Abstract
Escherichia coli sequence type ST131 (from phylogenetic group B2), often carrying the extended-spectrum-β-lactamase (ESBL) gene blaCTX-M-15, is an emerging globally disseminated pathogen that has received comparatively little attention in the United States. Accordingly, a convenience sample of 351 ESBL-producing E. coli isolates from 15 U.S. centers (collected in 2000 to 2009) underwent PCR-based phylotyping and detection of ST131 and blaCTX-M-15. A total of 200 isolates, comprising 4 groups of 50 isolates each that were (i) blaCTX-M-15 negative non-ST131, (ii) blaCTX-M-15 positive non-ST131, (iii) blaCTX-M-15 negative ST131, or (iv) blaCTX-M-15 positive ST131, also underwent virulence genotyping, antimicrobial susceptibility testing, and pulsed-field gel electrophoresis (PFGE). Overall, 201 (57%) isolates exhibited blaCTX-M-15, whereas 165 (47%) were ST131. ST131 accounted for 56% of blaCTX-M-15-positive- versus 35% of blaCTX-M-15-negative isolates (P < 0.001). Whereas ST131 accounted for 94% of the 175 total group B2 isolates, non-ST131 isolates were phylogenetically distributed by blaCTX-M-15 status, with groups A (blaCTX-M-15-positive isolates) and D (blaCTX-M-15-negative isolates) predominating. Both blaCTX-M-15 and ST131 occurred at all participating centers, were recovered from children and adults, increased significantly in prevalence post-2003, and were associated with molecularly inferred virulence. Compared with non-ST131 isolates, ST131 isolates had higher virulence scores, distinctive virulence profiles, and more-homogeneous PFGE profiles. blaCTX-M-15 was associated with extensive antimicrobial resistance and ST131 with fluoroquinolone resistance. Thus, E. coli ST131 and blaCTX-M-15 are emergent, widely distributed, and predominant among ESBL-positive E. coli strains in the United States, among children and adults alike. Enhanced virulence and antimicrobial resistance have likely promoted the epidemiological success of these emerging public health threats.
INTRODUCTION
Extraintestinal Escherichia coli infections, including urinary tract infections, sepsis, and neonatal meningitis, are important causes of morbidity, mortality, and increased health care costs (26). Their management is increasingly challenging due to the rising prevalence of resistance to first-line antimicrobial agents, including fluoroquinolones and extended-spectrum cephalosporins (14, 23). Contributing significantly to this increase is a recently emerged E. coli lineage designated sequence type ST131, which is characterized as belonging to serotype O25:H4 and is associated with fluoroquinolone resistance and CTX-M-15, an IncFII-plasmid-mediated extended-spectrum β-lactamase (ESBL) (21, 25).
The initial reports in 2008 of E. coli ST131 as an important new pathogen involved isolates from diverse non-U.S. international locales (3, 5, 18). Subsequently, case reports and small series documented the presence of ST131 also in the United States, with the ESBL-positive ST131 isolates usually producing CTX-M-15 (6, 7, 9, 11, 17, 20, 27, 29, 30). The only published large-scale survey to date for ST131 in the United States appeared in 2010 and was based on clinical isolates from 2007 from two national surveillance programs (8). There, ST131 accounted for approximately two-thirds of all ESBL-positive or fluoroquinolone-resistant E. coli study isolates, with 73% of ESBL-positive ST131 isolates exhibiting CTX-M-15 (8).
Limitations of that study included the single study year, restrictions with respect to the participating sites and selection criteria of the surveillance systems, and inclusion of only 59 total ESBL-positive isolates (8). Here we assessed the reproducibility of that survey's findings and explored new associations, using a 10-year study period and a much larger population that included isolates from both children and adults.
MATERIALS AND METHODS
Strains.
The 351 ESBL-positive E. coli study isolates, including some previously published isolates (16, 17, 27, 29), were obtained as a series of convenience samples from colleagues at 15 research or clinical laboratories across the United States (Table 1). Each center was invited to submit all available E. coli clinical isolates that met the study criteria, including (i) isolation from 2000 to 2009 and (ii) confirmed ESBL production according to Clinical and Laboratory Standards Institute (CLSI) criteria (4). Upon receipt in the study laboratory, all isolates were screened for blaCTX-M-15 by PCR (as described below). All blaCTX-M-15 PCR-negative isolates were further screened by disk diffusion for extended-spectrum-cephalosporin susceptibility (as described below). All extended-spectrum-cephalosporin-susceptible isolates so identified were further screened for ESBL production, using cefotaxime and ceftazidime disks with or without clavulanate (4). For study inclusion, isolates were required to be blaCTX-M-15 positive or, if blaCTX-M-15 negative, to be either extended-spectrum cephalosporin resistant or (if retested) to exhibit an ESBL phenotype.
Table 1.
