Abstract
With voriconazole (VRC) being approved as the first choice in treating invasive aspergillosis (IA) and its increasing use in treatment, a VRC-resistant strain of Aspergillus flavus, the second leading cause of IA after Aspergillus fumigatus, has emerged. The VRC-resistant strain of A. flavus was isolated for the first time from the surgical lung specimen of an IA patient with no response to VRC therapy. In order to ascertain the mechanism of VRC resistance, the azole target enzyme genes in this strain of A. flavus were cloned and sequenced, and 4 mutations generating amino acid residue substitutions were found in the cyp51C gene. To further determine the role of this mutated gene for VRC resistance in A. flavus, an Agrobacterium tumefaciens-mediated gene replacement approach was applied. Consequently, the mutated cyp51C gene from this A. flavus strain was proven to confer the VRC resistance. Finally, to discern the one out of the four mutations in the cyp51C gene that is responsible for contributing to VRC resistance, a site-directed gene mutagenesis procedure combined with a gene replacement method was performed. As a result, the T788G missense mutation in the cyp51C gene was identified as responsible for VRC resistance in A. flavus. These findings indicated that the detection of this mutation in A. flavus could serve as an indicator for physicians to avoid the use of VRC during IA treatment. Further comprehensive surveillance for antifungal susceptibility, as well as intensive study on the mechanism of azole resistance in A. flavus causing IA, would be required to fully understand this mechanism.
INTRODUCTION
Invasive aspergillosis (IA) mainly occurs in severely immunocompromised patients such as those with prolonged neutropenia, advanced AIDS, and chronic granulomatous disease. It is also a severe complication in patients with hematologic malignancies, allogeneic hematopoietic stem cell transplantation (HSCT), and solid organ transplantation (23). During IA treatment, voriconazole (VRC), an azole drug, is the first choice due to its favorable responses (11, 31). However, with the wide use of azoles in clinical practice, azole-resistant aspergillosis including that having multiple-triazole resistance is becoming a prominent clinical problem, resulting in the poor prognosis and high mortality (2, 28, 29). Azole resistance in Aspergillus fumigatus has become a new challenge in the management of IA (25). Although azole-resistant aspergillosis was reported for the first time in an A. fumigatus infection in 1997 (5), until now, there has been no report of azole resistance in Aspergillus flavus-infected patients, even though A. flavus is the second most common IA pathogen (7, 9, 14, 23, 31, 32). Recently, a patient with IA resistant to VRC therapy appeared in our practice. From the lung surgical specimen of this patient, we isolated a VRC-resistant A. flavus strain, from which we then determined that VRC resistance in this A. flavus strain resulted from the T788G missense mutation in the cyp51C gene encoding azole target enzyme. This mutation was different from that in the cyp51A gene of A. fumigatus (2), which was responsible for azole resistance.
MATERIALS AND METHODS
Isolation of VRC-resistant A. flavus strain from the patient.
The pathogenic fungal strain isolated from the culturing of the lung surgical specimen of the IA patient was used to extract the genomic DNA according to the procedure described previously (33). This strain was identified by macroscopic and microscopic characteristics after being grown on both Czapek agar and malt extract agar at 25°C for 10 days (20). The actin and calmodulin genes and the internal transcribed spacer of this strain were then amplified (12, 22) and identified by alignment online using the recognized Aspergillus species database (http://www.cbs.knaw.nl). This strain was identified and named A. flavus BMU29791. To verify whether A. flavus BMU29791 is resistant to the commonly used antifungal drugs, the Clinical and Laboratory Standards Institute (CLSI) M38-A2 method (4, 30) and Etest (27) were used to assay antifungal susceptibility in A. flavus BMU29791.
Expression level and sequencing analysis of the azole target enzyme genes in A. flavus BMU29791.
Since overexpression in azole target enzyme gene cyp51A could contribute to azole resistance in A. fumigatus (2), we hypothesized that this is also the case in this VRC-resistant strain, A. flavus BMU29791. We searched the homologue of the A. fumigatus cyp51A gene in A. flavus using the nucleotide databank NCBI BLAST (http://www.ncbi.nlm.nih.gov). Three gene homologues of the A. fumigatus cyp51A gene, namely, cyp51A (NCBI accession number XM_002375082.1), cyp51B (NCBI accession number XM_002379089.1), and cyp51C (NCBI accession number XM_002383890.1), were found in A. flavus. We then downloaded the whole sequences of the open reading frames (ORFs), flanked by both the upstream and downstream regions with a size of about 1,000 bp, of these three genes from the A. flavus genome (http://www.broadinstitute.org). According to these sequences, the primers for cloning as well as the TaqMan probes (Table 1) for measuring the expression levels of the cyp51A, cyp51B, and cyp51C genes by quantitative real-time PCR were designed. Relative quantitative real-time PCR (19) to measure expression levels of genes for the azole target enzyme in A. flavus BMU29791 was performed by using an ABI 7500 PCR device. The primers and TaqMan probes are listed in Table 1.
Table 1.
