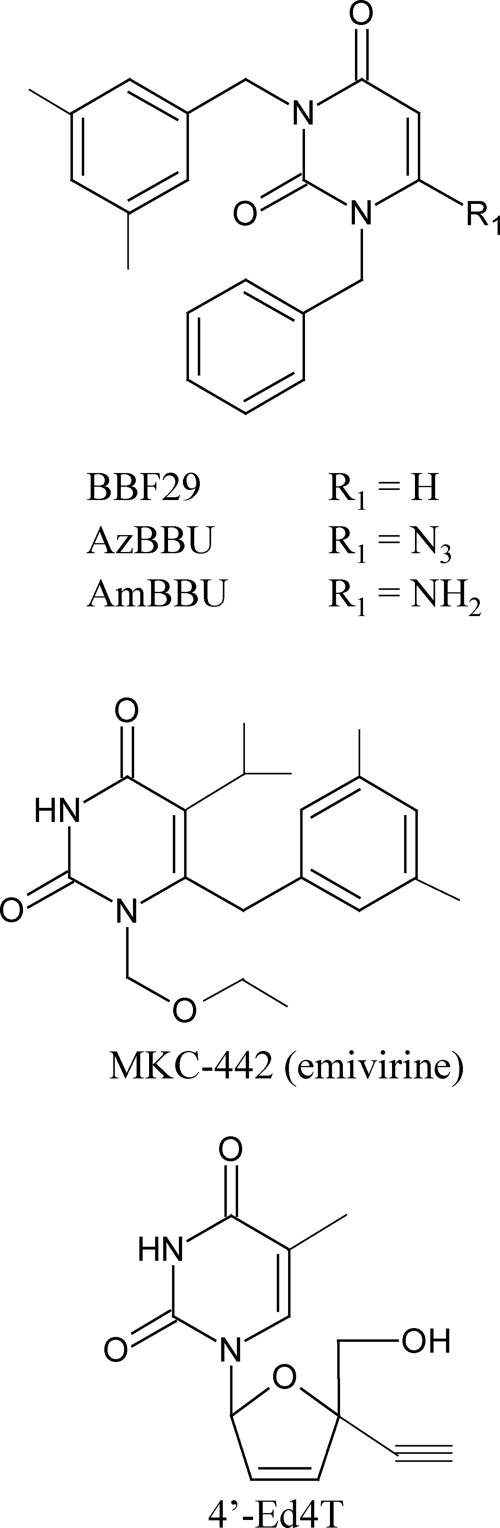
AzBBU and AmBBU were synthesized as previously described (28). The lead compound 1-benzyl-3-(3,5-dimethylbenzyl) uracil (BBF-29), 6-benzyl-1-ethoxymethyl-5-isopropyluracil (MKC-442), and the nucleoside analog 2′,3′-didehydro-3′-deoxy-4′-ethynylthymidine (4′-Ed4T) were prepared according as previously described (25, 34, 52). All compounds were dissolved in dimethyl sulfoxide at 100 mM and stored at −20°C until use. The chemical structures of AzBBU, AmBBU, BBF-29, MKC-442, and 4′-Ed4T are shown in Fig. 1.
Fig 1.
Structures of BBF-29, AzBBU, AmBBU, MKC-442, and 4′-Ed4T.
Cells and viruses.
MT-4 and M8166 cells were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U of penicillin G/ml, and 100 μg of streptomycin/ml. The HIV-1 strain IIIB, the HIV-1 resistance strain IIIB-R, and the HIV-2 strain ROD were used throughout the antiviral experiments (Table 1). IIIB-R is a NNRTI-resistant mutant established by a serial passage of infected cells in the presence of increasing concentrations of MKC-442 (5). Viruses were propagated and titrated in MT-4 cells (HIV-1) or M8166 cells (HIV-2). Virus stocks were stored at −80°C until use. Escape viruses obtained after long-term culture with AzBBU and AmBBU were used for the following anti-HIV-1 experiments (Table 2).
Table 1.
Antiviral activity of AzBBU and AmBBU against HIV-1 and HIV-2
| Compound | Antiviral activity (mean EC50 [μM] and CC50 [μM] ± SD)a against: |
|||||
|---|---|---|---|---|---|---|
| HIV-1 IIIB |
HIV-1 IIIB-R |
HIV-2 ROD |
||||
| EC50 | CC50 | EC50 | CC50 | EC50 | CC50 | |
| AzBBU | 0.088 ± 0.009 | 40.5 ± 6.7 | >45.1 | 45.1 ± 0.5 | >40.0 | 40.0 ± 2.7 |
| AmBBU | 0.060 ± 0.011 | 50.1 ± 1.1 | >50.5 | 50.5 ± 6.3 | >45.9 | 45.9 ± 1.2 |
| BBF-29 | 0.26 ± 0.02 | 43.2 ± 9.1 | 13.9 ± 6.0 | >100 | >100 | >100 |
| MKC-442 | 0.015 ± 0.002 | >100 | 6.2 ± 1.4 | >100 | ||
| NVP | 0.057 ± 0.005 | >100 | 52.1 ± 23.6 | >100 | ||
| 4′-Ed4T | 0.029 ± 0.008 | >100 | 0.053 ± 0.038 | >100 | 0.019 ± 0.007 | >100 |
EC50, 50% effective concentration that inhibits virus-induced CPE of infected cells by 50%; CC50, 50% cytotoxic concentration that reduces the viability of uninfected cells by 50%.The data represent the means for three independent experiments.
Table 2.