Sources and ST131 and blaCTX-M-15 status of 351 extended-spectrum-β-lactamase-producing Escherichia coli isolates collected in the United States in 2000 to 2009
| City or center source (reference, if published) | Yr of isolation | Patient population | Total no. of isolates | No. (%) of isolates in indicated categorya,b |
No. of ST131 isolates/total no. of isolates (%)a |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait |
Subseta,b |
||||||||||
| M15+ | ST131+ | ST131− M15− | ST131− M15+ | ST131+ M15− | ST131+ M15+ | M15− | M15+ | ||||
| Miami, FL | 2009 | Adultsc | 23 | 19 (83) | 7 (30) | 1 (4) | 15 (65) | 3 (13) | 4 (17) | 3/4 (75) | 4/19 (21) |
| Chicago, IL | 2007–2008 | Adults | 8 | 6 (75) | 5 (63) | 1 (13) | 2 (25) | 1 (13) | 4 (50) | 1/2 (50) | 4/6 (67) |
| Evanston, IL | 2007 | Adults | 27 | 16 (59) | 17 (63) | 4 (15) | 6 (22) | 7 (26) | 10 (37) | 7/11 (64) | 10/16 (63) |
| Portland, ME | 2008 | Adult | 1 | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | (none) | 1/1 (100) |
| Minneapolis, MN (Hennepin County Medical Center) | 2008–2009 | Adults | 31 | 17 (55) | 17 (55) | 9 (29) | 5 (16) | 5 (16) | 12 (39) | 5/14 (36) | 12/17 (71) |
| Minneapolis, MN (Children's Hospital) | 2002–2009 | Children | 33 | 12 (36) | 12 (36) | 16 (48) | 5 (15) | 5 (15) | 7 (21) | 5/21 (24) | 7/12 (58) |
| St. Paul, MN | 2007–2008 | Adults | 20 | 12 (60) | 7 (35) | 5 (25) | 8 (40) | 3 (15) | 4 (20) | 3/8 (38) | 4/12 (33) |
| Winston-Salem, NC | 2008–2009 | Adults | 11 | 8 (73) | 8 (73) | 3 (27) | 0 (0) | 0 (0) | 8 (73) | 0 (0) | 8/8 (100) |
| Omaha, NE | 2005 | Adults | 4 | 1 (25) | 2 (50) | 2 (50) | 0 (0) | 1 (25) | 1 (25) | 1/3 (33) | 1/1 (100) |
| Philadelphia, PA (17) | 2007 | Adults | 31 | 14 (45) | 15 (48) | 10 (32) | 6 (19) | 7 (23) | 8 (26) | 7/17 (41) | 8/14 (57) |
| Pittsburgh, PA (27) | 2006–2007 | Adults | 17 | 7 (41) | 10 (59) | 5 (29) | 2 (12) | 5 (29) | 5 (29) | 5/10 (50) | 5/7 (71) |
| New York City (Queens), NY (29) | 2004–2008 | Adult | 63 | 37 (59) | 38 (60) | 15 (24) | 10 (16) | 11 (17) | 27 (43) | 11/26 (42) | 27/37 (73) |
| Houston, TX (30) | 2004–2009 | Adults | 38 | 30 (79) | 10 (26) | 7 (18) | 21 (53) | 1 (3) | 9 (24) | 1/8 (13) | 9/30 (30) |
| San Antonio, TX (16) | 2000–2006 | Adults | 20 | 11 (55) | 7 (35) | 7 (35) | 6 (30) | 2 (10) | 5 (25) | 2/9 (22) | 5/11 (45) |
| Seattle, WA | 2004–2007 | Childrend | 24 | 10 (42) | 9 (38) | 13 (54) | 2 (8) | 1 (4) | 8 (33) | 1/14 (7) | 8/10 (80) |
| Total | 2000–2009 | Mixed | 351 | 201 (57) | 165 (47) | 97 (28) | 89 (25) | 51 (15) | 114 (33) | 51/148 (34) | 114/203 (56) |
M15+, positive for blaCTX-M-15; M15−, negative for blaCTX-M-15.
ST131+, ST131 sequence type; ST131−, non-ST131 sequence type.
Includes 1 pediatric isolate.
Includes 1 adult isolate.
Detection of blaCTX-M-15.
The PCR screen for blaCTX-M-15 involved a combination of published and newly devised primers. After an initial screen using primers that detect blaCTX-M-15 plus related blaCTX-M variants (15), PCR-positive isolates were retested using a novel extended “CTX-M-15-specific” forward primer (15extF [ATAAAACCGGCAGCGGTGG]), which hybridizes with blaCTX-M-15 and, per the data shown at http://www.lahey.org/Studies/other.asp#table1, with several other group 1 blaCTX-M variants (including blaCTX-M-28, -29, 3-3, -53, -82 but not blaCTX-M-1, blaCTX-M-3, or several other blaCTX-M-3-like variants detected by the blaCTX-M-15 screening primers). They also were retested using a novel extended “CTX-M-3-specific” forward primer (3extF [ATAAAACCGGCAGCGGTGA]), which hybridizes with blaCTX-M-3 and a different set of group 1 blaCTX-M variants (including blaCTX-M-1, -10, -11, -12, -22, -23, -30, -32, -34, -36, -37, -42, -52). Isolates reacting with the extended blaCTX-M-15 primer but not the extended blaCTX-M-3 primer were further screened using a novel blaCTX-M-28-specific forward primer (28for [GGTTAAAAAATCACTGCGT]), in combination with CTX-M group 1 reverse primer M13L (15), since blaCTX-M-28 is the most frequent of the potential cross-reacting blaCTX-M variants.
Conditions for the novel PCRs were largely as described elsewhere (13). Reaction mixtures included 1× buffer, 4 mM MgCl2, 0.8 mM deoxynucleoside triphosphate (dNTP) mix, 1.25 U of AmpliTaq Gold, 0.6 μM specific primers, 2.0 μl of boiled lysate DNA, and H2O to reach a final volume of 25 μl. Cycling was performed as follows: 95°C activation for 10 min; 25 cycles of 30 s of denaturation at 94°C and then 3 min of annealing and extension at 76°C (for blaCTX-M-15 and blaCTX-M-3 PCR [amplicon, 483 bp]) or at 68°C (for blaCTX-M-28 PCR [amplicon, 863 bp]); 10′ extension at 72°C; and then a hold at 4°C. Isolates that gave positive test results with the initial and extended blaCTX-M-15 primers but negative test results with the blaCTX-M-3 and blaCTX-M-28 primers were regarded as blaCTX-M-15 positive.
Validation of this tiered PCR approach using 257 reference isolates containing sequenced blaCTX-M variants (181 reportedly blaCTX-M-15, 76 reportedly non-blaCTX-M-15) yielded only 9 discrepancies, all but three of which were resolved in favor of the PCR results by repeat PCR and sequencing. The persisting discrepancies involved 3 blaCTX-M-53-positive isolates that were falsely identified by PCR as containing blaCTX-M-15, consistent with the predicted cross-reactivity of blaCTX-M-15 primers with the (rarely encountered) blaCTX-M-53 variant. This yielded an estimated overall accuracy rate of 98.8% (95% confidence interval, 96.6% to 99.8%) for detection of blaCTX-M-15 by tiered PCR.
Phylogenetic group and ST131 status.