Primers and probes used in the present study
| Primer/TaqMan probe | Sequencee | Target |
|---|---|---|
| CYP51AFa | 5′-ATCCCGGGATAAACTCTATACATCATAG-3′ (SmaI) | cyp51A gene of A. flavus |
| CYP51ARa | 5′-ATGCGGCCGCGACCTTACATGGAACGCCTT-3′ (NotI) | |
| CYP51BFa | 5′-ATGCGGCCGCGCCCGTGGGTTGCTGTCTTT-3′ (NotI) | cyp51B gene of A. flavus |
| CYP51BRa | 5′-ACGCATGCTTTTCGGTTCCTATCCACCC-3′ (SphI) | |
| CYP51CF1a | 5′-ATCCCGGGCACGTACACCAAACACTGCT-3′ (SmaI) | cyp51C gene of A. flavus |
| CYP51CR1a | 5′-ATGCGGCCGCAGCTGGAAGCTCTTCCGCTA-3′ (NotI) | |
| AflaF1b | 5′-GCGCGCATGAGGGAGAT-3′ | cyp51A gene of A. flavus |
| AflaR1b | 5′-CAATGCATGAGGTTCCAGATCA-3′ | |
| ALF1-TaqManb | FAM-5′-TCATTAACGAGCGCCGCAAGAACC-3′-TAMRA | |
| AflaF2b | 5′-ATTCGACTCGACATTTGCTGAA-3′ | cyp51B gene of A. flavus |
| AflaR2b | 5′-GCATCACGCTTGCGGTTAT-3′ | |
| ALF2-TaqManb | FAM-5′-CATGATCTCGACATGGGTTTTGCCC-3′-TAMRA | |
| AflaF3b | 5′-GTGACAACGTCCCGGAGAAG-3′ | cyp51C gene of A. flavus |
| AflaR3b | 5′-ATCAGCAGCGTGATCATGATG-3′ | |
| ALF3-TaqManb | FAM-5′-ATGCAGTGCACCTACAAGAACGGACAGC-3′-TAMRA | |
| AflaActinFb | 5′-TGGTTCCAATCTACGAAGGTTTC-3′ | actin gene of A. flavus |
| AflaActinRb | 5′-ATCTCGTGCTCGGCAGATGT-3′ | |
| ALFActin-TaqManb | FAM-5′-CTATGCCACACGCTATCGCTCGGATG-3′-TAMRA | |
| AflaCYP51CF2c | 5′-TCGAGCTCCACGTACACCAAACACTGCT-3′ (SacI) | cyp51C gene of A. flavus |
| AflaCYP51CR2c | 5′-AACCCGGGAGCTGGAAGCTCTTCCGCTA-3′ (SmaI) | |
| F-161d | 5′-AGCACAATCCACTATGGAACGGATCCGTAC-3′ | Recombinant plasmid pDHt/sk-3357C-161 |
| R-161d | 5′-GTTCCATAGTGGATTGTGCTGCCGATGAAT-3′ | |
| F-788d | 5′-CTGCCCATGCTCGGATGCGTGCTATCTACAT-3′ | Recombinant plasmid pDHt/sk-3357C-788 |
| R-788d | 5′-CACGCATCCGAGCATGGGCAGCATCGCGTTT-3′ | |
| F-1325d | 5′-CCGGTGGGAAACACAGGCAACTCAGGAAAA-3′ | Recombinant plasmid pDHt/sk-3357C-1325 |
| R-1325d | 5′-TTGCCTGTGTTTCCCACCGGTGCGGGTCCCA-3′ | |
| F-1337d | 5′-CACAGGCACCTCAGGAAAACGATAAGGATGA-3′ | Recombinant plasmid pDHt/sk-3357C-1337 |
| R-1337d | 5′-CGTTTTCCTGAGGTGCCTGTGTTTCCCACCG-3′ |
Primers used in cloning of the genes cyp51A, cyp51B, and cyp51C for sequencing analysis and construction of the A. flavus transformants with extra copies of these genes from A. flavus.
Primers and probes used for real-time PCR assaying.
Primers used for amplification of cyp51C for both gene replacement and site-directed mutagenesis.
Primers used for construction of plasmids pDHt/sk-3357C-161, pDHt/sk-3357C-788, pDHt/sk-3357C-1325, and pDHt/sk-3357C-1337 during site-directed mutagenesis experiment.
Underlining indicates the sites for the restriction enzymes indicated in parentheses or site-directed mutations (single underlined bases). FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
Construction of A. flavus transformants with extra copies of azole target enzyme genes.
In an attempt to assess whether VRC resistance in A. flavus was the result of increased copies of the cyp51A, cyp51B, or cyp51C gene, we used an approach described for A. fumigatus (16) to study gene-based dose-dependent resistance to antifungals. Primers were designed (Table 1) to amplify the cyp51A, cyp51B, and cyp51C genes, including the whole coding region with 1 kb of genomic DNA upstream and downstream of each gene, from the genomic DNA (33) of A. flavus NRRL3357 (obtained from the Fungal Genetics Stock Center [17]). The PCR amplicons were cloned into the pRG3-AMA1-NotI plasmid (16), and the recombinant plasmids (Table 2) pRG3-AMA1-NotI-A, pRG3-AMA1-NotI-B, and pRG3-AMA1-NotI-C were further transformed into the protoplast of the uracil prototroph A. flavus strain 3357-5 (kind gift of Zhumei He, School of Life Sciences, Sun Yat-Sen University, Guangzhou, People's Republic of China) (Table 2) (8) to obtain the corresponding transformants, namely, AF-A, AF-B, and AF-C. The transformant AF-E with the empty plasmid pRG3-AMA1-NotI was used as a control.