Anti-HIV-1 activity of AzBBU and AmBBU against escape viruses
| Compound | Anti-HIV-1 activity (mean EC50 [μM] and CC50 [μM] ± SD)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| Passage 12 |
Passage 24 |
|||||||
| IIIB-AZ12 |
IIIB-AM12 |
IIIB-AZ24 |
IIIB-AM24 |
|||||
| EC50 | CC50 | EC50 | CC50 | EC50 | CC50 | EC50 | CC50 | |
| AzBBU | >44.5* (>500) | 44.5 ± 0.3 | >42.6* (>480) | 42.6 ± 3.0 | >43.6* (>500) | 43.6 ± 0.5 | >44.0* (>500) | 44.0 ± 1.0 |
| AmBBU | >60.7* (>1,000) | 60.7 ± 6.5 | >56.2* (>940) | 56.2 ± 5.7 | >48.1* (>800) | 48.1 ± 4.8 | >47.0* (>780) | 47.0 ± 7.6 |
| BBF-29 | >46.4* (>180) | 46.4 ± 9.5 | >42.5* (>160) | 42.5 ± 3.9 | >49.4* (>190) | 49.4 ± 3.0 | >45.9* (>170) | 45.9 ± 7.2 |
| MKC-442 | 3.8 ± 1.2* (257) | >100 | 2.5 ± 0.6* (168) | >100 | 5.4 ± 3.5 (369) | >100 | 3.8 ± 1.2* (257) | >100 |
| NVP | 49.9 ± 10.9* (875) | >100 | 32.6 ± 4.6* (572) | >100 | 37.2 ± 3.5* (652) | >100 | 33.6 ± 6.7* (589) | >100 |
| 4′-Ed4T | 0.038 ± 0.029 (1.3) | >100 | 0.092 ± 0.011* (3.2) | >100 | 0.028 ± 0.012 (1) | >100 | 0.023 ± 0.003 (0.8) | >100 |
EC50, 50% effective concentration that inhibits virus-induced CPE of infected cells by 50%; CC50, 50% cytotoxic concentration that reduces the viability of uninfected cells by 50%. Fold changes, based on the EC50 for the wild type, are indicated in parentheses. The data represent the means for three independent experiments. Statistical analysis (Student t test) was performed for each EC50 compared to that of the wild type (*, P < 0.01).
Antiviral assays.
The antiviral activity of test compounds against HIV-1 and HIV-2 was determined by the inhibition of virus-induced cytopathic effect (CPE) (4). Briefly, MT-4 or M8166 cells (105 cells/ml) were infected with HIV-1 or HIV-2, respectively, at a multiplicity of infection (MOI) of 0.1 and were cultured in the presence of various concentrations of the tested compounds. After a 4-day incubation at 37°C, the number of viable cells was monitored by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) method (39). The cytotoxicity of the compounds was evaluated in parallel with their antiviral activity, based on the viability of mock-infected cells, as determined by the MTT method. All experiments were performed in triplicate.
Long-term culture of infected MT-4 cells.
MT-4 cells were infected with the HIV-1 strain IIIB and incubated at 37°C for 4 days in the presence of compounds. The initial concentration of AzBBU and AmBBU corresponded to 2-fold higher than their 50% effective concentrations (EC50s), i.e., 0.175 and 0.119 μM, respectively. As control cultures, exactly identical passages of the infected MT-4 cells in the absence of the compounds were carried out in parallel with the cultures exposed to the compounds. At each passage, virus-induced CPE of the cells was monitored to confirm virus replication. The concentration of each compound was escalated 2-fold, when the CPE in the compound-treated culture exceeded 70%. The escape viruses as well as the control viruses were propagated once in MT-4 cells, and they were used for further experiments.
Sequence analysis of RT.
Genomic DNA was extracted from the infected MT-4 cells with a DNA extraction kit (Wako, Tokyo, Japan). The extracted DNA was quantified using a NanoDrop spectrophotometer ND-1000 (NanoDrop Technologies, Wilmington, DE) and subjected to nested PCR. Two regions of RT were amplified (RT1 and RT2). The first PCR consisted of an initial denaturation at 95°C for 2 min, followed by 40 cycles (95°C for 1 min, 46°C for 1 min, and 72°C for 1 min) and a final extension at 72°C for 5 min. The primers rt-1-f (5′-AGGGGGAATTGGAGGTTT-3′) and rt-1-r (5′-TCCCACAACTTCTGTATGTC-3′) were used to amplify the region RT1, and the primers rt-2-f (5′-ATGAACTCCATCCTGATAAATG-3′) and rt-2-r (5′-TGTACAATCTAGTTGCCATAT-3′) were used to amplify the region RT2. The second PCR consisted of an initial denaturation at 95°C for 2 min, followed by 40 cycles (95°C for 1 min, 48°C for 1 min, and 72°C for 1 min) and a final extension at 72°C for 5 min. The primers rt-12-f (5′-CCAGTAAAATTAAAGCCAG-3′) and rt-12-r (5′-TCCCACTAACTTCTGTATGTC-3′) were used to amplify the region RT1, and the primers rt-22-f (5′-CCAGAAAAAGACAGCTGGACT-3′) and rt-22-r (5′-TGGCAGGTTAAAATCACTAGCC-3′) were used to amplify the region RT2. The second PCR generated fragments encompassing nucleotides 2120 to 2881 (RT1) and nucleotides 2834 to 3862 (RT2) of the RT gene corresponding to the HIV-1 complete genome (GenBank accession number AF033819). The amplified products were confirmed by capillary gel electrophoresis using the Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA). The PCR products were sequenced directly with a cycle sequence kit BigDye Terminator version 3.1 (Applied Biosystems, Foster City, CA), using both forward and reverse primers on an automated DNA analyzer model 3730 (Applied Biosystems), according to the manufacturer's instructions.
Docking study of compounds against HIV-1 RT.
All in silico studies were performed using Molecular Operating Environment (MOE) software (Chemical Computing Group, Montreal, Quebec, Canada). The X-ray crystal structure of HIV-1 RT (PDB code 3m8p) (40) was downloaded from PDB at the Research Collaboration for Structural Bioinfomatics (http://www.rcsb.org/pdb/home/home.do) and optimized for the docking study by removing ligand and water, adding hydrogen atoms, assigning atomic charges, and minimizing using the Merck molecular force field 94X (MMFF94x) (22, 23, 55). Based on this structure, the structure of the HIV-1 RT (Y181C) was constructed. The docking site at the HIV-1 RT structure was searched by Alpha Site Finder, a function of MOE. Partial charges were added to the compound and a maximum of 250 conformers were generated using MMFF94x. MOE-ASEDock 2005 (Ryoka Systems, Tokyo, Japan) was then used for the docking of the compound to HIV-1 RT, and docking scores were calculated (19).
Statistical analysis.
Statistical analysis for the EC50s of the test compounds against the wild-type and resistant viruses was performed using an unpaired two-tailed Student t test. P values of <0.01 were considered statistically significant.
RESULTS
Antiviral activity of AzBBU and AmBBU against HIV-1 and HIV-2.