The major E. coli phylogenetic groups (A, B1, B2, and D) were determined by triplex PCR (1). ST131 isolates were identified by PCR-based detection of ST131-specific single-nucleotide polymorphisms (SNPs) in mdh and gyrB (10). Seventeen randomly selected presumptive ST131 isolates underwent confirmatory 7-locus multilocus sequence typing (MLST) based on partial sequences for adk, fumC, gyrB, icd, mdh, purA, and recA (http://mlst.ucc.ie/mlst/dbs/Ecoli); all were confirmed as ST131. All isolates were screened by PCR for ST131-associated traits, including the F10 papA allele (P-antigen-recognizing fimbria [P fimbria] structural subunit variant) and the O25b rfb (lipopolysaccharide synthesis) variant (2, 10).
Antimicrobial susceptibility.
A total of 200 isolates, comprising 4 groups of 50 randomly selected isolates each that were (i) blaCTX-M-15 negative non-ST131; (ii) blaCTX-M-15 positive non-ST131; (iii) blaCTX-M-15 negative ST131; or (iv) blaCTX-M-15-positive ST131 underwent additional testing. Antimicrobial susceptibility to 21 agents was determined by disk diffusion using Clinical and Laboratory Standards Institute (CLSI)-specified procedures, control strains, and (2011) interpretive criteria. Agents tested included amikacin (AMK), ampicillin (AMP), amoxicillin-clavulanate (AML), aztreonam (ATM), cefepime (FEP), ceftazidime (CAZ), ceftriaxone (CRO), cephalothin (KF), chloramphenicol (C), ciprofloxacin (CIP), gentamicin (CN), imipenem (IMP), nalidixic acid (NA), nitrofurantoin (F), piperacillin (PIP), piperacillin-tazobactam (TZP), streptomycin (S), sulfonamides (SF), tetracycline (TE), trimethoprim (W), and trimethoprim-sulfamethoxazole (SXT). Intermediate results were analyzed as representing resistance. The resistance score represented the number of agents to which an isolate was resistant, excluding SXT (due to redundancy with SF and W).
Extended virulence genotypes.
The 200 isolates were tested for the presence of 50 extraintestinal pathogenic E. coli (ExPEC)-associated virulence genes and housekeeping gene uidA (β-glucuronidase) by multiplex PCR (10). Isolates were defined as ExPEC if positive for ≥2 of papA and/or papC (P fimbria major subunit and assembly), sfa/focDE (S and F1C fimbriae), afa/draBC (Dr-binding adhesins), kpsM II (group 2 capsule), and iutA (aerobactin receptor) (12). The virulence score represented the number of virulence genes detected, adjusted for multiple detection of the pap, sfa, foc, and kpsM II operons.
PFGE analysis.
The 200 isolates underwent pulsed-field gel electrophoresis (PFGE) analysis of XbaI-restricted total DNA per the PulseNet protocol (24). By using BioNumerics (Applied Maths) and band-based Dice similarity coefficients, pulsotype designations were assigned at the ≥94% profile similarity level, corresponding to an approximately 3-band difference (28).
Statistical methods. Comparisons of proportions were tested using Fisher's exact test (two-tailed) or a chi-square test if unpaired and McNemar's test if paired. Comparisons of virulence scores were tested using the Mann-Whitney U test (two-tailed). To simplify the virulence genotype and antimicrobial susceptibility data for comparisons among the four subgroups as defined by combined CTX-M-15 status and ST131 status, principal coordinate analysis (PCoA) was performed using GENALEX 6 software (19). This multidimensional scaling method captures within the first 2 axes most of the total variance contained in a multivariable, multispecimen data set. Analysis of molecular variance (AMOVA) was used to determine the percentage of diversity within populations versus among populations (19).
RESULTS
Origins of isolates.
The 351 ESBL-producing E. coli study isolates were from 15 centers distributed broadly across the United States (Table 1). Centers contributed a median of 23 isolates each (range, 1 to 63 isolates). Years of isolation ranged from 2000 to 2009 (median, 2007). Source patients included both adults (n = 295 [84%]) and children (n = 57 [16%]). Additional clinical data, including specimen type, were unavailable.
CTX-M-15 and ST131 status.
According to tiered PCR-based testing, 201 (57%) of the 351 study isolates contained CTX-M-15 (median per center, 59%; range, 25% to 100%), whereas 165 (47%) represented ST131 (median per center, 50%; range, 26% to 100%) (Table 1). Overall, ST131 was statistically significantly associated with CTX-M-15, accounting for 56% of CTX-M-15-positive isolates versus 34% of CTX-M-15-negative isolates (P < 0.001). This trend was evident at 12 of the 15 individual centers, including both children's hospitals (Table 1).
The 4 possible combinations of ST131 status and CTX-M-15 status defined 4 subgroups (Table 1). Of these, the dual-positive subgroup (i.e., ST131+ CTX-M-15+) was the most prevalent (33%), followed sequentially by the ST131− CTX-M-15− subgroup (28%), the ST131− CTX-M-15+ subgroup (25%), and the ST131+ CTX-M-15− subgroup (15%) (Table 1). All but three centers contributed one or more representatives of each of these 4 subgroups, and the 3 centers that did not do so provided only 1 to 11 isolates each.
CTX-M-15 and ST131 versus phylogenetic group, F10 papA allele, and O25b rfb variant.
In the total population, group B2 was the dominant phylogenetic group, accounting for 50% of isolates, followed sequentially in prevalence by groups D (25%), A (18%), and B1 (7%) (Table 2). Whereas by definition the ST131 isolates were all from group B2, regardless of CTX-M-15 status, the non-ST131 isolates were least likely to be from group B2 (versus other groups), and their phylogenetic group distribution varied significantly with CTX-M-15 status. That is, the CTX-M-15-negative non-ST131 isolates were predominantly from group D, followed by groups B1, A, and B2. In contrast, the CTX-M-15-positive non-ST131 isolates were predominantly from group A, followed by groups D and B1, with none from group B2.
Table 2.