Table 2.
Strains and plasmids used in the present study
| Strain or plasmid | Relevant characteristic | Source |
|---|---|---|
| Strains | ||
| A. flavus 3357-5 | A. flavus lacking pyrG | 8 |
| NRRL3357 | Standard A. flavus strain | 17 |
| BMU29791 | VRC-resistant A. flavus | Present study |
| AF-A | A. flavus 3357-5 transformed with plasmid pRG3-AMA1-NotI-A | Present study |
| AF-B | A. flavus 3357-5 transformed with plasmid pRG3-AMA1-NotI-B | Present study |
| AF-C | A. flavus 3357-5 transformed with plasmid pRG3-AMA1-NotI-C | Present study |
| AF-E | A. flavus 3357-5 transformed with plasmid pRG3-AMA1-NotI | Present study |
| AflavC | A. flavus NRRL3357 transformant with cyp51C gene from A. flavus BMU29791 | Present study |
| AflavC-3357 | A. flavus BMU29791 transformant with cyp51C gene from A. flavus NRRL3357 | Present study |
| AflavC-161 | A. flavus NRRL3357 transformant with T161C mutation in cyp51C gene | Present study |
| AflavC-788 | A. flavus NRRL3357 transformant with T788G mutation in cyp51C gene | Present study |
| AflavC-1325 | A. flavus NRRL3357 transformant with C1325A mutation in cyp51C gene | Present study |
| AflavC-1337 | A. flavus NRRL3357 transformant with A1337G mutation in cyp51C gene | Present study |
| E. coli DH10B | E. coli | Invitrogen |
| A. tumefaciens EHA105 | A. tumefaciens | 26 |
| Plasmids | ||
| pRG3-AMA1-NotI | Shuttle plasmid allowing autonomous nonintegrating replication of itself in life cycle of Aspergillus spp. | 16 |
| pRG3-AMA1-NotI-A | Recombinant plasmid of pRG3-AMA1-NotI with complete cyp51A gene of A. flavus | Present study |
| pRG3-AMA1-NotI-B | Recombinant plasmid of pRG3-AMA1-NotI with complete cyp51B gene of A. flavus | Present study |
| pRG3-AMA1-NotI-C | Recombinant plasmid of pRG3-AMA1-NotI with complete cyp51C gene of A. flavus | Present study |
| pDHt/sk | Binary plasmid | 26 |
| pDHt/sk-29791C | Recombinant plasmid of pDHt/sk with complete cyp51C gene of A. flavus BMU29791 | Present study |
| pDHt/sk-3357C | Recombinant plasmid of pDHt/sk with complete cyp51C gene of A. flavus NRRL3357 | Present study |
| pDHt/sk-3357C-161 | Recombinant plasmid of pDHt/sk with A. flavuscyp51C gene carrying T161C mutation | Present study |
| pDHt/sk-3357C-788 | Recombinant plasmid of pDHt/sk with A. flavuscyp51C gene carrying T788G mutation | Present study |
| pDHt/sk-3357C-1325 | Recombinant plasmid of pDHt/sk with A. flavuscyp51C gene carrying C1325A mutation | Present study |
| pDHt/sk-3357C-1337 | Recombinant plasmid of pDHt/sk with A. flavuscyp51C gene carrying A1337G mutation | Present study |
| pMD20-T vector | For T-A clone | TaKaRa Biotechnology |
We further tested the antifungal susceptibility in these 3 transformants with 2 independent methods. First, a broth microdilution method according to the CLSI M38-A2 guidelines was used. Selective liquid minimal medium (MM; 70 mM NaNO3, 7 mM KCl, 2 mM MgSO4, 12 mM KPO4 [pH 6.8], trace elements, 1% glucose) minus uracil was used to replace the recommended RPMI 1640 broth medium in order to maintain the selection of the pyrG+ plasmid in these transformants. AF-E, the A. flavus transformant with extra copies of the empty plasmid pRG3-AMA1-NotI, was used as a control. Second, the Etest was performed. Similarly, MM plates minus uracil were used to maintain the selection of the pyrG+ plasmid in these transformants.
Sequence analysis of azole target enzyme genes in A. flavus BMU29791.
Because azole resistance in A. fumigatus is mainly the result of mutations and a 34-bp duplicate in the promoter region in cyp51A (2), we speculated that this would also be the case in A. flavus BMU29791.