AzBBU and AmBBU were tested for their inhibitory effects on the replication of HIV-1 IIIB, HIV-1 IIIB-R, and HIV-2 ROD. The NNRTIs BBF-29, MKC-442, and NVP, as well as the nucleoside analog 4′-Ed4T, were also tested for comparison. Their activities are given in Table 1. AzBBU and AmBBU showed high activity against HIV-1 IIIB with similar EC50s (0.088 ± 0.009 and 0.060 ± 0.011 μM, respectively) and 50% cytotoxic concentrations (CC50s) (40.5 ± 6.7 and 50.1 ± 1.1 μM, respectively). AmBBU showed a higher selectivity index (SI) than AzBBU (SI = 835 versus SI = 460). These results are in accordance with those in the previous report (28). Although these compounds showed higher anti-HIV-1 activity against IIIB compared to the lead compound BBF-29 (0.26 ± 0.02 μM), they were not active against the NNRTI-resistant HIV-1 stain IIIB-R. In addition, AzBBU and AmBBU did not show any activity against HIV-2 ROD. In contrast, NRTI 4′-Ed4T was equally active against HIV-1 IIIB, HIV-1 IIIB-R, and HIV-2 ROD.
Isolation of escape viruses.
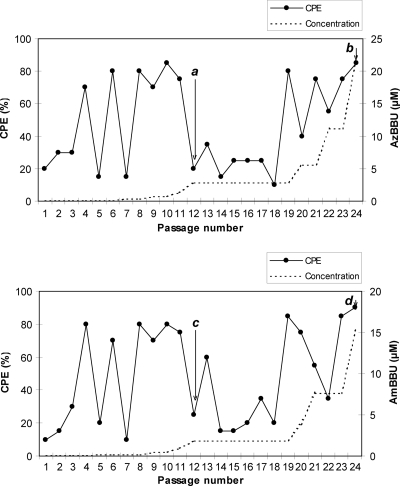
Long-term cultures of HIV-1 (IIIB strain)-infected MT-4 cells were started in the absence or presence of AzBBU and AmBBU (Fig. 2). The concentration of each compound was escalated 2-fold, when the CPE in the compound-treated culture exceeded 70%. At passage 24, the concentrations of AzBBU and AmBBU could reach 256-fold their EC50s (22.4 and 15.2 μM, respectively). Viruses were isolated from culture supernatants at passages 12 and 24 (Fig. 2, points a, b, c, and d) and subjected to phenotypic and genotypic analyses.
Fig 2.
Long-term culture of infected MT-4 cells with escalating concentrations of AzBBU and AmBBU. MT-4 cells were infected with HIV-1 (IIIB strain) and passaged every 4 days. Viral replication was monitored by determining the CPE of the cells at each passage. Culture supernatants from passages 12 and 24 were used for further experiments (CPE > 70%). Isolated viruses (points a, b, c, and d) were subjected to phenotypic and genotypic analyses. Points: a, passage 12, AzBBU (IIIB-AZ12); b, passage 24, AzBBU (IIIB-AZ24); c, passage 12, AmBBU (IIIB-AM12); d, passage 24, AmBBU (IIIB-AM24).
Anti-HIV-1 activity of AzBBU and AmBBU against escape viruses.
When AzBBU and AmBBU were examined for their activity against the escape viruses obtained at passages 12 and 24, the compounds did not show any significant inhibition at their nontoxic concentrations (Table 2). Thus, the isolates were more than 500-fold resistant to AzBBU and AmBBU compared to the wild type. The lead compound BBF-29 was also inactive against the escape viruses. The viruses had partial cross-resistance to MKC-442, probably due to its structural similarity (Fig. 1 and Table 2). NVP marginally inhibited the replication of the escape viruses. In contrast, 4′-Ed4T was equally inhibitory to the replication of the escape viruses and the wild type (Table 2).
Amino acid changes of escape viruses.
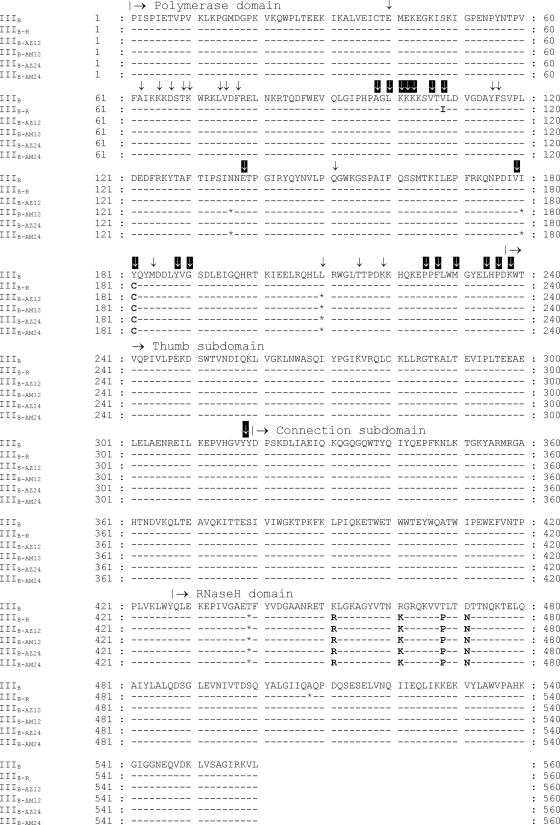
To determine what amino acid changes are associated with resistance to AzBBU and AmBBU, sequence analysis of full-length RT genes from escape viruses (IIIB-AZ12, IIIB-AM12, IIIB-AZ24, IIIB-AM24), as well as the wild-type control without treatment (IIIB), was further performed at passages 12 and 24. The sequences of the escape viruses, IIIB-AzBBU and IIIB-AmBBU, were deposited in a public database (GenBank accession numbers JQ070415 and JQ070416). In addition, the sequence of the resistant strain IIIB-R was also analyzed for comparison (GenBank accession number JQ070417). Figure 3 shows the RT (subunit p66) amino acid sequences of escape viruses. Several synonymous mutations were observed in the RT gene. Overall, 5 amino acid changes were observed. One was in the polymerase domain, and four were in the RNase H domain. The same amino acid changes were detected at passages 12 and 24 (Fig. 3). Mutation Y181C in the polymerase domain was identified in all escape viruses. The resistant strain IIIB-R displayed the mutation V108I in addition to Y181C. Four additional amino acid changes (K451R, R461K, T468P, and D471N) were detected in the RNase H domain of all escape viruses.