Characteristics of 351 extended-spectrum-β-lactamase-producing Escherichia coli clinical isolates from the United States, 2000 to 2009
| Characteristica | No. (%) of isolatesb |
P valuec |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Total (n = 351) | Subset |
||||||||
| ST131− M15− (group 1; n = 98) | ST131− M15+ (group 2; n = 89) | ST131+ M15− (group 3; n = 52) | ST131+ M15+ (group 4; n = 113) | Group 1 vs 2 | Group 3 vs 4 | Group 1 vs 3 | Group 2 vs 4 | ||
| Group A | 63 (18) | 17 (17) | 46 (52) | 0 (0) | 0 (0) | < 0.001 | 0.001 | < 0.001 | |
| Group B1 | 24 (7) | 20 (20) | 4 (5) | 0 (0) | 0 (0) | 0.002 | < 0.001 | 0.035 | |
| Group B2 | 175 (50) | 10 (10) | 0 (0) | 52 (100) | 113 (100) | 0.002 | < 0.001 | < 0.001 | |
| Group D | 89 (25) | 51 (52) | 38 (43) | 0 (0) | 0 (0) | < 0.001 | < 0.001 | ||
| F10 papA allele | 168 (48) | 9 (9) | 8 (9) | 42 (81) | 109 (97) | 0.002 | < 0.001 | < 0.001 | |
| O25b rfb allele | 159 (45) | 0 (0) | 0 (0) | 48 (92) | 110 (97) | < 0.001 | < 0.001 | ||
| Post-2003 origin | 338 (96) | 89 (91) | 85 (97) | 51 (98) | 113 (100) | 0.10 | 0.08 | ||
Group, major E. coli phylogenetic group; F10 papA allele, P fimbria structural subunit variant; O25b rfb allele, O25 lipopolysaccharide synthesis.
M15+, positive for blaCTX-M-15; M15−, negative for blaCTX-M-15; ST131+, ST131 sequence type; ST131−, non-ST131 sequence type.
P values (by Fisher's exact test; two-tailed) are shown where P ≤ 0.10. For all variables, an initial 4-group comparison yielded P ≤ 0.003.
The F10 papA allele and O25 rfb variant were significantly more prevalent among ST131 isolates than non-ST131 isolates, regardless of CTX-M-15 status (Table 1). Additionally, for the ST131 isolates, the F10 papA allele was significantly more prevalent among CTX-M-15-positive than CTX-M-15-negative isolates, although the absolute difference was modest (81% versus 97%) (Table 1).
CTX-M-15 and ST131 versus year of isolation.
When analyzed as a continuous variable, the year of isolation was borderline significantly more recent for CTX-M-15-positive isolates than CTX-M-15-negative isolates (P = 0.099) and for ST131 isolates than non-ST131 isolates (P = 0.053). The distribution of isolation years by CTX-M-15 and ST131 status suggested that, for both variables, 2003 to 2004 would be the optimal breakpoint for dichotomous stratification (not shown). With the years categorized using this breakpoint, CTX-M-15 and ST131 were both significantly more prevalent among isolates from 2004 to 2009 than among those from 2000 to 2003 (for CTX-M-15, 77% versus 23% [P = 0.019]; for ST131, 49% versus 8% [P = 0.004]), consistent with the emergence of both traits over the decade.
Among the four subgroups defined by combined CTX-M-15 status and ST131 status, the proportion of isolates from 2004 to 2009 (versus isolates from 2000 to 2003) increased progressively across subgroups, from a low of 91% (CTX-M-15-negative, non-ST131 isolates) to a high of 100% (CTX-M-15-positive, ST131 isolates) (P = 0.003 for the 4-group comparison) (Table 2). In pairwise comparisons among the 4 subgroups, ST131 isolates were borderline significantly more likely than non-ST131 isolates to be from 2004 to 2009, regardless of CTX-M-15 status (Table 2).
Extended virulence genotypes.
Genotypes for 51 ExPEC-associated virulence markers were determined for 50 randomly selected representatives per subgroup for the 4 subgroups as defined by CTX-M-15 status and ST131 status (total n = 200). The selected isolates exhibited the same phylogenetic group distribution as their respective source subgroups, evidence that they were a representative subset (Table 3). All but 8 of the virulence markers sought were detected in ≥1 isolate each, with 18 (42%) of the detected markers exhibiting a significant between-subgroup prevalence difference in relation to CTX-M-15 status, ST131 status, or both (Table 3).
Table 3.
Molecular characteristics of 200 selected extended-spectrum-β-lactamase-producing Escherichia coli isolates (collected in 2000 to 2009) in relation to ST131 and blaCTX-M-15 status
| Trait categorya,b | Specific traita | No. (%) of isolates |
P valuec |
||||||
|---|---|---|---|---|---|---|---|---|---|
| ST131− M15− (group 1; n = 50) | ST131− M15+ (group 2; n = 50) | ST131+ M15− (group 3; n = 50) | ST131+ M15+ (group 4; n = 50) | Group 1 vs 2 | Group 3 vs 4 | Group 1 vs 3 | Group 2 vs 4 | ||
| Phy. group | Group A | 10 (20) | 28 (56) | 0 (0) | 0 (0) | <0.001 | <0.001 | <0.001 | |
| Group B1 | 7 (14) | 1 (2) | 0 (0) | 0 (0) | 0.01 | ||||
| Group B2 | 5 (10) | 0 (0) | 50 (100) | 50 (100) | <0.001 | <0.001 | |||
| Group D | 28 (56) | 21 (42) | 0 (0) | 0 (0) | <0.001 | <0.001 | |||
| Adhesins | F10 papA | 4 (8) | 5 (10) | 40 (80) | 50 (100) | 0.001 | <0.001 | <0.001 | |
| afa/draBC | 3 (6) | 2 (4) | 2 (4) | 13 (26) | 0.03 | 0.004 | |||
| iha | 8 (16) | 3 (6) | 39 (78) | 48 (96) | 0.02 | <0.001 | <0.001 | ||
| fimH | 42 (84) | 30 (60) | 50 (100) | 50 (100) | 0.01 | 0.006 | <0.001 | ||
| hra | 10 (20) | 6 (12) | 1 (2) | 4 (8) | 0.008 | ||||
| Toxins | sat | 6 (12) | 3 (6) | 39 (78) | 48 (96) | 0.02 | <0.001 | <0.001 | |