We hence amplified the whole coding regions of the cyp51A, cyp51B, and cyp51C genes as well as the 1-kb genomic DNA fragment upstream and downstream of each gene in A. flavus BMU29791 by using primers (Table 1) designed according to the azole target enzyme gene sequences in A. flavus. After cloning, each PCR product was sequenced. The PCR amplicons were then cloned into the pMD20-T vector (TaKaRa Biotechnology Co., Ltd., Dalian, China) (21) for sequencing. These DNA sequences were aligned online (http://www.ebi.ac.uk) with those from A. flavus NRRL3357, and four mutations were found in the cyp51C gene of A. flavus BMU29791.
cyp51C gene replacement in A. flavus.
In order to ascertain whether the mutated cyp51C gene in A. flavus BMU29791 accounts for VRC resistance, we used in A. flavus an Agrobacterium tumefaciens-mediated gene transformation system as described previously for A. fumigatus (18) with minor modifications to replace the cyp51C gene of A. flavus NRRL3357 with the corresponding gene from A. flavus BMU29791. Primers were designed (Table 1) to amplify the cyp51C gene from the genomic DNA (33) of A. flavus BMU29791, including the whole coding region, 1 kb of genomic DNA upstream of the gene, and 1 kb of genomic DNA downstream of the gene. The PCR amplicon was cloned into the pDHt/sk vector (Table 2) (18, 26) to obtain the recombinant plasmid pDHt/sk-29791C, which was further transformed into A. tumefaciens EHA105 (a gift from K. J. Kwon-Chung, Laboratory of Clinical Infection, National Institutes of Health) (Table 2) (26) to obtain the resulting A. tumefaciens strain EHA105-29791C. Then, the A. tumefaciens EHA105-29791C suspension (200 μl) was mixed with an equal volume of A. flavus NRRL3357 conidia (1 × 107/ml), and the mixture was spread on the induction medium plates (26) containing 0.5 μg/ml VRC for incubation at 24°C for 48 h followed by another 48 h at 37°C. In our pilot experiment, it was proven that 200 μl of the A. flavus NRRL3357 conidium suspension (1 × 107/ml) could be killed on the induction medium with VRC at concentration of 0.5 μg/ml, without any VRC-resistant A. flavus strains grown. The colonies of the A. flavus transformants were selected up and cultured on the MM plates containing 200 μg/ml cefotaxime sodium and 0.5 μg/ml VRC at 37°C to verify the purity. After amplification by PCR, the cyp51C gene in the A. flavus transformant (named AflavC) was sequenced to confirm the successful replacement of the cyp51C gene. Finally, the expression levels of the cyp51A, cyp51B, and cyp51C genes of the transformant AflavC preexposed to VRC at 0.25 μg/ml were assayed by using real-time PCR as described above.
At the same time, the mutated cyp51C gene in A. flavus BMU29791 was also replaced with the one from A. flavus NRRL3357 per the procedure described above with a minor modification: primers were designed (Table 1) to amplify the cyp51C gene from the genomic DNA (33) of A. flavus NRRL3357, including the whole coding region, 1 kb of genomic DNA upstream of the gene, and 1 kb of genomic DNA downstream of the gene. The PCR amplicon was cloned into the pDHt/sk vector (Table 2) (18, 26) to obtain the recombinant plasmid pDHt/sk-3357C, which was further transformed into A. tumefaciens EHA105 (Table 2) (26) to obtain the resulting A. tumefaciens strain, EHA105-3357C. Then, the A. tumefaciens EHA105-3357C suspension (200 μl) was mixed with an equal volume of the A. flavus BMU29791 conidia (1 × 107/ml), and the mixture was spread on the induction medium plates (26) for incubation at 24°C for 48 h followed by another 48 h at 37°C. The A. flavus transformant colonies were selected and cultured on the MM plates containing 200 μg/ml cefotaxime sodium to kill A. tumefaciens. Each of the A. flavus transformant colonies was further subcultured on MM both without and with 0.5 μg/ml VRC at 37°C. Then, 48 to 72 h later, A. flavus transformant colonies which were not able to grow on MM with 0.5 μg/ml VRC despite being grown on MM without VRC were used for sequencing analysis after PCR amplification. Finally, the A. flavus transformant (named AflavC-3357) containing the cyp51C gene from A. flavus NRRL3357 was confirmed.
Site-directed mutagenesis of the cyp51C gene in A. flavus.