Fig 3.
RT (subunit p66) amino acid sequences of the escape viruses. The control sequence without treatment was used as reference (WT). Asterisks (*) indicate the positions for synonymous mutations. Black arrows indicate common NRTI resistance mutations. White arrows indicate common NNRTI resistance mutations.
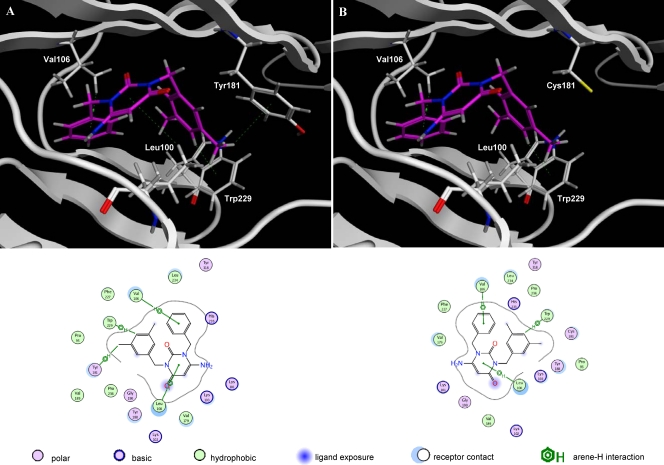
Docking studies of AmBBU.
Docking of the metabolically relevant derivative AmBBU to the binding-pocket of HIV-1 RT was performed. Figure 4 shows the proposed interactions between HIV-1 RT and AmBBU for HIV-1 RT wild type (Fig. 4A) and the mutant Y181C (Fig. 4B). AmBBU interacted with the amino acid residues Leu100, Val106, Tyr181, and Trp229 of HIV-1 RT (wt) through arene-H interactions (arene-H) (Fig. 4A), such as the hydrogen in the side chain of Leu100 with the central benzene ring (2-pyrimidine) of AmBBU, the hydrogen in the side chain of Val106 with 1-benzyl of AmBBU, the phenyl ring in the side chain of Tyr181 with hydrogen (3-methyl) at 3-(3,5-dimethylbenzyl) of AmBBU, and the indol ring in the side chain of Trp229 with 4′-hydrogen of 3-(3,5-dimethylbenzyl) of AmBBU (D2 representation in Fig. 4A). The compound lost the interaction at position 181 when replaced by cysteine (Y181C) (Fig. 4B). According to these data, the docking score between HIV-1 RT (wt) and AmBBU (−13.5110 kcal/mol) was higher than that between HIV-1 RT (Y181C) and AmBBU (−11.1648 kcal/mol).
Fig 4.
Docking of AmBBU to the binding pocket of HIV-1 RT. Docking structures between HIV-1 RT and AmBBU are shown for HIV-1 RT wt (A) and the mutant Y181C (B). The backbone is represented by white ribbons, and the side chains (residues 100, 106, 181, and 229) are represented by colored wire style. AmBBU is represented by magenta wire style. Dotted lines show the interactions between HIV-1 RT and AmBBU. A D2 representation of the interactions is presented below the docking.
DISCUSSION
Initially, we evaluated the antiviral activities of AzBBU and AmBBU against the HIV-1 strains IIIB and IIIB-R and the HIV-2 strain ROD. AzBBU and AmBBU were highly active against IIIB, and the EC50s were similar for both compounds (Table 1). This is explainable, since the 6-azido uracil derivative (AzBBU) may be reduced metabolically to its 6-amino congener (AmBBU) in cell cultures (28). These compounds showed higher activity against IIIB than the 1-substituted 3-(3,5-dimethylbenzyl)uracils previously reported (33). In contrast, AzBBU and AmBBU were not active against IIIB-R, although the lead compound BBF-29 had weak activity. This may be due to the azido and amino groups introduced at 6-position of the 1-benzyl moiety, which is not present in BBF-29 (Fig. 1). Thus, 6-azido and 6-amino substitutions on the uracil ring increased the antiviral activity against IIIB but decreased the antiviral activity against IIIB-R. In addition, AzBBU and AmBBU did not show any activity against HIV-2 ROD. This is consistent with previous reports showing that HIV-2 is intrinsically resistant to most NNRTIs (14, 57). As expected, the nucleoside analog 4′-Ed4T was almost equally active against HIV-1 IIIB, IIIB-R, and HIV-2 ROD.
We isolated two HIV-1 strains highly resistant to AzBBU and AmBBU through long-term culture of HIV-1 IIIB-infected MT-4 cells (Fig. 2). The phenotypic analysis revealed that escape viruses had partial cross-resistance to MKC-442, probably due to its structural similarity (Table 2 and Fig. 1). A similar finding has been reported for 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine (HEPT) derivatives, where the HEPT-resistant virus also displayed cross-resistant to virtually all of other HIV-1-specific inhibitors, including NVP (6). 4′-Ed4T was equally active against the AzBBU- and AmBBU-resistant viruses compared to the wild type because of its different mechanism of action.
Our genotypic analysis revealed amino acid changes associated with resistance to AzBBU and AmBBU (Fig. 3). Several mutations were observed in the RT gene of resistant viruses, but some of them corresponded to synonymous mutations (Fig. 3). Amino acid changes were accumulated within a short period of cultivation (from passage 12, 48 days). Five amino acid changes were identified in all escape viruses at passages 12 and 24 (Fig. 3). They were attributable to the selection pressure by the compounds and were not the consequences of in vitro passage of infected cells, since these changes could be identified only for the escape viruses but not for the corresponding control viruses (data not shown).