| vat | 6 (12) | 0 (0) | 1 (2) | 0 (0) | 0.03 | ||||
| astA | 9 (18) | 5 (10) | 0 (0) | 0 (0) | 0.03 | ||||
| Siderophores | fyuA | 30 (60) | 31 (62) | 49 (98) | 50 (100) | <0.001 | <0.001 | ||
| iutA | 21 (42) | 39 (78) | 43 (86) | 49 (98) | <0.001 | <0.001 | <0.001 | ||
| Capsule | kpsM II | 18 (36) | 11 (22) | 35 (70) | 45 (90) | 0.02 | 0.001 | <0.001 | |
| K2 | 3 (6) | 4 (8) | 5 (10) | 17 (34) | 0.007 | 0.003 | |||
| K5 | 3 (6) | 0 (0) | 20 (40) | 18 (36) | <0.001 | <0.001 | |||
| kpsMT III | 3 (6) | 9 (18) | 0 (0) | 0 (0) | 0.003 | ||||
| Miscellaneous | usp | 7 (14) | 1 (2) | 49 (98) | 50 (100) | <0.001 | <0.001 | ||
| ompT | 15 (30) | 7 (14) | 48 (96) | 50 (100) | <0.001 | <0.001 | |||
| traT | 34 (68) | 42 (84) | 44 (88) | 46 (92) | 0.03 | ||||
| malX | 17 (34) | 20 (40) | 48 (96) | 50 (100) | <0.001 | <0.001 | |||
| ExPEC | n.a.d | 15 (30) | 7 (14) | 33 (66) | 47 (94) | 0.001 | 0.001 | <0.001 | |
| PFGE statuse | ≥2 per type | 9 (19)f | 22 (44) | 27 (54) | 41 (82) | 0.009 | 0.005 | <0.001 | <0.001 |
| ≥5 per type | 0 (0) | 0 (0) | 18 (36) | 33 (66) | <0.001 | <0.001 | |||
Traits shown are those that yielded P < 0.05 in an initial four-way comparison (not shown) plus at least one pairwise comparison (as shown). These included afa/draBC (Dr-binding adhesins), astA (enteroaggregative E. coli heat-stable toxin), the F10 papA allele (P fimbria structural subunit variant), fimH (type 1 fimbria adhesin), fyuA (yersiniabactin system), hra (heat-resistant agglutinin), iha (adhesin siderophore), iutA (aerobactin system), K2 (group 2 capsule variant), K5 (group 2 capsule variant), kpsMII (group 2 capsule), kpsMTIII (group 3 capsule), malX (fitness island marker), ompT (outer membrane protease), sat (secreted autotransporter toxin), usp (uropathogenic specific protein), and vat (vacuolating toxin). Traits detected in ≥1 isolate but without significant by-group prevalence differences (definition: overall prevalence) included afaE8 (afimbrial adhesin: 1.5%), bmaE (M fimbriae: 1.5%), cdtB (cytolethal distending toxin B: 0.5%), clbB and clbN (polyketide synthesis: 1% and 2%), clpG (mannose-resistant adhesin: 0.5%), cnf1 (cytotoxic necrotizing factor: 2%), cvaC (microcin V: 1%), H7 fliC allele (flagellin: 0.5%), hlyD (alpha hemolysin: 3.5%), hlyF (hemolysin variant: 4%), ibeA (invasion of brain endothelium: 2%), ireA (siderophore receptor: 1.5%), iroN (salmochelin receptor: 3%), iss (increased serum survival: 3%), K1 (capsule variant: 3%), papAH (P fimbria major subunit: 4.5%), papC (P fimbria assembly: 7.0%), papEFG (P fimbria tip pilins: 5.5%), papG alleles II and III (P adhesin variants: 4.5% and 1%), pic (autotransporter protease: 0.5%), rfc (O4 lipopolysaccharide synthesis: 1%), traT (serum resistance associated: 82.5%), and tsh (temperature-sensitive hemagglutinin: 2%). Traits sought but not detected included F17 (mannose-resistant adhesin), focG (F1C adhesin), gafD (G fimbriae), K15 (capsule variant), papG allele I (P adhesin variant), pic (protein associated with intestinal colonization), sfa/foc (S and F1C fimbriae), and sfaS (S fimbria adhesin).
ExPEC, extraintestinal pathogenic E. coli; PFGE, pulsed-field gel electrophoresis; Phy., phylogenetic. ExPEC was defined as the presence of ≥2 of papA and/or papC and of sfa/foc, afa/dra, kpsMII, and iutA.
P values (by Fisher's exact test, two-tailed) are shown where P < 0.05.
n.a., not applicable.
The data correspond to isolates belonging to pulsotype groups consisting of ≥2 isolates or ≥5 isolates each.
The denominator was 48 (rather than 50), since 2 isolates were refractory to XbaI PFGE analysis.
Several general trends were apparent from these differences (Table 3). First, statistically significant prevalence differences were more than twice as frequent in relation to ST131 status as they were in relation to CTX-M-15 status. Second, whereas most of the ST131 versus non-ST131 prevalence differences favored the ST131 isolates (e.g., for the F10 papA allele, afa/dra, iha, fimH, sat, fyuA, kpsMII, K2, K5, usp, ompT, traT, and malX), several favored the non-ST131 isolates (e.g., for hra, astA, and kpsMT III). Third, whereas quite similar sets of genes exhibited ST131-associated prevalence differences among CTX-M-15-positive compared with CTX-M-15-negative isolates, distinct sets of genes exhibited CTX-M-15-associated prevalence differences among ST131 isolates compared with non-ST131 isolates.
Consistent with the greater prevalence of many ExPEC-associated virulence genes among the ST131 isolates, a significantly greater proportion of ST131 isolates than non-ST131 isolates fulfilled molecular criteria for ExPEC, regardless of CTX-M-15 status (Table 3). Furthermore, among ST131 (but not non-ST131) isolates, CTX-M-15 was positively associated with ExPEC status. Thus, of the 4 subgroups, the CTX-M-15-positive ST131 isolates were the most likely to qualify as ExPEC (94%). Likewise, aggregate virulence scores were significantly greater among ST131 isolates compared with non-ST131 isolates and were greatest of all within the CTX-M-15-positive ST131 subgroup (see Table 5).
Table 5.