To further discern the one out of the four mutations in cyp51C which was responsible for contributing to VRC resistance in A. flavus BMU29791, we performed a site-directed gene mutagenesis procedure in A. flavus. Primers for site-directed mutagenesis were designed (10) (Table 1) to amplify the recombinant plasmid pDHt/sk-3357C, a derivative of the plasmid pDHt/sk, which contained the A. flavus NRRL3357 cyp51C gene. The PCR amplicon, an entire circular plasmid anticipated to be about 9,500 bp in size, was then transformed into bacterial competent cells (Beijing TransGen Biotech Co., Ltd.) for sequencing. The corresponding plasmids pDHt/sk-3357C-161, pDHt/sk-3357C-788, pDHt/sk-3357C-1325, and pDHt/sk-3357C-1337 (Table 2), containing the A. flavus cyp51C gene with respective mutated sites T161C (M54T), T788G (S240A), C1325A (P419T), and A1337G (N423D), were confirmed by alignment online (http://www.ebi.ac.uk). These 4 recombinant plasmids were then transformed into A. tumefaciens EHA105, and the suspension of resulting A. tumefaciens transformants (named EHA105-3357C-161, EHA105-3357C-788, EHA105-3357C-1325, and EHA105-3357C-1337, respectively) was further coincubated with A. flavus NRRL3357 conidia as described above. The cyp51C gene in the corresponding A. flavus transformants was further analyzed by PCR sequencing and alignment online (http://www.ebi.ac.uk) to confirm the success of replacement. Consequently, 4 transformants carrying each of the 4 mutations in the cyp51C gene, namely, AflavC-161, AflavC-788, AflavC-1325, and AflavC-1337, were obtained. The expression levels of the cyp51A, cyp51B, and cyp51C genes of the transformants AflavC-161, AflavC-788, AflavC-1325, and AflavC-1337 preexposed to VRC at 0.25 μg/ml were also assayed by using real-time PCR as described above.
RESULTS
IA patient unresponsive to VRC treatment.
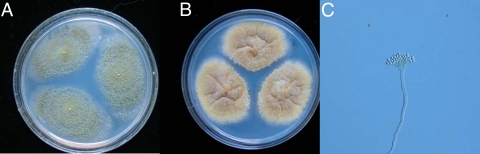
The patient, a 14-year-old girl, was diagnosed with acute myeloid leukemia (AML-M2) in December 2007, and she was treated with chemotherapy consisting successively of dexamethasone and epirubicin; arabinosylcytosine, dexamethasone, and epirubicin; harringtonine and arabinosylcytosine; idarubicin, arabinosylcytosine, and epirubicin; and idarubicin and arabinosylcytosine. Twelve months later, her bone marrow examination indicated a relapse of leukemia. Allogeneic HSCT was performed in February 2009. Two days after the transplantation, the patient coughed and had expectoration with back pain, and Staphylococcus aureus was isolated from her sputum. Then, she was treated with vancomycin and azythromycin. After a short period of remission, symptoms emerged again during antibiotic treatment. VRC was given orally 200 mg every 12 h to control the possible fungal infection. Her symptoms were alleviated only for a short period of time, and her condition did not improve after antifungal treatment for 2 months. Radiological examination showed localized masses in the upper lobe of her right lung, and pulmonectomy was conducted on 23 September 2009. Her infection and respiratory symptoms disappeared after the operation. Histopathological examination showed branched hyphae with septa in the lung tissue. Culture of the surgical specimen found mold, which was further identified as A. flavus by morphological (Fig. 1) and molecular experiments and was named A. flavus BMU29791. Therefore, this patient was diagnosed with IA.
Fig 1.
Morphological observation for A. flavus BMU29791 following 10 days of culture at 25°C. (A) Macroscopic appearance on MEA plate: velvet-like, yellow-green colony. (B) Macroscopic appearance on CZA plate: cottony and yellow colony. (C) Microscopy: radiate conidial heads with uniseriate conidiogenous cells.
Etiologic organism A. flavus BMU29791 was resistant to VRC.
The MICs of VRC, itraconazole (ITC), and amphotericin B (AMB) against A. flavus BMU29791 were 8 μg/ml, 2 μg/ml, and 1 μg/ml (Table 3), respectively. According to the proposed interpretative breakpoints (4, 30), A. flavus BMU29791 was defined as resistant to VRC, intermediate to ITC, and susceptible to AMB. The minimal effective concentration (MEC) of caspofungin (CAS) for A. flavus BMU29791 was 0.25 μg/ml. An Epsilometer test (Etest; AB Biodisk, Solna, Sweden) for antifungal susceptibility in this strain showed similar results (data not shown).
Table 3.
MIC or MEC of antifungal drugs against Aspergillus sp. strainsl
| Strain | MIC/MEC (μg/ml) of: |
Portion of sequence of cyp51C gene containing mutationk | |||
|---|---|---|---|---|---|
| ITC | VRC | AMB | CASj | ||
| BMU29791a | 2 | 8 | 1 | 0.25 | ATGCGTGCTATCTAC (240A) |
| AflavCb | 2 | 8 | 1 | 0.25 | ATGCGTGCTATCTAC (240A) |
| AflavC-3357c | 1 | 0.25 | 1 | 0.25 | ATGCGTTCTATCTAC (240S) |
| AflavC-788d | 2 | 8 | 1 | 0.25 | ATGCGTGCTATCTAC (240A) |
| NRRL3357e | 1 | 0.25 | 1 | 0.25 | ATGCGTTCTATCTAC (240S) |
| AF-Af | ≥16 | 4 | 0.5 | 0.03 | NA |
| AF-Bg | ≥16 | 4 | 0.5 | 0.03 | NA |
| AF-Ch | ≥16 | 1 | 0.5 | 0.03 | NA |
| AF-Ei | 0.5 | 0.5 | 0.5 | 0.03 | NA |
VRC-resistant A. flavus strain isolated from the lung tissue of the patient.