We firstly identified the mutation Y181C within the polymerase domain of RT. Mutations responsible for NNRTI resistance occur in the hydrophobic inhibitor-binding pocket of RT, which includes the residue Y181 (46). A single mutation in this pocket can lead to high-level resistance to earlier NNRTIs, including EFV and NVP. However, two or more mutations are required to cause high-level resistance to ETR, RPV, and other recent NNRTIs (3, 35, 53, 61). Y181C alone can cause multidrug resistance to NNRTIs, such as high-level resistance to NVP and DLV, and low-level resistance to EFV (18). In addition, Y181C provides the mutational foundation for the development of higher levels of ETR resistance (49). The Y181C mutation has emerged as an initial mutation in previous studies in patients failing an NNRTI-containing regimen, including NVP monotherapy (15, 43) and DLV monotherapy (17). K103N is frequently observed among NNRTI resistance mutations, but it is usually followed by Y181C and G190A (27). Resistance to HEPT derivatives is generated by mutations at multiple sites in the HIV-1 RT (8), while 1-(3-cyclopenten-1-yl)methyl-6-(3,5-dimethylbenzoyl)-5-ethyl-2,4-pyrimidinedione (SJ-3366) selected for a virus with the mutation Y181C after five tissue culture passages (9). In the present study, the initial mutation Y181C was sufficient for the acquisition of HIV-1 resistance to AzBBU and AmBBU. Other common NNRTI resistance mutations, such as K101E, K103N, and Y188C, were not identified.
Using docking studies, we examined the binding sites for the metabolically relevant derivative AmBBU on the allosteric pocket of HIV-1 RT (Fig. 4). According to the docking, AmBBU binds the allosteric pocket through arene-H interactions with the amino acid residues Leu100, Val106, Tyr181, and Trp229 (Fig. 4A). When the RT mutant Y181C was used for docking, AmBBU lost the interaction at position 181 (Fig. 4B). The docking score between HIV-1 RT (wt) and AmBBU was higher than that between HIV-1 RT (Y181C) and AmBBU, indicating the importance of the residue Tyr181 in the binding to the allosteric pocket. Most NNRTIs are engaged in the H-bond with the backbone of the residues Lys101 and/or Lys103 of RT (13, 26, 41). Previous docking studies suggested an H-bond between the amide of Lys101 and nitrogen of the cyanomethyl and picolyl group of 1-substituted 3-(3,5-dimethylbenzyl)uracils (33). In our previous study, we showed that 6-subtitutions on the uracil ring resulted in elevation of the anti-HIV-1 activity of the uracil derivatives (28). The structure-activity relationship among these uracil derivatives suggested that the strong anti-HIV-1 activity of the 6-amino derivative AmBBU is due to the H-bond formed between the 6-amino group of AmBBU and the amide group of the residue Lys101, as well as hydrophobic interactions (28). Here, our docking showed arene-H interactions of AmBBU with Leu100, Val106, Tyr181, and Trp229 (Fig. 4A). However, we could not identify the precise role of the 6-amino substitution in the binding to the allosteric pocket and how it leads to an increased anti-HIV-1 activity. Thus, although several interactions occur within the allosteric pocket, the interaction with the Tyr181 residue appears essential for docking of 6-substituted 1-benzyl-3-(3,5-dimethylbenzyl)uracils. In addition, Y181C loses important aromatic ring interactions in the core of the NNRTI-binding pocket, decreasing binding of NNRTIs (42). Taken together, the results of sequence analysis and docking studies indicate that AmBBU requires the interaction(s) with Tyr181 for its inhibitory effect on HIV-1.
In addition to Y181C, we identified four mutations in the RNase H domain of RT: K451R, R461K, T468P, and D471N (Fig. 3). Since we do not know exactly when these mutations emerged in relation to Y181C, it is difficult to elucidate their precise role in acquisition of HIV-1 resistance to AzBBU and AmBBU. One possibility is that mutations in the RNase H domain are merely the result of polymorphisms of RT and are not related to Y181C, as indicated by several HIV-1 (subtype B) prototype strains containing some of the identified mutations in the RNase H domain (Table 3). The strains MCK1_LAI_IIIB and PV22_LAI_IIIB, for instance, display all four amino acid changes but are not associated with any common NNRTI-resistant mutation. However, some strains display one or more NNRTI-resistant mutations in association with the mutation T468P/S, suggesting that mutations in the RNase H domain could act in coordination with NNRTI-resistant mutations to favor drug resistance. It has been postulated that drug resistance mutations reduce the replication fitness of HIV-1 (10). NNRTI-resistant RTs with the Y181C mutation have been shown to alter the rate of one or both modes of RNase H cleavage with no significant effects on RNA- or DNA-dependent DNA polymerization, and a decrease in RNase H activity has been associated with greater reductions in replication fitness (2).
Table 3.
NNRTI-resistant mutations in the polymerase domain and amino acid changes in the RNase H domain of escape virus
| GenBank accession no. | Name | RT domaina |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Polymerase |
RNase H |
|||||||||
| A98 | K102 | K103 | V179 | Y181 | K451 | R461 | T468 | D471 | ||
| A04321 | IIIB LAI | – | – | – | – | – | – | – | P | – |
| A07867 | LAI-J19 | – | – | – | – | – | – | – | – | – |
| AB221005 | Ba-L | Q | – | – | – | – | – | – | S | – |
| AF033819 | HXB2_copy LAI | – | – | – | – | – | – | – | – | – |
| AF070521 | NL43E9 LAI IIIB_NY5 | – | Q | – | – | – | – | – | P | – |
| D86068 | MCK1 LAI IIIB | – | – | – | – | – | R | K | P | N |
| D86069 | PM213 LAI IIIB | – | – | – | – | – | – | – | – | – |
| EU541617 | pIIIB | – | Q | – | – | – | – | – | P | – |
| HB388803 | MN patent seq | – | – | – | – | – | – | – | S | – |
| K02007 | SF2 LAV2 ARV2 | – | – | – | – | – | – | – | S | – |
| K02013 | LAI BRU | – | – | – | – | – | – | – | – | – |
| K02083 | PV22 LAI IIIB | – | – | – | – | – | R | K | P | N |
| K03455 | HXB2-LAI-IIIB-BRU | – | – | – | – | – | – | – | – | – |
| M17449 | MNCG MN | – | – | – | – | – | – | – | S | – |
| NC_001802 | HXB2-LAI-HXB2R | – | – | – | – | – | – | – | – | – |
| U26942 | NL4_3 LAI_NY5 pNL43 NL43 | – | Q | – | – | – | – | – | P | – |
| U63632 | JRFL JR_FL | – | – | R | I | – | – | – | S | – |
| X01762 | REHTLV3 LAI IIIB | – | – | – | – | – | – | K | P | N |
| JQ070415 | AzBBU IIIB-AZ | – | – | – | – | C | R | K | P | N |
| JQ070416 | AmBBU IIIB-AM | – | – | – | – | C | R | K | P | N |
The default amino acids and position numbers are specified in each column subheading. –, No change.