Aggregate virulence and antimicrobial resistance scores of 200 selected extended-spectrum-β-lactamase-producing Escherichia coli isolates (collected in 2000 to 2009) in relation to ST131 and blaCTX-M-15 status
| Type of score | Score median (range) |
P valuea |
||||||
|---|---|---|---|---|---|---|---|---|
| ST131− M15− (group 1; n = 50) | ST131− M15+ (group 2; n = 50) | ST131+ M15− (group 3; n = 50) | ST131+ M15+ (group 4; n = 50) | Group 1 vs 2 | Group 3 vs 4 | Group 1 vs 3 | Group 2 vs 4 | |
| Virulence | 4 (0-18) | 4 (1-10) | 10 (2-13) | 10 (6-14) | 0.01 | < 0.001 | < 0.001 | |
| Resistance | 13 (4-19) | 15 (7-19) | 12 (4-19) | 13 (9-17) | 0.04 | 0.003 | ||
P values (Mann-Whitney U test) are shown where P < 0.05.
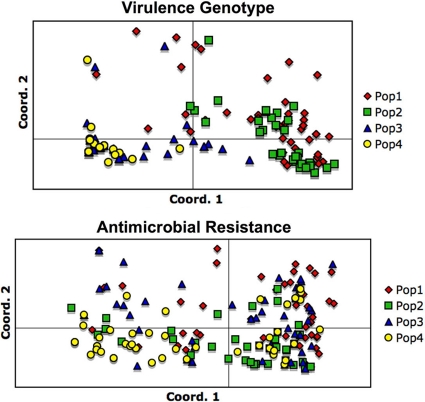
According to PCoA, which was used to summarize the total virulence gene data set for a simplified comparison among subgroups, the ST131 isolates and non-ST131 isolates were clearly separated on the coordinate 1-coordinate 2 plane (Fig. 1). In contrast, among ST131 and non-ST131 isolates alike, CTX-M-15-positive and CTX-M-negative isolates were largely intermingled, without obvious differences. AMOVA indicated that 43% of the total variance was among populations, whereas 57% was within populations.
Fig 1.
Principal coordinate analysis of virulence genotype data (upper panel) and antimicrobial resistance data (lower panel) among 200 Escherichia coli isolates (collected in 2000 to 2009) in relation to ST131 and blaCTX-M-15 status. Populations 1 to 4 correspond to the groups shown in the tables (i.e., Table 1, non-ST131 blaCTX-M-15 negative; Table 2, non-ST131 blaCTX-M-15 positive; Table 3, ST131 blaCTX-M-15 negative; Table 4, ST131 blaCTX-M-15 positive). Upper panel (virulence genotypes): coordinates 1 and 2 capture 58% and 12% of total variation, respectively. Note the marked separation of (overlapping) populations 1 and 2 from (overlapping) populations 3 and 4. Lower panel (antimicrobial resistance): coordinates 1 and 2 capture 32% and 26% of total variation, respectively. Note the marked overlap of all four populations.
Antimicrobial resistance profiles.
Within the 200-isolate subset, resistance to each tested antimicrobial agent except imipenem was detected in ≥1 isolate (Table 4). Significant resistance prevalence differences among the 4 isolate subgroups, as defined by combined ST131 and CTX-M-15 status, were noted for 13 agents. Compared with CTX-M-15-negative isolates, CTX-M-15-positive isolates had a significantly higher prevalence of resistance to multiple β-lactam agents, regardless of ST131 status, but a significantly lower prevalence of resistance to streptomycin (if non-ST131) or sulfonamides (if ST131). In contrast, ST131 and non-ST131 isolates exhibited comparatively few significant resistance prevalence differences for β-lactam agents, but non-ST131 isolates (especially if CTX-M-15 positive) exhibited a significantly greater prevalence of resistance to numerous non-β-lactam agents (Table 4). Accordingly, aggregate resistance scores, although differing only slightly among the 4 subgroups, were highest among the CTX-M-15-positive non-ST131 isolates (median, 15) and lowest among the CTX-M-15-negative ST131 isolates (median, 12) (Table 5).
Table 4.
Antimicrobial resistance phenotypes of 200 selected extended-spectrum-β-lactamase-producing Escherichia coli isolates (collected in 2000 to 2009) in relation to ST131 and blaCTX-M-15 status
| Antimicrobial class | Specific agenta | No. (%) of isolates showing resistance to the indicated agent |
P valueb |
||||||
|---|---|---|---|---|---|---|---|---|---|
| ST131− M15− (group 1; n = 50) | ST131− M15+ (group 2; n = 50) | ST131+ M15− (group 3; n = 50) | ST131+ M15+ (group 4; n = 50) | Group 1 vs 2 | Group 3 vs 4 | Group 1 vs 3 | Group 2 vs 4 | ||
| β-Lactams | ATM | 33 (66) | 47 (94) | 29 (58) | 41 (82) | 0.001 | 0.002 | ||
| CAZ | 35 (70) | 47 (94) | 34 (68) | 39 (78) | 0.003 | 0.04 | |||
| CRO | 44 (88) | 50 (100) | 39 (78) | 48 (96) | 0.03 | 0.02 | |||
| FEP | 11 (22) | 27 (54) | 11 (22) | 30 (60) | 0.002 | <0.001 | |||
| Quinolones | NA | 36 (72) | 49 (98) | 43 (86) | 50 (100) | <0.001 | 0.02 | ||
| CIP | 30 (60) | 49 (98) | 43 (86) | 50 (100) | <0.001 | 0.01 | 0.006 | ||
| Aminoglycosides | CN | 23 (46) | 31 (62) | 25 (50) | 17 (34) | 0.009 | |||
| SF | 38 (76) | 20 (40) | 36 (72) | 26 (52) | 0.001 | ||||
| Phenicols | C | 23 (46) | 14 (28) | 8 (16) | 2 (4) | 0.002 | 0.002 | ||
| Tetracyclines | TE | 45 (90) | 44 (88) | 30 (60) | 38 (76) | 0.001 | |||
| Folate antagonists | SF | 44 (88) | 39 (78) | 39 (78) | 28 (56) | 0.03 | 0.03 | ||
| W | 36 (72) | 39 (78) | 32 (64) | 23 (46) | 0.002 | ||||
| SXT | 37 (74) | 37 (74) | 31 (62) | 22 (44) | 0.004 | ||||
Agents shown are those that yielded P < 0.05 in an initial four-way comparison (not shown), plus at least one pairwise comparison (as shown). These included ATM (aztreonam), C (chloramphenicol), CAZ (ceftazidime), CIP (ciprofloxacin), CN (gentamicin), CRO (ceftriaxone), FEP (cefepime), NA (nalidixic acid), S (streptomycin), SF (sulfonamide), SXT (trimethoprim-sulfamethoxazole), TE (tetracycline), and W (trimethoprim). Agents to which resistance was detected in ≥1 isolate but without significant by-group prevalence differences (definition: overall prevalence) included AMK (amikacin: 10%), AML (amoxicillin-clavulanate: 69%), AMP (ampicillin: 100%), FOX (cefoxitin: 19%), F (nitrofurantoin: 8%), KF (cefazolin: 97%), PIP (piperacillin: 96%), and TZP (piperacillin-tazobactam: 5%). No resistance was detected to IMP (imipenem).