A. flavus NRRL3357 transformant carrying the cyp51C gene of A. flavus BMU29791.
A. flavus BMU29791 transformant carrying the cyp51C gene of A. flavus NRRL3357.
A. flavus NRRL3357 transformant carrying the cyp51C gene with T788G missense mutation.
VRC-susceptible A. flavus strain obtained from the Fungal Genetics Stock Center.
A. flavus transformant with recombinant plasmid pRG3-AMA1-NotI-A.
A. flavus transformant with recombinant plasmid pRG3-AMA1-NotI-B.
A. flavus transformant with recombinant plasmid pRG3-AMA1-NotI-C.
A. flavus transformant with the empty plasmid pRG3-AMA1-NotI.
The antifungal activity of CAS was determined by using MEC.
Codons (underlined) and corresponding amino acid residues (in parentheses) are shown. NA, not available.
MEC, minimal effective concentration; ITC, itraconazole; VRC, voriconazole; AMB, amphotericin B; CAS, caspofungin.
VRC resistance in A. flavus BMU29791 was not related to the expression level of azole target enzyme genes.
The expression level of the cyp51C gene, excluding those for cyp51A and cyp51B, was slightly increased in A. flavus BMU29791 in comparison with what was seen for the standard strain, A. flavus NRRL3357. Compared to control transformant AF-E, however, the A. flavus transformant AF-C with extra copies of the A. flavus cyp51C gene showed that it was susceptible to VRC despite being resistant to ITC (Table 3). Moreover, A. flavus transformant AF-A or AF-B with extra copies of the A. flavus cyp51A or cyp51B gene exhibited in vitro resistance to both ITC and VRC (Table 3). The Etest assay showed similar results (data not shown).
Mutations in the cyp51C gene in A. flavus BMU29791 were detected.
Comparing with the standard strain, A. flavus NRRL3357, no abnormal sequences in the upstream regions of the coding sequences in these 3 genes were observed. The cyp51A and cyp51B genes in A. flavus BMU29791 were also intact. However, four missense mutations, the T161C, T788G, C1325A, and A1337G mutations, were found in the cyp51C gene in the A. flavus BMU29791 strain. These four mutations result in amino acid residue substitutions of M54T, S240A, P419T, and N423D, respectively, in the CYP51C protein.
The mutated cyp51C gene from A. flavus BMU29791 contributed to VRC resistance.
Using an A. tumefaciens-mediated gene transformation system, the cyp51C gene in A. flavus NRRL3357, the standard A. flavus strain susceptible to VRC, was replaced with the one from A. flavus BMU29791. At the same time, the cyp51C gene in A. flavus BMU29791 was also replaced with the one from A. flavus NRRL3357. The antifungal susceptibility assay showed that the A. flavus transformant AflavC containing the mutated cyp51C gene from A. flavus BMU29791 achieved the specific resistance to VRC, while being intermediate to ITC and susceptible to AMB and CAS (Table 3). Moreover, the A. flavus transformant AflavC-3357 containing the cured allele of the cyp51C gene (from A. flavus NRRL3357) showed the same antifungal susceptibility spectrum as A. flavus NRRL3357 (Table 3).
In the transformants AflavC, AflavC-161, AflavC-788, AflavC-1325, and AflavC-1337 preexposed to VRC at 0.25 μg/ml, no significant change was observed in the expression levels of the cyp51A, cyp51B, and cyp51C genes, compared to those in A. flavus NRRL3357 (data not shown).
The T788G mutation in the cyp51C gene confers VRC resistance in A. flavus.
Using a site-directed gene mutagenesis procedure combined with the gene replacement method mentioned above, the cyp51C gene in A. flavus NRRL3357 was respectively replaced by each mutated cyp51C gene containing individual mutations observed in A. flavus BMU29791. Consequently, 4 transformants of A. flavus carrying each of the 4 mutations in the cyp51C gene were identified, which were named AflavC-161, AflavC-788, AflavC-1325, and AflavC-1337. After the antifungal susceptibility assay, we found that an A. flavus transformant harboring a mutated cyp51C gene with the T788G missense mutation (Table 3, strain AflavC-788) was resistant to VRC, intermediate to ITC, and susceptible to AMB, while the transformants carrying other point mutations, including T161C, C1325A, and A1337G, were all still susceptible to these antifungal drugs, suggesting that the T788G missense mutation (amino acid change of S240A) in the cyp51C gene endows A. flavus including the strain A. flavus BMU29791 with the ability to resist VRC treatment.
DISCUSSION
According to the clinical practice guideline by the Infectious Diseases Society of America (31) and the definitions of the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group/National Institute of Allergy and Infectious Diseases Mycoses Study Group (6), the patient described in the present study was diagnosed as having IA. She failed to respond to antifungal treatment with VRC. After the isolation of fungus from the surgical specimen of her pulmonary tissue, we first ascertained that the colonies originated from a single Aspergillus strain by a random-amplification polymorphism DNA approach (data not shown), since mixed infections by different microbes, such as fungi and bacteria (1), Candida albicans and non-albicans Candida (13), or A. fumigatus and A. flavus (3, 11), happen occasionally in the clinical environment. This strain was further identified as A. flavus by using the standard molecular and morphological approaches for Aspergillus species identification and was named A. flavus BMU29791.