In 2005, Nikolenko et al. suggested that mutations in the RNase H domain could significantly contribute to an increase of RT resistance to NRTIs (37). Later, the same group proposed the RNase H-dependent NNRTI-resistant model, which suggests that mutations in the RNase H domain that reduce RNase H cleavage will allow more time for the NNRTIs to dissociate from the NNRTI-RT-template/primer complex, allowing the reinitiation of polymerization and thereby resulting in enhanced NNRTI resistance (36). Thus, combining mutations in RT that reduce NNRTI affinity with mutations that reduce RNase H cleavage should further increase NNRTI resistance (24, 36). In an RNase H-independent mechanism, NNRTIs themselves can increase RNase H activity, so that mutations reducing RNase H activity are selected in response to NNRTI therapy, because they restore the balance between RNase H activity and polymerization (16). An alternative explanation is that NNRTIs may inhibit HIV-1 replication by increasing RT dimer stability (51). Thus, NNRTI-binding pocket mutants that confer drug resistance should decrease the stability of RT heterodimers. In general, the nucleic acid structure-dependent interplay between polymerase and RNase H domains is likely to affect overall efficacy of NNRTIs against HIV-1 replication, as well as the selection of mutations in the NNRTI-binding site associated with NNRTI resistance.
The effects of mutations in the RNase H domain on NNRTI resistance have been confirmed in vitro, and yet their clinical impact is still unclear. Current HIV-1 genotypic analyses of patients generally focus on the N terminus of the polymerase domain, thus missing important information on mutations in the thumb-connection subdomains and RNase H domain that might be related to resistance either alone or in combination with other RT mutations. Yap et al. showed that N348I in the connection subdomain was highly prevalent in a patient cohort and was highly associated with thymidine analogue-associated mutations (TAMs) and the NNRTI mutations K103N and Y181C (59). Hachiya et al. also examined N348I in treatment-experienced clinical isolates from Japan and found that N348I was prevalent in AZT and/or ddI therapy and that several mutations in the connection subdomain and RNase H domain typically acted as pretherapy polymorphisms (20, 21). Waters et al. found N348I prevalent in treatment-experienced patients (54). In addition, these researchers found that the genotypic profiles of patients with or without the K451R mutation within a treatment-experienced group showed a higher incidence of NNRTI mutations in patients with the K451R mutation. Santos et al. analyzed 450 sequences from Brazilian subtype B isolates and public databases and found nine mutations in the connection subdomain and six mutations in the RNase H domain that were associated with NRTI therapy (45). Positions K451 and D471 were less conserved in NRTI-experienced patients, while R461 and T468 were equally variable in both naive and experienced patients. A comparison of RNase H sequences in naive versus NRTI-experienced patients in a French cohort showed that mutations L469T/I/M/H, T470P/S/E/K, A554T/L/K, and K558R/G/E were more prevalent among treatment-experienced patients (44). However, Ntemgwa et al. analyzed RNase H mutations in NRTI-experienced patients from a Canadian and an Italian cohort and found positions D460, P468, H483, K512, and S519 to be extensively polymorphic in both naive and experienced patients (38). Recently, an analysis of patient sequences from databases showed that several mutations in the connection subdomain were significantly higher for sequences that contained one or more RTI resistance mutations compared to sequences without RTI resistance mutations (16). Moreover, subtype B-infected patient database analysis showed that RNase H mutations, including K451R, increased in frequency with the number of TAMs in a dose-dependent fashion (31). That study demonstrated that distinct RT C-terminal mutations can act as primary or secondary drug resistance mutations and are associated in a complex array of phenotypes with RT polymerase domain mutations.
In this in vitro study, we identified four mutations in the RNase H domain that might be related to the NNRTI resistance mutation Y181C. Biochemical studies are needed to understand the molecular mechanism of the associations and interactions between mutations within the polymerase and RNase H domains of RT. Further experiments are under consideration to validate the role of these mutations in the acquisition of resistance to AzBBU and AmBBU. First, viral strains containing the identified mutations in the RNase H domain, such as MCK1_LAI_IIIB and PV22_LAI_IIIB, will be used for testing the uracil derivatives, since these strains have all four amino acid changes but are not associated with any common NNRTI-resistant mutation. Second, recombinant RT enzymes containing the identified mutations will be used to determine the inhibitory effects of AzBBU and AmBBU on their catalytic activity. Third, an experiment on site-directed mutagenesis will be performed to elucidate the precise role of the RNase H mutations in the acquisition of resistance.
In terms of drug development, we found useful information for the future design of 6-substituted uracil derivatives with enhanced chemical properties that improve anti-HIV-1 activity and resistance profiles. Although the chemical properties of AzBBU and AmBBU suggest a good drug-likeness profile, further studies are required to assess the toxicity and pharmacokinetics of 6-substituted uracil derivatives in vivo. A limitation of our study relates to the use of laboratory strains. However, the mutations that we identified have been reported in clinical isolates (38, 45, 54). Another limitation of our study relates to the lack of information on the mutations in the RNase H domain of NNRTI-treated patients, since these mutations have been reported mostly in NRTI-treated patients. Furthermore, in a clinical setting, NNRTIs must be used in combination therapy, which may alter the pattern for resistance. Thus, the emergence of drug resistance should be further investigated and confirmed in clinical trials.
In conclusion, our results provide important information on the acquisition of resistance to the novel uracil derivatives AzBBU and AmBBU. Although the initial mutation Y181C can be sufficient in the acquisition of HIV-1 resistance, additional mutations in the RNase H domain of RT could additionally be associated to the mechanisms of resistance. Docking studies using the crystal structure of RT showed that AmBBU requires the amino acid residues Leu100, Val106, Tyr181, and Trp229 for its inhibitory effect on HIV-1. Further studies are necessary to determine the precise role of each mutation in the acquisition of HIV-1 resistance to the present compounds.
ACKNOWLEDGMENT
This study was supported in part by a research grant from the Ministry of Health, Labor, and Welfare of Japan.