P values (by Fisher's exact test, two-tailed) are shown where P < 0.05.
In a PCoA based on the 21 studied antimicrobial agents, considerable diversity of aggregate resistances profiles was evident from the overall spread of points on the coordinate 1-coordinate 2 plane (Fig. 1). However, the 4 subgroups as defined by combined CTX-M-15 status and ST131 status were considerably intermingled, without obvious among-subgroup differences. According to AMOVA, only 9% of total variance was among populations; fully 91% was within populations.
PFGE.
All but two of the 200 selected isolates yielded interpretable XbaI PFGE profiles. The 94% similarity criterion resolved 122 pulsotypes, each containing from 1 isolate (99 pulsotypes) to 27 isolates (1 pulsotype [type 968]). Twenty-three pulsotypes contained multiple isolates (99 isolates total); of these, 5 contained ≥5 isolates each (51 isolates total). Membership in a multiple-isolate pulsotype was significantly associated with both blaCTX-M-15 (regardless of ST131 status) and ST131 (regardless of blaCTX-M-15 status) (Table 3). Similarly, membership in a high-prevalence (≥5 isolates) pulsotype was confined to ST131 isolates, and among ST131 isolates it was significantly associated with blaCTX-M-15 status (Table 3) and ExPEC status (92% ExPEC for high-frequency pulsotypes versus 67%; P = 0.002).
DISCUSSION
In this analysis of ESBL-positive E. coli isolates from across the United States (collected in 2000 to 2009), we defined the overall, by-center, and temporal prevalences of blaCTX-M-15 and ST131, identified associations of these two traits with one another and with virulence genotype and resistance profiles, and assessed the clonal structures of the populations. Our findings extend the results of a previous national survey (8) and offer novel insights into the basis for the striking epidemiological success of both blaCTX-M-15 and E. coli ST131 strains.
We found a marked predominance of both blaCTX-M-15 and ST131 among ESBL-producing E. coli isolates collected across the United States and a very strong (albeit incomplete) association of these two traits with one another. This confirms, in a very different study population, the findings of a previous, smaller, single-year national survey (8). Notably, both blaCTX-M-15 and ST131 occurred at each of the 15 participating centers, representing evidence of widespread distribution, but with by-center prevalence differences suggesting possible geographical or host-group specificity.
A novel finding in the present study was that both blaCTX-M-15 and ST131 appeared to have emerged and expanded in distribution during the past decade, since they were both significantly more prevalent from 2004 onward. Likewise, both occurred among isolates from children and adults alike. The somewhat lower overall prevalences of blaCTX-M-15 and ST131 in the present study (57% and 47%, respectively), compared with the previous national survey (78% and 67%, respectively) (8), may reflect in part the present study's inclusion of “all-comer” and pediatric isolates from 2000 through 2009, compared with the previous survey's focus on blood isolates from adults from 2007.
Our findings provide insights into possible reasons for the striking epidemiological success of blaCTX-M-15 and ST131 and their association with one another. First, despite the obvious horizontal mobility of blaCTX-M-15 (as demonstrated by its occurrence in diverse phylogenetic backgrounds), blaCTX-M-15 showed evidence of clonal spread and expansion. That is, among the non-ST131 isolates, blaCTX-M-15 was concentrated within major phylogenetic groups A and D and within high-prevalence pulsotypes within those groups. Similarly, among ST131 isolates, blaCTX-M-15 was significantly associated with the highest-prevalence pulsotypes. To what degree blaCTX-M-15 has contributed directly to the expansion of these lineages, or is an opportunistic hitchhiker within intrinsically successful clones, warrants further study.
Second, blaCTX-M-15 was significantly associated with resistance to key first-line antimicrobials, notably β-lactams and fluoroquinolones, and (among non-ST131 isolates) with higher aggregate resistance scores. This predictably would favor expansion of blaCTX-M-15-containing strains in preference to other ESBL-producing E. coli strains in the presence of selection pressure from the corresponding antimicrobials, which is abundant and likely increasing.
Third, the large molecularly inferred virulence advantage of ST131 over non-ST131 isolates (regardless of blaCTX-M-15 status) and, within ST131, the similar (albeit smaller) advantage of blaCTX-M-15-positive over blaCTX-M-15-negative isolates would tend to favor expansion of ST131 and, specifically, its blaCTX-M-15-positive subset. The marked inferred virulence advantage of ST131 strains may more than compensate for their modest resistance disadvantage compared with other ESBL-producing E. coli strains. In this regard, the high prevalence of certain ST131 pulsotypes also may relate in part to differential virulence, since isolates from high-frequency pulsotypes were significantly more likely to qualify as ExPEC than were other ST131 isolates (92% versus 67%; P = 0.0002). These highly successful lineages appear to represent an extreme version of the combined threats of virulence and resistance (22).
Study limitations included the convenience sample (with its attendant potential biases), the undefined clinical background of the isolates, and a lack of information regarding the identity of the non-CTX-M-15 ESBLs and non-ST131 clonal groups. Strengths included the large sample size relative to other studies of ESBL-positive E. coli strains from the United States, long study interval, broad geographic range, inclusion of pediatric and adult isolates, and extensive molecular characterization of the isolates.