A. flavus, the second leading cause of IA (7, 9, 14, 31, 32), is widely distributed in the environment, including soil, water, and air (7, 9, 14, 32). Individuals with severely impaired immune systems are prone to IA after inhaling A. flavus spores (9, 14, 23, 31). A. flavus is usually susceptible to the aforementioned antifungal drugs, including VRC. However, A. flavus strain BMU29791, isolated from the IA patient in the present study, was in vitro resistant to VRC, intermediate to ITC, and susceptible to both AMB and CAS (Table 3). To the best of our knowledge, this is the first description of VRC-resistant IA caused by the A. flavus strain with specific resistance to VRC.
Since ITC resistance was first described by Denning and colleagues in 1997 (5) in an IA patient infected by A. fumigatus, azole-resistant IA, all of which is caused by A. fumigatus as reported previously, has become a serious clinical issue (2, 24, 28, 29). Azole resistance in A. fumigatus has become a new challenge for IA treatment (25). Studies on the resistance mechanisms have shown that amino acid residue substitution derived from mutations in the azole target enzyme gene cyp51A, overexpression of this gene and drug efflux genes, and upregulation of homeostatic stress response pathways contribute to azole resistance in A. fumigatus (2, 24, 28, 29). Different from these reports, our data from the VRC-resistant A. flavus BMU29791 demonstrated that no abnormal sequences in the cyp51A and cyp51B genes and the upstream region of the coding sequence in the cyp51C gene were observed, whereas 4 missense mutations causing amino acid residue substitutions were detected in the cyp51C gene, a member of the azole target enzyme gene family in A. flavus. These findings are also different from the report by Krishnan-Natesan et al. (15) that the VRC-resistant strain of A. flavus, selected in vitro by preexposure with VRC in the laboratory, had mutations in both the cyp51A and cyp51B genes but not in the cyp51C gene. In addition, we also demonstrated that the expression levels of the azole target enzyme genes cyp51A, cyp51B, and cyp51C were not related to VRC resistance in A. flavus BMU29791, even though extra copies of the A. flavus cyp51A or cyp51B gene conferred reliable azole resistance in A. flavus (Table 3).
We then confirmed that the mutated cyp51C gene from A. flavus BMU29791 was responsible for VRC resistance by an A. tumefaciens-mediated gene replacement method. Furthermore, we determined that the T788G (S240A) missense mutation in the cyp51C gene in A. flavus accounted for the specific resistance to VRC (Table 3) by using site-directed gene mutagenesis combined with gene replacement approaches. However, other mutations in genes cyp51C, cyp51A, and cyp51B, in addition to the T788G (S240A) mutation in the cyp51C gene as we presented here, may also correlate with VRC resistance in A. flavus.
In conclusion, with the wide use of VRC as the first choice in IA treatment (11, 31), VRC-resistant IA caused by an A. flavus strain with a T788G missense mutation in the cyp51C gene is now emerging as a potentially common cause of IA in clinics. Detection of this mutation in A. flavus will be a good indicator to avoid VRC use during IA treatment. Further comprehensive surveillance for antifungal susceptibility, as well as intensive study on the mechanism of azole resistance in A. flavus causing IA, is certainly required to gain full understanding.
ACKNOWLEDGMENTS
This work was supported by National Natural Science Foundation of China grants 30970131 (to Wei Liu) and 30930006 (to Ruoyu Li), by Beijing Natural Science Foundation grant 7102149 (to Wei Liu), and by the Program for New Century Excellent Talents in University grants NCET-10-0198 (to Wei Liu) and BMU20110158 (to Wei Liu). The funders had no role in the study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 6 February 2012
REFERENCES
- 1. Adam B, Baillie GS, Douglas LJ. 2002. Mixed species biofilms of Candida albicans and Staphylococcus epidermidis. J. Med. Microbiol. 51:344–349 [DOI] [PubMed] [Google Scholar]
- 2. Chamilos G, Kontoyiannis DP. 2005. Update on antifungal drug resistance mechanisms of Aspergillus fumigatus. Drug Resist. Updat. 8:344–358 [DOI] [PubMed] [Google Scholar]
- 3. Cimerman M, Gunde-Cimerman N, Zalar P, Perkovic T. 1999. Femur osteomyelitis due to a mixed fungal infection in a previously healthy man. J. Clin. Microbiol. 37:1532–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clinical and Laboratory Standards Institute 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard. CLSI document M38-A2. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. Denning DW, et al. 1997. Itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 41:1364–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Pauw B, et al. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 46:1813–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao L, Yu J, Li R. 2010. Epidemiology of aspergillosis in mainland China. Chin. J. Mycol. 5:245–247 [Google Scholar]