Footnotes
Published ahead of print 30 January 2012
REFERENCES
- 1. Agarwal AK, Fishwick CW. 2010. Structure-based design of anti-infectives. Ann. N. Y. Acad. Sci. 1213:20–45 [DOI] [PubMed] [Google Scholar]
- 2. Archer RH, et al. 2000. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J. Virol. 74:8390–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Azijn H, et al. 2010. TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob. Agents Chemother. 54:718–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baba M, et al. 1991. Potent and selective inhibition of human immunodeficiency virus type 1 (HIV-1) by 5-ethyl-6-phenylthiouracil derivatives through their interaction with the HIV-1 reverse transcriptase. Proc. Natl. Acad. Sci. U. S. A. 88:2356–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baba M, et al. 1994. Preclinical evaluation of MKC-442, a highly potent and specific inhibitor of human immunodeficiency virus type 1 in vitro. Antimicrob. Agents Chemother. 38:688–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balzarini J, Karlsson A, De Clercq E. 1993. Human immunodeficiency virus type 1 drug-resistance patterns with different 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine derivatives. Mol. Pharmacol. 44:694–701 [PubMed] [Google Scholar]
- 7. Boyer PL, Currens MJ, McMahon JB, Boyd MR, Hughes SH. 1993. Analysis of nonnucleoside drug-resistant variants of HIV-1 reverse transcriptase. J. Virol. 67:2412–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buckheit RW, Jr, et al. 1995. Resistance to 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine derivatives is generated by mutations at multiple sites in the HIV-1 reverse transcriptase. Virology 210:186–193 [DOI] [PubMed] [Google Scholar]
- 9. Buckheit RW, Jr, et al. 2001. SJ-3366, a unique and highly potent nonnucleoside reverse transcriptase inhibitor of human immunodeficiency virus type 1 (HIV-1) that also inhibits HIV-2. Antimicrob. Agents Chemother. 45:393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coffin JM. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483–489 [DOI] [PubMed] [Google Scholar]
- 11. Coffin JM, Hughes SH, Varmus HE. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, New York, NY: [PubMed] [Google Scholar]
- 12. Corbau R, et al. 2010. Lersivirine, a nonnucleoside reverse transcriptase inhibitor with activity against drug-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 54:4451–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Das K, Jr, et al. 2004. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J. Med. Chem. 47:2550–2560 [DOI] [PubMed] [Google Scholar]
- 14. De Clercq E. 1993. HIV-1-specific RT inhibitors: highly selective inhibitors of human immunodeficiency virus type 1 that are specifically targeted at the viral reverse transcriptase. Med. Res. Rev. 13:229–258 [DOI] [PubMed] [Google Scholar]
- 15. Delaugerre C, et al. 2001. Resistance profile and cross-resistance of HIV-1 among patients failing a non-nucleoside reverse transcriptase inhibitor-containing regimen. J. Med. Virol. 65:445–448 [PubMed] [Google Scholar]
- 16. Delviks-Frankenberry KA, Nikolenko GN, Pathak VK. 2010. The “connection” between HIV drug resistance and RNase H. Viruses 2:1476–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Demeter LM, et al. 2000. Delavirdine susceptibilities and associated reverse transcriptase mutations in human immunodeficiency virus type 1 isolates from patients in a phase I/II trial of delavirdine monotherapy (ACTG 260). Antimicrob. Agents Chemother. 44:794–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fontaine E, Vaerenbergh KV, Vandamme AM, Schmit JC. 1999. Multidrug resistant human immunodeficiency virus type 1. AIDS Rev. 1:231–237 [Google Scholar]
- 19. Goto J, Kataoka R, Muta H, Hirayama N. 2008. ASEDock-docking based on alpha spheres and excluded volumes. J. Chem. Infect. Model. 48:583–590 [DOI] [PubMed] [Google Scholar]
- 20. Hachiya A, et al. 2008. Amino acid mutation N348I in the connection subdomain of human immunodeficiency virus type 1 reverse transcriptase confers multiclass resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. J. Virol. 82:3261–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hachiya A, et al. 2009. Clinical relevance of substitutions in the connection subdomain and RNase H domain of HIV-1 reverse transcriptase from a cohort of antiretroviral treatment-naive patients. Antivir. Res. 82:115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Halgren TA. 1999. MMFF VI. MMFF94s option for energy minimization studies. J. Comput. Chem. 20:720–729 [DOI] [PubMed] [Google Scholar]
- 23. Halgren TA. 1999. MMFF VII. Characterization of MMFF94, MMFF94s, and other widely available force fields for conformational energies and for intermolecular-interaction energies and geometries. J. Comput. Chem. 20:730–748 [DOI] [PubMed] [Google Scholar]
- 24. Hang JQ, et al. 2007. Substrate-dependent inhibition or stimulation of HIV RNase H activity by non-nucleoside reverse transcriptase inhibitors (NNRTIs). Biochem. Biophys. Res. Commun. 352:341–350 [DOI] [PubMed] [Google Scholar]
- 25. Haraguchi K, et al. 2003. Synthesis of a highly active new anti-HIV agent 2′,3′-didehydro-3′-deoxy-4′-ethynylthymidine. Bioorg. Med. Chem. Lett. 13:3775–3777 [DOI] [PubMed] [Google Scholar]
- 26. Hopkins AL, et al. 1996. Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors. J. Med. Chem. 39:1589–1600 [DOI] [PubMed] [Google Scholar]
- 27. Ibe S, Sugiura W. 2011. Clinical significance of HIV reverse-transcriptase inhibitor-resistance mutations. Future Microbiol. 6:295–315 [DOI] [PubMed] [Google Scholar]
- 28. Isono Y, et al. 2011. Synthesis of 1-benzyl-3-(3,5-dimethylbenzyl)uracil derivatives with potential anti-HIV activity. Antivir. Chem. Chemother. 22:57–65 [DOI] [PubMed] [Google Scholar]
- 29. Johnson VA, et al. 2006. Update of the drug resistance mutations in HIV-1: Fall 2006. Top. HIV Med. 14:125–130 [PubMed] [Google Scholar]
- 30. Klibanov OM, Kaczor RL. 2010. IDX-899, an aryl phosphinate-indole non-nucleoside reverse transcriptase inhibitor for the potential treatment of HIV infection. Curr. Opin. Invest. Drugs 11:237–245 [PubMed] [Google Scholar]