In summary, in this large molecular-epidemiological analysis of ESBL-producing E. coli clinical isolates from the United States (collected in 2000 to 2009), we found a predominance of blaCTX-M-15 and ST131 strains, which were widely dispersed geographically, involved pediatric as well as adult patients, and increased significantly in prevalence during the study period. Both entities were strongly associated with one another and, especially in combination, with enhanced molecularly inferred virulence, extensive antimicrobial resistance, and decreased clonal diversity, suggesting recent clonal expansion and dissemination. These findings confirm the importance in the United States of the ST131 clonal group and its characteristic blaCTX-M-15 gene, suggest possible reasons for their emergence, and underscore the need for effective predictive and preventive measures.
ACKNOWLEDGMENTS
The AMERECUS (Assessing the Molecular Epidemiology of Resistant E. coli in the United States) investigators include the following: Robert L. Bergsbaken (Health Partners and Regions Medical Center, St. Paul, MN), Thomas M. Hooton (University of Miami, Miami, FL), Michelle Hulse (Childrens Hospital, Minneapolis, MN), Karen Lolans (Rush University, Chicago, IL), Rob Owens (Cubist Pharmaceuticals, Falmouth, ME), Elizabeth Palavecino (Wake Forest University Baptist Medical Center, Winston-Salem, NC), and Karen Vigil (University of Texas Health Sciences Center at Houston, Houston, TX).
Dave Prentiss (Minneapolis Veterans Affairs Medical Center) helped prepare the figure.
This material is based upon work supported by Office of Research and Development, Medical Research Service, Department of Veterans Affairs (J.R.J.).
J.R.J. has received research funding from Merck and Rochester Medical Group. J.S.L. has received research funding and/or honoraria from Merck, Ortho-McNeil, and Pfizer. J.Q. is an employee and shareholder in Pfizer Global Research. The other authors report no conflicts of interest.
Footnotes
Published ahead of print 21 February 2012
REFERENCES
- 1. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clermont O, et al. 2009. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J. Antimicrob. Chemother. 64:274–277 [DOI] [PubMed] [Google Scholar]
- 3. Clermont O, et al. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 61:1024–1028 [DOI] [PubMed] [Google Scholar]
- 4. Clinical Laboratory Standards Institute 2009. Performance standards for antimicrobial susceptibility testing; nineteenth informational supplement (M100-S19). Clinical Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. Coque TM, Novais Â, et al. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg. Infect. Dis. 14:195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ender PT, et al. 2009. Transmission of extended-spectrum-beta-lactamase-producing Escherichia coli (sequence type ST131) between a father and daughter resulting in septic shock and emphysematous pyelonephritis. J. Clin. Microbiol. 47:3780–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson JR, Anderson JT, Clabots C, Johnston B, Cooperstock M. 2010. Within-household sharing of a fluoroquinolone-resistant Escherichia coli sequence type ST131 strain causing pediatric osteoarticular infection. Pediatr. Infect. Dis. J. 29:473–475 [DOI] [PubMed] [Google Scholar]
- 8. Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States (2007). Clin. Infect. Dis. 51:286–294 [DOI] [PubMed] [Google Scholar]
- 9. Johnson JR, et al. 2010. Escherichia coli sequence type ST131 as an emerging fluoroquinolone-resistant uropathogen among renal transplant recipients. Antimicrob. Agents Chemother. 54:546–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson JR, et al. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob. Agents Chemother. 53:2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson JR, Miller S, Johnston B, Clabots C, Debroy C. 2009. Sharing of Escherichia coli sequence type ST131 and other multidrug-resistant and urovirulent E. coli strains among dogs and cats within a household. J. Clin. Microbiol. 47:3721–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson JR, et al. 2003. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 47:2161–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261–272 [DOI] [PubMed] [Google Scholar]
- 14. Johnson L, et al. 2008. Emergence of fluoroquinolone resistance in outpatient urinary Escherichia coli isolates. Am. J. Med. 121:876–884 [DOI] [PubMed] [Google Scholar]
- 15. Leflon-Guibout V, et al. 2004. Emergence and spread of three clonally related virulent isolates of CTX-M-15-producing Escherichia coli with variable resistance to aminoglycosides and tetracycline in a French geriatric hospital. Antimicrob. Agents Chemother. 48:3736–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lewis JS, II, Herrera M, Wickes B, Patterson JE, Jorgensen JH. 2007. First report of the emergence of CTX-M-type extended-spectrum beta-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Antimicrob. Agents Chemother. 51:4015–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McGettigan SE, Hu B, Andreacchio K, Nachamkin I, Edelstein PH. 2009. Prevalence of CTX-M beta-lactamases in Philadelphia, Pennsylvania. J. Clin. Microbiol. 47:2970–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nicolas-Chanoine M-H, et al. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273–281 [DOI] [PubMed] [Google Scholar]
- 19. Peakall R, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6:288–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peirano G, Costello M, Pitout JDD. 2010. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli from the Chicago area: high prevalence of ST131 producing CTX-M-15 in community hospitals. Int. J. Antimicrob. Agents 36:19–23 [DOI] [PubMed] [Google Scholar]
- 21. Peirano G, Pitout JDD. 2010. Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: the worldwide emergence of clone ST131 O25:H4. Int. J. Antimicrob. Agents 35:316–321 [DOI] [PubMed] [Google Scholar]
- 22. Pitout JD. 2012. Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front. Microbiol. 3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pitout JD, Nordmann P, Laupland KB, Poirel L. 2005. Emergence of Enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 56:52–59 [DOI] [PubMed] [Google Scholar]
- 24. Ribot EM, et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 25. Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14 [DOI] [PubMed] [Google Scholar]
- 26. Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: an overlooked epidemic. Microbes Infect. 5:449–456 [DOI] [PubMed] [Google Scholar]
- 27. Sidjabat HE, et al. 2009. Molecular epidemiology of CTX-M-producing Escherichia coli isolates at a tertiary medical center in western Pennsylvania. Antimicrob. Agents. Chemother. 53:4733–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tenover FC, et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Urban C, et al. 2010. Identification of CTX-M β-lactamases in Escherichia coli from hospitalized patients and residents of long-term care facilities. Diagn. Microbiol. Infect. Dis. 66:402–406 [DOI] [PubMed] [Google Scholar]
- 30. Vigil KJ, et al. 2009. Escherichia coli pyomyositis: an emerging entity among patients with hematologic malignancies. Clin. Infect. Dis. 50:374–380 [DOI] [PubMed] [Google Scholar]