- 8. He Z, Price MS, OBrian GR, Georgianna DR, Payne GA. 2007. Improved protocols for functional analysis in the pathogenic fungus Aspergillus flavus. BMC Microbiol. 7:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hedayati MT, Pasqualotto AC, Warn PA, Bowyer P, Denning DW. 2007. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology 153:1677–1692 [DOI] [PubMed] [Google Scholar]
- 10. Hemsley A, Arnheim N, Toney MD, Cortopassi G, Galas DJ. 1989. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic. Acids Res. 17:6545–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Herbrecht R, et al. 2002. Voriconazole versus amphotericin B for primary therapy of invasive aspergillosis. N. Engl. J. Med. 347:408–415 [DOI] [PubMed] [Google Scholar]
- 12. Hong SB, Go SJ, Shin HD, Frisvad JC, Samson RA. 2005. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 97:1316–1329 [DOI] [PubMed] [Google Scholar]
- 13. Jensen J, et al. 2007. Mixed fungemia: incidence, risk factors, and mortality in a general hospital. Clin. Infect. Dis. 44:e109–e114 [DOI] [PubMed] [Google Scholar]
- 14. Krishnan S, Manavathu EK, Chandrasekar PH. 2009. Aspergillus flavus: an emerging non-fumigatus Aspergillus species of significance. Mycoses 52:206–222 [DOI] [PubMed] [Google Scholar]
- 15. Krishnan-Natesan S, Chandrasekar PH, Alangaden GJ, Manavathu EK. 2008. Molecular characterisation of cyp51A and cyp51B genes coding for P450 14alpha-lanosterol demethylases A (cyp51Ap) and B (cyp51Bp) from voriconazole-resistant laboratory isolates of Aspergillus flavus. Int. J. Antimicrob. Agents. 32:519–524 [DOI] [PubMed] [Google Scholar]
- 16. Liu W, May GS, Lionakis MS, Lewis RE, Kontoyiannis DP. 2004. Extra copies of the Aspergillus fumigatus squalene epoxidase gene confer resistance to terbinafine: a genetic approach to study gene-dose dependent resistance to antifungals in Aspergillus fumigatus. Antimicrob. Agents Chemother. 48:2490–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCluskey K. 2003. The Fungal Genetics Stock Center: from molds to molecules. Adv. Appl. Microbiol. 52:245–262 [DOI] [PubMed] [Google Scholar]
- 18. Michielse CB, Hooykaas PJ, van den Hondel CA, Ram AF. 2005. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 48:1–17 [DOI] [PubMed] [Google Scholar]
- 19. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qi Z. 1997. Flora fungorum sinicorum, vol 5. Aspergillus et teleomorphi cognati, p 76–82 Science Press, Beijing, China [Google Scholar]
- 21. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 22. Samson RA, Varga J. 2009. What is a species in Aspergillus? Med. Mycol. 47:S13–S20 [DOI] [PubMed] [Google Scholar]
- 23. Segal BH. 2009. Aspergillosis. N. Engl. J. Med. 360:1870–1884 [DOI] [PubMed] [Google Scholar]
- 24. Snelders E, et al. 2008. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 5:e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Snelders E, Melchers WJ, Verweij PE. 2011. Azole resistance in Aspergillus fumigatus: a new challenge in the management of invasive aspergillosis? Future Microbiol. 6:335–347 [DOI] [PubMed] [Google Scholar]
- 26. Sugui JA, Chang YC, Kwon-Chung KJ. 2005. Agrobacterium tumefaciens-mediated transformation of Aspergillus fumigatus: an efficient tool for insertional mutagenesis and targeted gene disruption. Appl. Environ. Microbiol. 71:1798–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Szekely A, Johnson EM, Warnock DW. 1999. Comparison of E-test and broth microdilution methods for antifungal drug susceptibility testing of molds. J. Clin. Microbiol. 37:1480–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Verweij PE, Mellado E, Melchers WJ. 2007. Multiple-triazole-resistant aspergillosis. N. Engl. J. Med. 356:1481–1483 [DOI] [PubMed] [Google Scholar]
- 29. Verweij PE, Snelders E, Kema GH, Mellado E, Melchers WJ. 2009. Azole resistance in Aspergillus fumigatus: a side-effect of environmental fungicide use? Lancet Infect. Dis. 9:789–795 [DOI] [PubMed] [Google Scholar]
- 30. Verweij PE, Howard SJ, Melchers WJ, Denning DW. 2009. Azole-resistance in Aspergillus: proposed nomenclature and breakpoints. Drug Resist. Updat. 12:141–147 [DOI] [PubMed] [Google Scholar]
- 31. Walsh TJ, et al. 2008. Treatment of aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America. Clin. Infect. Dis. 46:327–360 [DOI] [PubMed] [Google Scholar]
- 32. Yu J, Cleveland TE, Nierman WC, Bennett JW. 2005. Aspergillus flavus genomics: gateway to human and animal health, food safety, and crop resistance to diseases. Rev. Iberoam. Micol. 22:194–202 [DOI] [PubMed] [Google Scholar]
- 33. Zhang D, Yang Y, Castlebury LA, Cerniglia CE. 1996. A method for the large scale isolation of high transformation efficiency fungal genomic DNA. FEMS Microbiol. Lett. 145:261–265 [DOI] [PubMed] [Google Scholar]