- 31. Lengruber RB, et al. 2011. Phenotypic characterization of drug resistance-associated mutations in HIV-1 RT connection and RNase H domains and their correlation with thymidine analogue mutations. J. Antimicrob. Chemother. 66:702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lightfoote MM, et al. 1986. Structural characterization of reverse transcriptase and endonuclease polypeptides of the acquired immunodeficiency syndrome retrovirus. J. Virol. 60:771–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maruyama T, et al. 2006. Synthesis and anti-HIV-1 and anti-HCMV activity of 1-substituted 3-(3,5-dimethylbenzyl)uracil derivatives. Chem. Pharm. Bull. 54:325–333 [DOI] [PubMed] [Google Scholar]
- 34. Maruyama T, et al. 2003. Synthesis and antiviral activity of 1,3-disubstituted uracils against HIV-1 and HCMV. Antivir. Chem. Chemother. 14:271–279 [DOI] [PubMed] [Google Scholar]
- 35. Moyle G, et al. 2010. Phase 2a randomized controlled trial of short-term activity, safety, and pharmacokinetics of a novel nonnucleoside reverse transcriptase inhibitor, RDEA806, in HIV-1-positive, antiretroviral-naive subjects. Antimicrob. Agents Chemother. 54:3170–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nikolenko GN, Delviks-Frankenberry KA, Pathak VK. 2010. A novel molecular mechanism of dual resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. J. Virol. 84:5238–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nikolenko GN, et al. 2005. Mechanism for nucleoside analog-mediated abrogation of HIV-1 replication: balance between RNase H activity and nucleotide excision. Proc. Natl. Acad. Sci. U. S. A. 102:2093–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ntemgwa M, et al. 2007. Variations in reverse transcriptase and RNase H domain mutations in human immunodeficiency virus type 1 clinical isolates are associated with divergent phenotypic resistance to zidovudine. Antimicrob. Agents Chemother. 51:3861–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pauwels R, et al. 1988. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J. Virol. Methods 20:309–312 [DOI] [PubMed] [Google Scholar]
- 40. Ren J, et al. 1995. High resolution structures of HIV-1 RT from four RT-inhibitor complexes. Nat. Struct. Biol. 2:293–302 [DOI] [PubMed] [Google Scholar]
- 41. Ren J, et al. 2000. Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase. Structure 8:1089–1094 [DOI] [PubMed] [Google Scholar]
- 42. Ren J, et al. 2001. Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors. J. Mol. Biol. 312:795–805 [DOI] [PubMed] [Google Scholar]
- 43. Richman DD, et al. 1994. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J. Virol. 68:1660–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roquebert B, et al. 2007. Relationship between mutations in HIV-1 RNase H domain and nucleoside reverse transcriptase inhibitors resistance mutations in naive and pretreated HIV-infected patients. J. Med. Virol. 79:207–211 [DOI] [PubMed] [Google Scholar]
- 45. Santos AF, et al. 2008. Conservation patterns of HIV-1 RT connection and RNase H domains: identification of new mutations in NRTI-treated patients. PLoS One 3:e1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sarafianos SG, et al. 2009. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 385:693–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smith JS, Roth MJ. 1992. Specificity of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H in removal of the minus-strand primer, tRNA(Lys3). J. Biol. Chem. 267:15071–15079 [PubMed] [Google Scholar]
- 48. Spence R, Kati W, Anderson K, Johnson K. 1995. Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors. Science 267:988–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stanford University HIV drug resistance database: NNRTI resistance notes. Stanford University, Stanford, CA: http://hivdb.stanford.edu/cgi-bin/NNRTIResiNote.cgi [Google Scholar]
- 50. Szczech GM, et al. 2000. Safety assessment, in vitro and in vivo, and pharmacokinetics of emivirine, a potent and selective nonnucleoside reverse transcriptase inhibitor of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tachedjian Orlova GM, Sarafianos SG, Arnold E, Goff SP. 2001. Nonnucleoside reverse transcriptase inhibitors are chemical enhancers of dimerization of the HIV type 1 reverse transcriptase. Proc. Natl. Acad. Sci. U. S. A. 98:7188–7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tanaka H, et al. 1995. Synthesis and antiviral activity of 6-benzil analogs of 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine (HEPT) as potent and selective anti-HIV-1 drugs. J. Med. Chem. 38:2860–2865 [DOI] [PubMed] [Google Scholar]
- 53. Vingerhoets J, et al. 2010. Resistance profile of etravirine: combined analysis of baseline genotypic and phenotypic data from the randomized, controlled Phase III clinical studies. AIDS 24:503–514 [DOI] [PubMed] [Google Scholar]
- 54. Waters JM, et al. 2009. Mutations in the thumb-connection and RNase H domain of HIV type-1 reverse transcriptase of antiretroviral treatment-experienced patients. Antivir. Ther. 14:231–239 [PubMed] [Google Scholar]
- 55. Weiner SJ, et al. 1984. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 106:765–784 [Google Scholar]
- 56. Wilkin A, et al. 17 October 2011. Long-term efficacy, safety and tolerability of rilpivirine (RPV, TMC278) in HIV type 1-infected antiretroviral-naive patients: week 192 results from a phase IIb randomized trial. AIDS Res. Hum. Retrovir. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 57. Witvrouw M, et al. 1999. Activity of non-nucleoside reverse transcriptase inhibitors against HIV-2 and SIV. AIDS 13:1477–1483 [DOI] [PubMed] [Google Scholar]
- 58. Wohrl BM, Moelling K. 1990. Interaction of HIV-1 ribonuclease H with polypurine tract containing RNA-DNA hybrids. Biochemistry 29:10141–10147 [DOI] [PubMed] [Google Scholar]
- 59. Yap SH, et al. 2007. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 4:e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhan P, et al. 2011. HIV-1 NNRTIs: structural diversity, pharmacophore similarity, and implications for drug design. Med. Res. Rev.. doi:10.1002/med [DOI] [PubMed] [Google Scholar]
- 61. Zhou XJ, et al. 2009. Single-dose escalation and multiple-dose safety, tolerability, and pharmacokinetics of IDX899, a candidate human immunodeficiency virus type 1 nonnucleoside reverse transcriptase inhibitor, in healthy subjects. Antimicrob. Agents Chemother. 53:1739–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]